Abstract
The cell-penetrating peptide (CPP) penetratin (PEN) has garnered attention for its potential to enter tumor cells. However, its translocation mechanism and lack of selectivity remain debated. This study investigated PEN’s insertion into healthy cells (H-) and cancer cells (C-) using micromolar concentrations and various techniques. Raman spectroscopy was used to determine PEN’s location in the lipid bilayer at different lipid-to-peptide ratios. Dynamic light scattering (DLS) and zeta potential analysis were used to measure the lipid–PEN complex’s size and charge. The results showed helical PEN particles directly inserted into C- membranes at a ratio of 110, while aggregated particles stayed on H- surfaces. Raman spectroscopy and scanning electron microscopy confirmed PEN insertion in C- membranes. Zeta potential studies revealed highly negative charges for PEN–C- complexes and neutral charges for PEN–H- complexes at pH 6.8. C- integrity remained unchanged at a ratio of 110. Specific lipid-to-peptide ratios with dipalmitoylphosphatidylserine (DPPS) were crucial for direct insertion. These results provide valuable insights into CPP efficacy for targeted drug delivery in cancer cells, considering membrane composition and lipid-to-peptide ratios.
1. Introduction
Cancer is a significant global health challenge and stands as the second leading cause of death worldwide, necessitating the development of innovative therapies [1]. The effectiveness of cancer treatment faces significant hurdles, including drug resistance, non-malignant cell toxicity, and inefficient drug delivery [1,2,3,4]. To address these challenges, researchers have turned their attention to cell-penetrating peptides (CPPs) as potential drug delivery systems. CPPs possess the unique ability to traverse cell membranes, either alone or with drugs, thereby enhancing drug uptake in tumor cells and potentially improving treatment efficacy [2,5].
Penetratin (PEN), a member of CPPs, has gained considerable attention due to its remarkable interactions with biological cells [6,7,8,9]. Liu et al. [6] investigated PEN for its cytotoxicity and cellular uptake in human conjunctival epithelial cells, revealing exceptional cellular uptake with minimal adverse effects on ocular cells and tissues. Jiao et al. [7] demonstrated that PEN partly enters cells, such as Chinese Hamster Ovary cells, through direct translocation.
Akari et al. [10] employed a droplet transfer method to create giant unilamellar vesicles (GUVs) with a lipid composition of DOPS/DOPC in a 1:1 molar ratio. These GUVs were subsequently exposed to PEN, and the results revealed that PEN in combination with asymmetric GUVs containing DOPS in the inner leaflet facilitated efficient DNase I translocation. PEN has also shown promise in traversing the rabbit cornea [7], crossing the blood–brain barrier [11,12,13], and in the development of multifunctional theragnostic agents for inhibiting Aβ fibrillogenesis [13].
Thoren et al. [9] conducted experiments suggesting an endocytic uptake mechanism, potentially in contrast to prior research findings. Nevertheless, comprehending the potential of PEN for tumor targeting and future cancer therapies necessitates a comprehensive understanding of its entry mechanisms [14] and selectivity [1,2,3,4].
The entry mechanisms of CPPs into cells have been a subject of extensive investigation, but uncertainties remain regarding the underlying processes [2,3,6,7,14,15]. Generally, CPPs can enter cells through two different mechanisms: endocytosis and direct translocation across the plasma membrane [3,7,11,14,16]. Direct translocation is particularly crucial for effective vectorization as it allows the CPP and its cargo to directly access the cytoplasm. Consequently, understanding the entry mechanisms of CPPs is vital for advancing their application as efficient vectors, transitioning from in vitro settings to clinical translation.
In this study, we focus on investigating the insertion mechanism of PEN for drug delivery [2,3,7,14]. PEN is a short CPP composed of a 16 amino acid sequence derived from the homeodomain of the antennapedia homeoprotein (RQIKIWFQNRRMKWKK). Model membranes that mimic mammalian cells have been suggested as valuable tools for studying CPP behavior and proposing new pathways for their insertion mechanisms [14]. Model membranes offer several advantages over live cells, including simplicity, ease of handling, and the ability to investigate conditions that live cells may not tolerate [17].
To investigate the specificity and insertion mechanism of PEN, we employed two types of cell membrane mimics representing healthy cells (H-) and cancer cells (C-). Both model membranes shared four different lipids, but the C- model membrane contained an additional lipid, phosphatidylserine (PS), which enhances membrane functionality and cellular uptake [18,19]. The interactions between PEN and these vesicles were examined at different lipid-to-peptide molar ratios.
To characterize the PEN–vesicle interactions, we employed a combination of techniques, including differential scanning calorimetry (DSC) and Raman spectroscopy, which have not been extensively utilized in previous studies investigating PEN–lipid interactions. Additionally, dynamic light scattering (DLS) and zeta potential measurements were employed to gain insights into the aggregation behavior and surface charge of PEN–vesicle complexes, as suggested by Alves et al. [20]. Furthermore, infrared (IR) spectroscopy studies and thermogravimetric analysis (TGA) were conducted to provide a comprehensive understanding of the physicochemical behavior of the dried PEN–vesicle complexes.
While many studies have focused on large unilamellar vesicles (LUVs) that possess a membrane curvature closer to that of cells [16,21], some authors have studied GUVs that are characterized by a curvature highly resembling that of biological cells and represent an invaluable model system for the investigation of various biological processes [8,10,15,22,23,24,25].
By investigating the specific lipid-to-peptide co-import triggering the direct insertion of PEN into cells [7] and utilizing model membranes, our study aims to advance the understanding of PEN’s entry mechanism. The knowledge gained from this investigation holds the potential to enhance the development of PEN-based drug delivery systems for targeted tumor therapy and improve the field of cancer treatment overall.
2. Results
2.1. Characterization of PEN–Vesicle Complexes by Raman Spectroscopy
Raman spectroscopy was employed to investigate the localization and insertion of PEN within the lipid bilayer. PEN can reside in the hydrophobic region alongside the lipid acyl chains, at the surface in contact with the phosphate head groups, or between these two regions. In the latter case, changes in the position of the carbonyl groups need to be monitored. Raman spectra of fresh samples of vesicles without PEN (H- and C-) were recorded and analyzed as reference peaks for experimental analysis. Subsequently, the effect of adding an appropriate amount of PEN on H- and C- was monitored by analyzing the intensity and frequency changes of the acyl, carbonyl, and phosphate group bands. The incubation time was 2 h at 37 °C. Freeze-dried samples were placed on a microscope slide. The Raman spectra revealed three characteristic peaks in the 1000–1200 cm−1 region (Figure 1), corresponding to vibrations of hydrocarbon chains and phosphate head groups [26,27,28,29]. The peaks at 1062 and 1126 cm−1 were assigned to the out-of-phase and in-phase skeletal C−C stretching modes of the all-trans segments, while the peak at 1096 cm−1 originated from the superposition of the symmetric PO2- stretching mode with C–C vibrations of gauche conformers [21,30]. The intensity ratios I1096/I1126 and I1096/I1062 were used to assess the proportion of trans/gauche conformation in the lipid chain, which characterizes lipid chain disorder [31,32]. The presence of PEN inside the lipid bilayer induced lipid chain disorder. The percentage error was between 1 and 2% for each experiment.
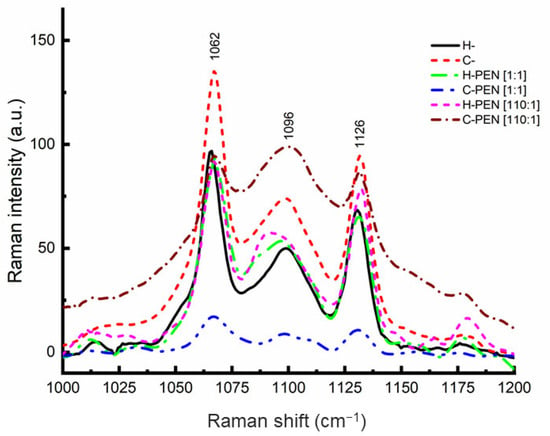
Figure 1.
Raman spectra of lyophilized samples of PEN (obtained from a 10 μM aqueous solution at physiological pH) and its complexes with H- and C- at 110:1 and 1:1 molar ratios.
The changes in the Raman intensity ratios I1096/I1126 and I1096/I1062 were monitored for H- and C-, and the insertion behavior of PEN was examined (Table 1). Across the entire range of lipid-to-peptide molar ratios up to 100, the Raman intensity ratios remained below 1, indicating a higher proportion of trans bonds than gauche bonds [33,34]. This suggests that PEN does not destabilize the C- lipid system. However, at the molar ratio of 110, the Raman intensity ratios I1096/I1126 and I1096/I1062 increased to more than 1, indicating a significant increase in the number of gauche bonds. This change in the insertion behavior of PEN suggests the presence of PEN inside the lipid bilayer at this specific molar ratio. In contrast, the Raman intensity ratios for H- remained below 1 throughout the entire lipid-to-peptide molar ratio range (Table 2), indicating the absence of lipid chain disorder and suggesting the absence of PEN within the H- lipid bilayer. The observed fluctuations in Raman intensities lack a comprehensive explanation at this juncture. It is noteworthy, however, that the Raman intensity values derived from three distinct and independent experiments exhibited a remarkably close degree of consistency. It is essential to highlight the methodology employed in this investigation: we utilized highly diluted solutions, subjected them to freeze drying through lyophilization, and subsequently analyzed the resulting powdered samples. Our working hypothesis posits that the relevance of these dried samples to the peptide insertion mechanism stems from the extreme dilution of the initial solutions, which were maintained at micromolar concentrations.

Table 1.
Physicochemical properties of PEN and its complexes with C- (cancer cells) at various lipid/peptide molar ratios.

Table 2.
Physicochemical properties of PEN and its complexes with H- (healthy cells) at various lipid/peptide molar ratios.
2.2. Characterization of PEN–Vesicle Complexes by DLS and Zeta Potential
To investigate the physicochemical properties of the PEN–vesicle complexes, we conducted dynamic light scattering (DLS) and zeta potential studies using dilute aqueous solutions of PEN at low micromolar levels (10 µM) and pH 6.8. Particle size and surface charge are crucial parameters for evaluating in vivo applications [11,33,35]. The aggregation of vesicles induced by peptide–membrane interactions was examined by DLS to measure the size of the vesicles. Electrostatic interactions between the charged residues in the peptide structure and the vesicles led to changes in the surface charge, which were monitored by zeta potential measurements at neutral pH. Table 1 and Table 2 present the physicochemical properties obtained from the DLS and zeta potential analyses, respectively. Figure S1 in the Supplemental Materials displays the corresponding graphs obtained from these experiments. The polydispersity values in our study spanned from 0.2 to 0.7, signifying that our samples were well suited for Dynamic Light Scattering (DLS) measurements [36]. As depicted in Figure S1, the widening of the distribution, serving as an indicator of the polydispersity of large unilamellar vesicles (LUVs), exhibited a notable increase primarily in the presence of PEN when considering H- and C- vesicles. This observation provides compelling evidence of a robust interaction between PEN and H- as well as C- vesicles.
From the data in the Tables, we assessed the influence of introducing DPPS into the vesicle host composition (H-). Pure H- vesicles exhibited a particle size of 395 nm and a surface charge of −0.75 mV (Table 2). Upon the addition of DPPS, the particle size decreased to 260 nm and the zeta potential shifted to −24.8 mV for the resulting vesicle composition (C-) (Table 1), which was consistent with previous findings in the literature [9,31,32]. The PEN solution displayed a particle size of 34 nm and a surface charge of +11.1 mV (Table 1 and Table 2).
In the investigated lipid-to-peptide molar ratio range, the particle size of PEN–H- complexes (Table 2) generally increased with higher lipid-to-peptide concentrations, except at the highest peptide concentration (1:1 molar ratio). The slightly negative surface charge of the pure H- lipid mixture was compensated for and became positive with increasing lipid-to-peptide molar ratio. Notably, at the 110:1 molar ratio, we observed a larger particle size, suggesting an interaction between PEN and H-. Additionally, the surface charge of H- became slightly positive, indicating that PEN was located at the surface of the vesicles [33,35]. The Raman data supported the observation that PEN maintained its insertion behavior at the 110:1 molar ratio, remaining associated with H-. Furthermore, DLS and zeta potential measurements indicated that H- increased in size and charge in the presence of PEN on its surface at this molar ratio, suggesting an impact of PEN on the structure of H-.
In contrast, the physicochemical properties of PEN–C- complexes (Table 1) varied differently within the same molar ratio range. The particle size of C- fluctuated slightly but ultimately returned to the size of the peptide-free vesicles at the 110:1 molar ratio. The surface charge exhibited a decrease in intensity but remained highly negative. These findings indicate that PEN did not interact significantly with C- and did not reside on the surface of the vesicles. The Raman data indicated a change in PEN’s insertion behavior at the 110:1 molar ratio, indicating its insertion into C-. Despite the presence of PEN within the C- bilayer, DLS and zeta potential measurements confirmed that C- maintained its initial size and charge. This suggests that PEN insertion did not induce damage to the vesicles at this specific molar ratio.
2.3. Thermal Analysis
We employed differential scanning calorimetry (DSC) to achieve two main objectives. Firstly, we aimed to monitor the changes in the main transition temperature (TM) of the lipid vesicles in the presence of PEN, as well as alterations in the transition enthalpy (∆H). DSC thermograms were recorded from room temperature to 100 °C, at a low scanning rate of 0.25 °C per minute (Figure 2), using rehydrated samples. Secondly, we sought to evaluate the ability of PEN to recruit and/or segregate the lipid vesicles. For this purpose, DSC thermograms were obtained from room temperature to 700 °C at a rate of 10 °C per minute, and enthalpy changes (∆H) were measured using dried samples (Figure 3). The specific focus was to gain further insights into the interactions between PEN and the lipid vesicles at the 110:1 lipid-to-peptide molar ratio [16,20,32]. Table 1 and Table 2 provide the thermodynamic data obtained for the pure lipid mixture and different complexes with PEN.
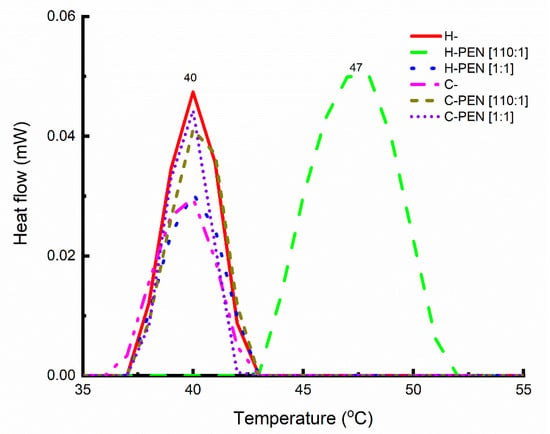
Figure 2.
Differential scanning calorimetry (DSC) thermograms for lyophilized samples of H- and C- (obtained from a 10 μM aqueous solution at physiological pH) and their complexes with PEN at 110:1 and 1:1 molar ratios. DSC thermograms were recorded from room temperature to 100 °C at a scanning rate of 0.25 °C per minute.

Figure 3.
DSC thermograms for lyophilized samples of H- and C- (obtained from a 10 μM aqueous solution at physiological pH) and their complexes with PEN at 110:1 and 1:1 molar ratios. DSC thermograms were recorded from room temperature to 700 °C at a scanning rate of 10 °C per minute.
The phase transition observed in H- (Table 2) exhibited a temperature very close to that of DPPC [33], one of the lipid components in the multi-component lipid system. Upon the addition of PEN at a 110:1 molar ratio, the phase transition temperature shifted to a higher value (from 40.1 °C to 47.5 °C), while the phase transition enthalpy remained unchanged. This upward shift in temperature indicates a change in the lateral organization of lipids with the segregation/recruitment of specific lipids, consistent with previous reports on PEN interactions with other cells [20].
To determine whether PEN could discriminate between the components of the phospholipid mixture mimicking healthy and cancer membranes, we compared the 250–400 °C regions, where lipid decomposition occurs. It is important to note that H- and C- share four lipids, with DPPS added to C-. Figure 3 shows that H- displayed a single prominent peak (∆H = 622 J/g), while C- exhibited one main peak (∆H = 558 J/g) along with a smaller peak (∆H = 44 J/g), likely induced by the presence of PS added to H-. In Figure 3, the main peak for C- is at 330 °C while a shoulder is seen at 355 °C. At a 1:1 lipid-to-peptide molar ratio, PEN recruited a lipid, as evidenced by the appearance of two distinct peaks in the DSC thermograms (Figure 3). At the 110:1 lipid-to-peptide molar ratio, C- displayed a single dissymmetric peak (∆H = 886 J/g) with a shoulder in the DSC thermogram, whereas H- exhibited two distinct peaks (with a total ∆H = 780 J/g). These two distinct peaks represent two domains: one peptide-poor domain and one peptide-rich domain [16,20,37,38,39,40]. Figure 4 displays the thermogravimetric analysis (TGA) curves for lyophilized samples of H- and C-. Importantly, the inclusion of PS in H- induced thermal instability, evident from C-’s (blue curve) decomposition occurring at a lower decomposition temperature than that of H- (red curve).

Figure 4.
Thermogravimetric analysis (TGA) curves for lyophilized samples of H- (healthy) and C- (cancer). The 50% mass loss (horizontal dotted line) occurred around 298 °C for C- (first vertical line) and 311 °C for H- (second vertical line).
2.4. Structural Bond Analysis
Infrared spectroscopy was employed to investigate the secondary structural bonding of PEN within the model membranes. The secondary structural bonding of PEN with and without H- and C- membranes was examined by comparing FTIR spectra in the amide I (1700–1600 cm−1) and amide II (1600–1500 cm−1) regions, which are characteristic of peptides (Figure 5) [37,38,39]. The objective was to observe whether the conformation of PEN changes in the presence of H- and C- model membranes. The FTIR spectrum of PEN exhibited a broad dissymmetric peak with two maxima at 1625 and 1653 cm−1 in the amide I region, indicating a mixture of β-sheet and α-helix structures (Figure 5). The position of the peak at 1625 cm−1 also suggests the presence of strong intermolecular sheet structures [41,42,43,44]. However, the broadness of the amide I band in these regions makes it challenging to determine the proportion of each secondary structure of the peptide [37,38,39,45].
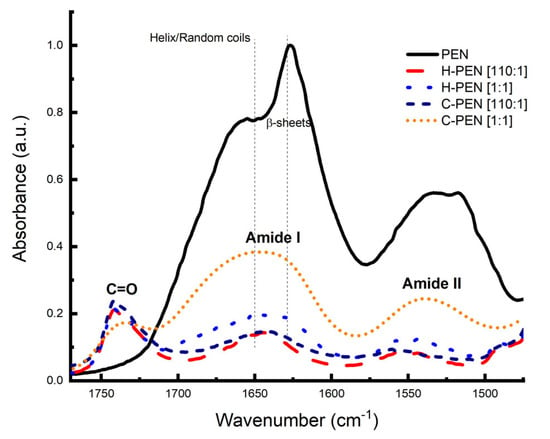
Figure 5.
Fourier transform infrared (FTIR) spectra of lyophilized samples of PEN (obtained from a 10 μM aqueous solution at physiological pH) and its complexes with H- and C- at 110:1 and 1:1 molar ratios. The dominant bands are the lipid ester C=O stretching vibration at 1738 cm−1, the amide I envelope at 1655 cm−1, and the remaining amide II band at 1549 cm−1. The vertical lines indicate the Helix/Random coils region and the β-sheet region.
A quantitative assessment of the secondary structural components of PEN and its complexes was conducted using a curve-fitting approach. Attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR) was employed to record the infrared spectra of PEN and its complexes. The spectral data were initially subjected to normalization, followed by the application of a linear baseline within the spectral range of 1700 to 1600 cm−1. Parameters such as peak frequency, the number of peaks for fitting, and a fixed half-width of 15 cm−1 for each peak were determined from the second derivative of the original IR spectra. Subsequently, the decomposition of the amide I band was carried out using OMNIC software 8.2.0.387 (Thermo Scientific, Waltham, MA, USA). This decomposition involved modeling the amide I band as a summation of Gaussian curves, with iterative optimization of amplitudes, band positions, half-widths, and the Gaussian composition of individual bands. The relative proportions of each secondary structural element were quantified as percentages by dividing the area of a specific amide I band component by the total area encompassing all amide band components.
Quantitative analysis of the peptide conformation based on FTIR spectra revealed that PEN comprises a mixture of helical (43 ± 2%) and β-sheet (57 ± 3%) structures. This observation aligned with expectations, as the presence of tryptophan and lysine residues in PEN is known to induce cation–π and π–π stacking structures, which are characteristic of ordered sheet structures.
Given the variability in the composition of model membranes, our objective was to investigate the impact of these differences on the peptide–membrane interface. In the presence of C- membranes, both at high and low concentrations, PEN predominantly adopted a helical conformation (61% ± 1%), with some instances of turns and intermolecular β-sheet structures. Conversely, in the presence of H- membranes, PEN at a high concentration favored a major β-sheet conformation (54 ± 2%), complemented by turns (29 ± 3%) and helical structures (17 ± 1%). However, at a low concentration in the presence of H- membranes, PEN predominantly assumed β-sheet (39 ± 2%) and random coil structures (61 ± 3%), with no presence of turn structures. The observed prevalence of a helical conformation in the presence of C- membranes is in accordance with prior reports, which have noted that the presence of anionic lipids, such as DPPS, induces helical conformations. Consequently, the absence of the anionic lipid DPPS in H- membranes at a low concentration led to the predominance of sheet-like structures.
3. Discussion
The present study aimed to investigate the interactions between PEN and H- and C- models, with the objectives of identifying a specific lipid-to-peptide molar ratio that triggers direct insertion of PEN into cells, as previously mentioned by Jiao et al. [7], and proposing a PEN insertion mechanism, as suggested by Gehan et al. [14].
3.1. Finding A Specific Lipid-to-Peptide Molar Ratio Triggering Direct Insertion of PEN into Cells
Raman data analysis (Table 1) revealed that within the molar range covered in this study, a lipid-to-peptide molar ratio of 110:1 caused a change in the populations of acyl chain trans and gauche conformers. Below the lipid-to-peptide molar ratio of 110, trans conformers outnumbered gauche conformers, while above this ratio, gauche segments were more abundant than trans ones. This inversion of gauche/trans populations indicated a disruption of the lipid acyl chain order. Only the presence of PEN induced this disorder in the lipid acyl chain of C-, indicating the direct insertion of PEN into the hydrophobic core [26,46]. Additionally, the unchanged frequency of the phosphate group at the 1096 cm−1 peak confirmed that PEN was not anchored to the phosphate head group of the lipid bilayer and did not reside on the membrane surface. Furthermore, no downshift in the frequency of the carbonyl groups was observed. Collectively, these findings suggest that PEN enters the cell directly. This study is the first, to the best of our knowledge, to observe the specific lipid-to-peptide molar ratio that indicates “direct” insertion and the location of PEN into a tumor-mimicking membrane. This 110:1 molar ratio could be the specific lipid-to-peptide molar ratio required to trigger translocation into cells, as mentioned but not specified by Jiao et al. [7]. Importantly, the inversion of acyl chain populations was not observed in PEN-H-. This study demonstrates that Raman techniques can provide unique insights into lipid–peptide interactions that are not easily obtained through other techniques.
It is worth noting that Raman analysis was performed using freeze-dried samples instead of solutions because concentrated samples are required to detect Raman bands. Dilute solutions were prepared, dried, and three independent samples were prepared for each lipid-to-peptide molar ratio.
Several researchers have reviewed the cancer applications of cell-penetrating peptides (CPPs) as drug delivery systems, and the lack of cell specificity has been identified as a major drawback for the clinical development of CPPs [2,47]. However, this study demonstrates that the CPP PEN, even at a relatively low concentration compared to the lipid concentration, can specifically recognize cancer cells and directly insert into them, highlighting the potential utility of PEN in cancer therapy [1]. A detailed characterization of the lipid-to-PEN interactions at the molar ratio of 110:1 is provided to determine the molecular mechanism responsible for PEN’s activity.
3.2. Mechanism of Interaction between C- Model and PEN at the Lipid-to Peptide Molar Ratio of 110
The entry mechanisms of cell-penetrating peptides (CPPs) or CPP/cargo complexes into cells have been extensively studied by researchers [3,7,14,16,20,47]. These studies have generally categorized cellular uptake pathways into two types based on the requirement of energy: direct translocation and endocytosis.
While previous studies have investigated the entry mechanisms of CPPs into cells using model membranes, there have been only a few studies on the entry mechanism of the CPP PEN itself [14,16,20,48]. These studies employed vesicles composed of various lipid compositions to understand the interaction between PEN and the lipid membrane. For instance, Gehan et al. [14] used a binary mixture of POPG and POPC and found that the translocation of PEN requires POPG, whereas Joanne et al. [16] observed recruitment and segregation of specific lipids by PEN in binary lipid mixtures. Alves et al. [20] investigated the internalization mechanism of PEN in cells using a binary mixture of lipids, including DPPS, and observed changes in membrane curvature and lipid supramolecular organization. These studies provide valuable insights into the interactions between PEN and model membranes, but they often used lipid mixtures containing only one or two lipids, which do not fully represent the complexity of actual biological membranes.
In contrast, our study focuses on the interactions between PEN and C- and H- models, which contain at least four different lipids and better represent the composition of living cells [17,49]. These advanced model membranes offer a more realistic representation of biological membranes compared to those composed of one or two lipids used in previous studies [4,7,20]. Additionally, our study utilizes a combination of Raman spectroscopy, dynamic light scattering (DLS), zeta potential analysis, and other techniques to investigate the aggregation behavior of PEN and monitor changes in the acyl chain and phosphate groups. This comprehensive approach adds to the uniqueness of our report.
Based on the Raman data, we propose that the lipid-to-peptide molar ratio of 110:1 (and above) triggers the “direct” insertion of PEN into complex model cancer cells containing DPPS. This finding was also supported by IR analysis. Interestingly, our study focuses on the lipid-to-peptide molar ratio rather than the peptide concentration itself. We observed that a high peptide concentration with respect to lipid did not induce direct insertion, as evidenced by the lipid-to-peptide molar ratio of 1:1, where trans conformers outnumbered gauche conformers and the presence of DPPS did not favor direct insertion.
Additional data from DLS, zeta potential analysis, TGA, and DSC further supported the direct insertion of PEN into C-. The DLS and zeta potential measurements indicated that the addition of PEN to C- does not alter the particle size and surface charge, suggesting that PEN is located deep inside the cell membrane, disrupting the acyl chain order. The TGA data showed an increase in ΔH (enthalpy change) of C- in the presence of PEN, indicating the presence of PEN inside the bilayer and disruption of acyl chain order. The DSC data confirmed the deep localization of PEN within the cell membrane, as no lipid recruitment was observed at the surface of the model cancer cells. The splitting of a peak in the DSC thermogram indicated the absence of lipid recruitment/segregation.
Furthermore, the IR curve-fitting data suggested that the direct insertion of PEN into cancer vesicles involves a conformational change in PEN itself, with the peptide adopting a major helical structure upon contact with DPPS. The presence of DPPS is crucial for direct insertion, but it is not sufficient alone. The specific lipid-to-peptide molar ratio is also necessary for inducing direct insertion, as observed in our study.
In summary, our study reveals that direct insertion of PEN into C- occurs through a series of events: attraction between the positively charged PEN and negatively charged DPPS at the membrane surface, conformational change of PEN from sheet to helical structure in contact with DPPS, and direct entry of PEN into the cell without affecting the particle size and surface charge of C-, as confirmed by DLS and zeta potential analysis. Importantly, the presence of DPPS alone is not enough to induce direct insertion; a specific lipid-to-peptide molar ratio is required. The particle size and surface charge analysis further indicated that PEN insertion at this specific molar ratio does not cause any damage to C-.
3.3. Mechanism of Interaction between H- and PEN at the Lipid-to Peptide Molar Ratio of 110
The interaction between the H- model and PEN at the lipid-to-peptide molar ratio of 110 exhibited distinct characteristics compared to the interaction with C-. The Raman data indicated that PEN is located at the surface of the H- model. The intensity ratios of Raman peaks corresponding to the acyl chain groups (I1098/I1126 and I1098/I1062) remained unchanged, suggesting that the acyl chain region is not significantly affected. However, a shift in the frequency of the phosphate groups to lower wavenumbers indicated substantial interactions between PEN and the phosphate groups at the H- surface (Table 2). Furthermore, comparison of the IR spectra of H- and H-PEN models at the 110:1 molar ratio showed that the frequencies of the carbonyl groups remained unchanged, suggesting that PEN does not interact with the negatively charged carbonyl groups. This implies that PEN is not located between the hydrophobic core and the membrane surface.
At the 110:1 molar ratio, the interaction between PEN and H- is primarily driven by electrostatic interactions with the headgroup region of the bilayer, without causing significant disturbance to the acyl chain region. Similar observations have been reported by Cabrera et al. [50], who showed that an aggregated peptide, jellein-I, accumulates on the headgroup region of a H- model composed of POPC/POPG (70:30 molar ratio). In their study, disturbance of the acyl chain region led to increased membrane permeability by forming pores or defects. In our case, there is no disturbance of the acyl chain groups. While our lipid mixture does not contain anionic lipids, a recent report by Graber et al. [51] suggests that interactions between different lipids can lead to significant ionization of the phosphate groups in the lipid mixture. In this scenario, PEN would bind to the ionized phosphate groups at the membrane surface, recruiting these lipids while segregating the remaining lipids [7]. Several other studies have investigated peptide–vesicle interactions using zwitterionic lipids [20,21,31,52], yielding varying results. In our study, we aimed to demonstrate the low accumulation of PEN on the surface of H- at the molar ratio of 110, indicating the selectivity of PEN at specific lipid-to-peptide molar ratios.
The DSC data (Figure 2) revealed that the addition of PEN to H- at the 110:1 molar ratio caused a shift in the phase transition temperature to a higher value, while the phase transition enthalpy remained unchanged. This shift suggests a change in the lateral lipid organization with the recruitment/segregation of specific lipids [20]. Similar increases in the temperature of the lipid bilayer have been observed by other researchers [16,53]. Consequently, we conclude that PEN can recruit a zwitterionic phospholipid, leading to the formation of a laterally heterogeneous membrane domain that could facilitate the passage of PEN into H-. The unchanged phase transition enthalpy indicates that the acyl chain order is not significantly affected, further supporting the surface localization of the peptide. At the 110:1 lipid-to-peptide molar ratio, the DSC data showed two distinct peaks in the H- thermogram (Figure 4), representing two domains: a peptide-poor domain and a peptide-rich domain [16,20,37,38,39,40]. This suggests that PEN recruits a lipid at the surface, resulting in domain formation. Notably, Alves et al. [20] reported that the cationic peptide used in their study did not interact with zwitterionic lipids at neutral pH. The differences in our findings may be attributed to three factors: the very high lipid-to-peptide molar ratio of 110 at pH 6.8 in our study; the use of a lipid mixture consisting of four different phospholipids with diverse head groups and acyl chains, unlike the single lipid used in their study [20]; and the sensitivity of the Raman technique, which enables the detection of weak interactions, particularly those involving phosphate groups [21,32].
The DLS data indicated that the addition of PEN to H- induces vesicle aggregation, resulting in an increase in particle size (from 395 nm to 563 nm in Table 2). The zeta potential data revealed a compensation of the negative charge, with the zeta potential changing from −0.75 to +1.313 after the addition of PEN.
The combination of various experimental techniques allowed us to propose a mechanism of PEN interaction with H-. At the lipid-to-peptide molar ratio of 110, aggregated PEN particles remain at the surface of H-. Although H- is influenced by the presence of PEN, we did not observe direct insertion of PEN into H- under our experimental conditions.
In this study, we acknowledge the limitations of our experimental conditions, where we deliberately simplified the system by conducting experiments without buffer and at lower ionic strength levels to focus on the fundamental behavior of the cell-penetrating peptide PEN with lipid membranes. For instance, the presence of 0.15 M NaCl would have brought more species (Na+ and Cl− ions) to be considered in the interpretation of results, and thus would have prevented us from seeing the interactions of PEN with phosphate and carbonyl groups of the lipid bilayer. While these conditions allowed us to gain foundational insights, we recognize their limited physiological relevance. To bridge this gap and facilitate the translation of our findings to more biologically representative settings, further investigations conducted under conditions that mimic physiological environments, including the use of buffers and appropriate ionic strengths, are necessary. Such studies will enhance our understanding of how PEN interacts with biological systems and its potential applications in drug delivery and therapeutics.
4. Materials and Methods
4.1. Materials
Dimyristoylphosphotidylethanolamine (DPPE), sphingomyelin (SM), dimyristoylglycerophosphocholine (DPPC), dipalmitoylphosphatidylserine (DPPS), and cholesterol were purchased from Avanti Polar Lipid Inc., Alabaster, AL, USA and used without further purification. The peptide PEN (RQIKIWFQNRRMKWKK) was obtained from GenScript (>97% purity) and used as received.
4.2. Preparation of Non-Cancer and Tumor Cell Membrane Mimetic Vesicles
Unilamellar vesicles were meticulously prepared using a specific lipid composition tailored for cancer cells (DPPC, SM, DPPE, DPPS, and cholesterol in a weight ratio of 4.35:4.35:1:0.3:1) and a distinct composition suitable for healthy cells (DPPC, SM, DPPE, and cholesterol in a weight ratio of 4.35:4.35:1:1). The procedure involved the precise weighing of the dried lipid constituents, followed by dissolution in chloroform and vigorous vortexing for 15 min. Subsequently, the samples were desiccated under a stream of nitrogen gas for a duration of 6 h, followed by 3 h vacuum desiccation period. This process yielded a thin lipid film coating the vial’s interior.
The resulting lipid film was rehydrated with water at physiological pH to achieve a final lipid concentration of 10 µM. LUVs were generated by subjecting the milky lipid suspension to sonication for approximately 1 h in an ice bath, continuing until the solution became transparent [26,32,33,54]. Additionally, stock solutions of the CPP PEN, with a defined concentration of 10 µM, were meticulously prepared.
For the PEN binding experiments, precise volumes of PEN were mixed with corresponding volumes of either cancer or healthy vesicles, resulting in various solutions with distinct proportions of cancer or healthy vesicles. The incubation period was set at 2 h and maintained at a temperature of 37 °C [17,18,20,31,37,49,55].
To validate the findings observed with lower molar ratios, a lipid–peptide mixture with an equimolar amount of DPPS and PEN (1:1 lipid-to-peptide molar ratio) was referred to as the “high peptide” condition. Conversely, “low peptide” samples, characterized by a molar ratio of 110 moles of DPPS to 1 mole of PEN (110:1 lipid-to-peptide molar ratio), were employed to investigate the mechanism of peptide insertion. Each lipid-to-peptide molar ratio was assessed using three independently prepared samples to ascertain the reproducibility of the experiments. The pH of the experimental environment was adjusted to 6.8 [1]. It is noteworthy that at this pH, the charge of PEN was estimated to be +7. Although the preparation process was initiated at room temperature, the subsequent sonication process elevated the temperature to approximately 48 °C.
4.3. Particle Size and Zeta Potential Measurements
The particle size distribution and nature of the aggregates of non-cancer (H-) and tumor cell (C-) membrane mimetic vesicles in solution were measured using dynamic light scattering (DLS) with a Zetasizer Nano Series (Nano-ZS) instrument (Malvern Instruments, Malvern, UK). The measurements were performed with a 633 nm laser diode [11,33]. Zeta potential studies were conducted using the same instrument to measure the surface charge at pH 6.8 [1]. Three measurements, each consisting of 13 runs, were carried out for all the samples. The reported results for particle size and surface charge represent the average of three independent measurements.
4.4. Raman Spectroscopy
Raman spectra were recorded using a confocal microscope Raman spectrometer (DXR Raman microscope from Thermo Scientific, Waltham, MA, USA) equipped with OMNIC software 8.2.0.387. The samples were in the freeze-dried state. The excitation wavelength was 532 nm, and the spectral coverage ranged from 4000 to 400 cm−1 with a spectral resolution of 2 cm−1. The output power was set to 8 mW. The Raman spectra were baseline-corrected, and the Raman intensities were measured as peak height [21,26,27,28,32,54]. The incubation time was 2 h at 37 °C. Freeze-dried samples were placed on a microscope slide.
4.5. Infrared Spectroscopy
Infrared spectra of PEN were obtained using a Fourier transform instrument (Thermo Scientific iS10, Waltham, MA, USA) equipped with a single reflection Ge attenuated total reflectance accessory. Samples in the freeze-dried state were used, and the changes in vibration modes and stretching bonds of PEN with and without the lipid mixture were monitored. The spectra were recorded at room temperature between 4000 and 600 cm−1 with a resolution of 4 cm−1. Sixty-four scans were accumulated, and routine smoothing and normalization were applied to all infrared spectra [30,34,56,57].
4.6. Thermogravimetric Analysis (TGA) and Differential Scanning Calorimetry (DSC)
Thermal stability and phase transitions of the non-cancer (H-) and tumor cell (C-) membrane mimetic vesicles and their complexes were recorded using a LINSEIS STA PT1600 Robbinsville, NJ, USA instrument, which allows simultaneous measurements of mass and caloric reactions of a sample. The instrument performs tests from ultrahigh vacuum (10−4 bar) to 5 bar overpressure. Transition parameters were obtained using the incorporated software. DSC thermograms were recorded from room temperature to 100 °C, at a low scanning rate of 0.25 °C per minute, using rehydrated samples. DSC thermograms were obtained from room temperature to 700 °C at a rate of 10 °C per minute, using samples in the freeze-dried state.
5. Conclusions
In conclusion, our study investigated the mechanism of the interactions between H- and C- (vesicles composed of a lipid mixture) and PEN (a peptide) at a lipid-to-peptide molar ratio of 110. Using complementary techniques such as Raman spectroscopy, IR spectroscopy, DSC, DLS, and zeta potential measurements, we obtained valuable insights into the behavior and effects of PEN on H-.
The results demonstrated that PEN primarily interacts with the surface of H-, as indicated by Raman data showing the localization of PEN at the lipid bilayer surface. The unchanged intensity ratios of acyl chain peaks suggested that PEN does not significantly disturb the acyl chain region. However, a shift in the frequency of phosphate groups to lower wavenumbers indicated significant interactions between PEN and the phosphate groups at the H- surface.
Furthermore, the addition of PEN to H- at the 110:1 molar ratio resulted in changes in the lateral lipid organization, as evidenced by the shift in the phase transition temperature in DSC data. This indicated the recruitment and segregation of specific lipids by PEN, leading to the formation of a laterally heterogeneous membrane domain. Importantly, the acyl chain order remained unaffected, supporting the notion that PEN remains localized at the membrane surface.
The DLS data revealed the aggregation of H- upon the addition of PEN, resulting in larger particle sizes. This was accompanied by a compensation of the negative charge, as indicated by the zeta potential measurements.
Considering the collective findings, we propose a mechanism in which PEN interacts with H- through electrostatic interactions with the headgroup region of the bilayer, specifically with ionized phosphate groups. The absence of disturbance in the acyl chain region suggests that PEN does not insert deeply into the H- structure.
Overall, our study provides valuable insights into the interactions between peptides and lipid vesicles, specifically emphasizing the behavior of PEN with H-. Understanding the mechanisms underlying such interactions is crucial for elucidating peptide–membrane interactions and their potential implications in various biological processes and applications, including drug delivery systems and biomaterial design.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biophysica3040042/s1.
Author Contributions
Conceptualization, N.P. and A.S.-M.; Data curation, B.A.; Formal analysis, Y.Z.H.; Project administration, A.S.-M.; Validation, N.P.; Writing—original draft, N.P.; Writing—review & editing, A.S.-M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to acknowledge the support provided by Tennessee State University under the NSF TIP project (HRD 1623085). Additionally, this research was made possible through funding from the National Science Foundation under grant numbers 1036147 and 1411209.
Data Availability Statement
The data presented in this study are openly available at https://osf.io/8v6q2/?view_only=6f83505181754d318c97c027288eb85d.
Acknowledgments
The authors gratefully acknowledge the support and resources provided by the Vanderbilt Institute of Nanoscale Science and Engineering (VINSE) for conducting Raman spectroscopy, dynamic light scattering (DLS), and zeta potential measurements. Nsoki Phambu would like to express his sincere appreciation to Tennessee State University for their support during this research.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Habault, J.; Poyet, J.-L. Recent Advances in Cell Penetrating Peptide-Based Anticancer Therapies. Molecules 2019, 24, 927. [Google Scholar] [CrossRef] [PubMed]
- Regberg, J.; Srimanee, A.; Langel, Ü. Applications of Cell-Penetrating Peptides for Tumor Targeting and Future Cancer Therapies. Pharmaceuticals 2012, 5, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.T.; Sayers, E.J. Cell entry of cell penetrating peptides: Tales of tails wagging dogs. J. Control. Release 2012, 161, 582–591. [Google Scholar] [CrossRef] [PubMed]
- González-Cruz, A.O.; Hernández-Juárez, J.; Ramírez-Cabrera, M.A.; Balderas-Rentería, I.; Arredondo-Espinoza, E. Peptide-based drug-delivery systems: A new hope for improving cancer therapy. J. Drug Deliv. Sci. Technol. 2022, 72, 103362. [Google Scholar] [CrossRef]
- Stiltner, J.; McCandless, K.; Zahid, M. Cell-Penetrating Peptides: Applications in Tumor Diagnosis and Therapeutics. Pharmaceutics 2021, 13, 890. [Google Scholar] [CrossRef]
- Liu, C.; Tai, L.; Zhang, W.; Wei, G.; Pan, W.; Lu, W. Penetratin, a Potentially Powerful Absorption Enhancer for Noninvasive Intraocular Drug Delivery. Mol. Pharm. 2014, 11, 1218–1227. [Google Scholar] [CrossRef]
- Jiao, C.-Y.; Delaroche, D.; Burlina, F.; Alves, I.D.; Chassaing, G.; Sagan, S. Translocation and Endocytosis for Cell-penetrating Peptide Internalization. J. Biol. Chem. 2009, 284, 33957–33965. [Google Scholar] [CrossRef]
- Zamotaiev, O.M.; Postupalenko, V.Y.; Shvadchak, V.V.; Pivovarenko, V.G.; Klymchenko, A.S.; Mély, Y. Monitoring penetratin interactions with lipid membranes and cell internalization using a new hydration-sensitive fluorescent probe. Org. Biomol. Chem. 2014, 12, 7036–7044. [Google Scholar] [CrossRef]
- Thorén, P.E.G.; Persson, D.; Isakson, P.; Goksör, M.; Önfelt, A.; Nordén, B. Uptake of analogs of penetratin, Tat(48–60) and oligoarginine in live cells. Biochem. Biophys. Res. Commun. 2003, 307, 100–107. [Google Scholar] [CrossRef]
- Miwa, A.; Kamiya, K. Control of Enzyme Reaction Initiation inside Giant Unilamellar Vesicles by the Cell-Penetrating Peptide-Mediated Translocation of Cargo Proteins. ACS Synth. Biol. 2022, 11, 3836–3846. [Google Scholar] [CrossRef]
- Nigatu, A.S.; Vupputuri, S.; Flynn, N.; Ramsey, J.D. Effects of cell-penetrating peptides on transduction efficiency of PEGylated adenovirus. Biomed. Pharmacother. 2015, 71, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Xie, W.; Sun, J.; Yang, L.; Liu, J. Penetratin Peptide-Functionalized Gold Nanostars: Enhanced BBB Permeability and NIR Photothermal Treatment of Alzheimer’s Disease Using Ultralow Irradiance. ACS Appl. Mater. Interfaces 2016, 8, 19291–19302. [Google Scholar] [CrossRef]
- Wang, W.; Liu, W.; Xu, S.; Dong, X.; Sun, Y. Design of Multifunctional Agent Based on Basified Serum Albumin for Efficient In Vivo β-Amyloid Inhibition and Imaging. ACS Appl. Bio Mater. 2020, 3, 3365–3377. [Google Scholar] [CrossRef] [PubMed]
- Gehan, P.; Kulifaj, S.; Soule, P.; Bodin, J.B.; Amoura, M.; Walrant, A.; Sagan, S.; Thiam, A.R.; Ngo, K.; Vivier, V.; et al. Penetratin translocation mechanism through asymmetric droplet interface bilayers. Biochim. Biophys. Acta BBA Biomembr. 2020, 1862, 183415. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Lindblom, G. A Molecular View on the Interaction of the Trojan Peptide Penetratin with the Polar Interface of Lipid Bilayers. Biophys. J. 2004, 87, 332–343. [Google Scholar] [CrossRef]
- Joanne, P.; Galanth, C.; Goasdoué, N.; Nicolas, P.; Sagan, S.; Lavielle, S.; Chassaing, G.; El Amri, C.; Alves, I.D. Lipid reorganization induced by membrane-active peptides probed using differential scanning calorimetry. Biochim. Biophys. Acta BBA Biomembr. 2009, 1788, 1772–1781. [Google Scholar] [CrossRef]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery. Mol. Pharm. 2009, 6, 1264–1276. [Google Scholar] [CrossRef]
- Kay, J.G.; Grinstein, S. Sensing Phosphatidylserine in Cellular Membranes. Sensors 2011, 11, 1744–1755. [Google Scholar] [CrossRef]
- Riedl, S.; Rinner, B.; Asslaber, M.; Schaider, H.; Walzer, S.; Novak, A.; Lohner, K.; Zweytick, D. In search of a novel target—Phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta BBA Biomembr. 2011, 1808, 2638–2645. [Google Scholar] [CrossRef]
- Alves, I.D.; Jiao, C.-Y.; Aubry, S.; Aussedat, B.; Burlina, F.; Chassaing, G.; Sagan, S. Cell biology meets biophysics to unveil the different mechanisms of penetratin internalization in cells. Biochim. Biophys. Acta 2010, 1798, 2231–2239. [Google Scholar] [CrossRef]
- Carrier, D.; Pézolet, M. Raman spectroscopic study of the interaction of poly-L-lysine with dipalmitoylphosphatidylglycerol bilayers. Biophys. J. 1984, 46, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Wodlej, C.; Riedl, S.; Rinner, B.; Leber, R.; Drechsler, C.; Voelker, D.R.; Choi, J.-Y.; Lohner, K.; Zweytick, D. Interaction of two antitumor peptides with membrane lipids—Influence of phosphatidylserine and cholesterol on specificity for melanoma cells. PLoS ONE 2019, 14, e0211187. [Google Scholar] [CrossRef] [PubMed]
- Dimova, R.; Aranda, S.; Bezlyepkina, N.; Nikolov, V.; Riske, K.A.; Lipowsky, R. A practical guide to giant vesicles. Probing the membrane nanoregime via optical microscopy. J. Phys. Condens. Matter. 2006, 18, S1151–S1176. [Google Scholar] [CrossRef]
- Liu, Q.; Bi, C.; Li, J.; Liu, X.; Peng, R.; Jin, C.; Sun, Y.; Lyu, Y.; Liu, H.; Wang, H.; et al. Generating Giant Membrane Vesicles from Live Cells with Preserved Cellular Properties. Research 2019, 2019, 6523970. [Google Scholar] [CrossRef]
- Persson, D.; Thorén, P.E.G.; Esbjörner, E.K.; Goksör, M.; Lincoln, P.; Nordén, B. Vesicle size-dependent translocation of penetratin analogs across lipid membranes. Biochim. Biophys. Acta BBA Biomembr. 2004, 1665, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Kitt, J.P.; Bryce, D.A.; Minteer, S.D.; Harris, J.M. Raman Spectroscopy Reveals Selective Interactions of Cytochrome c with Cardiolipin That Correlate with Membrane Permeability. J. Am. Chem. Soc. 2017, 139, 3851–3860. [Google Scholar] [CrossRef] [PubMed]
- Bryce, D.A.; Kitt, J.P.; Harris, J.M. Raman Microscopy Investigation of GLP-1 Peptide Association with Supported Phospholipid Bilayers. Langmuir 2021, 37, 14265–14274. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Pu, C.; Tang, W. Affinity and selectivity of anchovy antibacterial peptide for Staphylococcus aureus cell membrane lipid and its application in whole milk. Food Control. 2017, 72, 153–163. [Google Scholar] [CrossRef]
- Díaz-Caballero, M.; Navarro, S.; Ventura, S. Soluble Assemblies in the Fibrillation Pathway of Prion-Inspired Artificial Functional Amyloids are Highly Cytotoxic. Biomacromolecules 2020, 21, 2334–2345. [Google Scholar] [CrossRef]
- Krafft, C.; Neudert, L.; Simat, T.; Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2005, 61, 1529–1535. [Google Scholar] [CrossRef]
- Maherani, B.; Arab-Tehrany, E.; Rogalska, E.; Korchowiec, B.; Kheirolomoom, A.; Linder, M. Vibrational, calorimetric, and molecular conformational study on calcein interaction with model lipid membrane. J. Nanoparticle Res. 2013, 15, 1792. [Google Scholar] [CrossRef]
- Domingues, M.M.; Santiago, P.S.; Castanho, M.A.R.B.; Santos, N.C. What can light scattering spectroscopy do for membrane-active peptide studies? J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2008, 14, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Dandurand, J.; Samouillan, V.; Lacoste-Ferre, M.H.; Lacabanne, C.; Bochicchio, B.; Pepe, A. Conformational and thermal characterization of a synthetic peptidic fragment inspired from human tropoelastin: Signature of the amyloid fibers. Pathol. Biol. 2014, 62, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Nguyen, T.H.; Nguyen, D.H. Development and In Vitro Evaluation of Liposomes Using Soy Lecithin to Encapsulate Paclitaxel. Int. J. Biomater. 2017, 2017, 8234712. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Maity, P.; Halder, A. Charge-Driven Interaction of Antimicrobial Peptide NK-2 with Phospholipid Membranes. ACS Omega 2017, 2, 8859–8867. [Google Scholar] [CrossRef] [PubMed]
- Almarwani, B.; Phambu, E.N.; Alexander, C.; Nguyen, H.A.T.; Phambu, N.; Sunda-Meya, A. Vesicles mimicking normal and cancer cell membranes exhibit differential responses to the cell-penetrating peptide Pep-1. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Phambu, N.; Almarwani, B.; Garcia, A.M.; Hamza, N.S.; Muhsen, A.; Baidoo, J.E.; Sunda-Meya, A. Chain length effect on the structure and stability of antimicrobial peptides of the (RW)(n) series. Biophys. Chem. 2017, 227, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Phambu, N.; Almarwani, B.; Alwadai, A.; Phambu, E.N.; Faciane, N.; Marion, C.; Sunda-Meya, A. Calorimetric and Spectroscopic Studies of the Effects of the Cell Penetrating Peptide Pep-1 and the Antimicrobial Peptide Combi-2 on Vesicles Mimicking Escherichia coli Membrane. Langmuir 2017, 33, 12908–12915. [Google Scholar] [CrossRef] [PubMed]
- Willumeit, R.; Kumpugdee, M.; Funari, S.S.; Lohner, K.; Navas, B.P.; Brandenburg, K.; Linser, S.; Andrä, J. Structural rearrangement of model membranes by the peptide antibiotic NK-2. Biochim. Biophys. Acta BBA Biomembr. 2005, 1669, 125–134. [Google Scholar] [CrossRef]
- Roeters, S.J.; Iyer, A.; Pletikapić, G.; Kogan, V.; Subramaniam, V.; Woutersen, S. Evidence for Intramolecular Antiparallel Beta-Sheet Structure in Alpha-Synuclein Fibrils from a Combination of Two-Dimensional Infrared Spectroscopy and Atomic Force Microscopy. Sci. Rep. 2017, 7, 41051. [Google Scholar] [CrossRef]
- Chen, S.W.; Drakulic, S.; Deas, E.; Ouberai, M.; Aprile, F.A.; Arranz, R.; Ness, S.; Roodveldt, C.; Guilliams, T.; De-Genst, E.J.; et al. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci. USA 2015, 112, E1994–E2003. [Google Scholar] [CrossRef]
- Celej, M.S.; Sarroukh, R.; Goormaghtigh, E.; Fidelio, G.D.; Ruysschaert, J.-M.; Raussens, V. Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure. Biochem. J. 2012, 443, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Okubo, K.; Ikeda, K.; Yano, Y.; Hoshino, M.; Hayashi, Y.; Kiso, Y.; Itoh-Watanabe, H.; Naito, A.; Matsuzaki, K. Toxic Amyloid Tape: A Novel Mixed Antiparallel/Parallel β-Sheet Structure Formed by Amyloid β-Protein on GM1 Clusters. ACS Chem. Neurosci. 2019, 10, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Sayegh, R.S.R.; de Fátima Correia Batista, I.; de Melo, R.L.; Riske, K.A.; Daffre, S.; Montich, G.; da Silva Junior, P.I. Longipin: An Amyloid Antimicrobial Peptide from the Harvestman Acutisoma longipes (Arachnida: Opiliones) with Preferential Affinity for Anionic Vesicles. PLoS ONE 2016, 11, e0167953. [Google Scholar] [CrossRef] [PubMed]
- Schultz, Z.D.; Levin, I.W. Vibrational Spectroscopy of Biomembranes. Annu. Rev. Anal. Chem. 2011, 4, 343–366. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020, 11, 697. [Google Scholar] [CrossRef]
- Alves, I.D.; Correia, I.; Jiao, C.Y.; Sachon, E.; Sagan, S.; Lavielle, S.; Tollin, G.; Chassaing, G. The interaction of cell-penetrating peptides with lipid model systems and subsequent lipid reorganization: Thermodynamic and structural characterization. J. Pept. Sci. 2009, 15, 200–209. [Google Scholar] [CrossRef]
- Li, G.; Huang, Y.; Feng, Q.; Chen, Y. Tryptophan as a Probe to Study the Anticancer Mechanism of Action and Specificity of α-Helical Anticancer Peptides. Molecules 2014, 19, 12224–12241. [Google Scholar] [CrossRef]
- dos Santos Cabrera, M.P.; Baldissera, G.; da Costa Silva-Gonçalves, L.; de Souza, B.M.; Riske, K.A.; Palma, M.S.; Ruggiero, J.R.; Arcisio-Miranda, M. Combining experimental evidence and molecular dynamic simulations to understand the mechanism of action of the antimicrobial octapeptide jelleine-I. Biochemistry 2014, 53, 4857–4868. [Google Scholar] [CrossRef]
- Graber, Z.T.; Thomas, J.; Johnson, E.; Gericke, A.; Kooijman, E.E. Effect of H-Bond Donor Lipids on Phosphatidylinositol-3,4,5-Trisphosphate Ionization and Clustering. Biophys. J. 2018, 114, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, P.; Epand, R.F.; Heidenreich, N.; Burck, J.; Ulrich, A.S.; Epand, R.M. Membrane-Active Peptides and the Clustering of Anionic Lipids. Biophys. J. Vol. 2012, 103, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Mangelschots, J.; Bibian, M.; Gardiner, J.; Waddington, L.; Van Wanseele, Y.; Van Eeckhaut, A.; Acevedo, M.M.D.; Van Mele, B.; Madder, A.; Hoogenboom, R.; et al. Mixed α/β-Peptides as a Class of Short Amphipathic Peptide Hydrogelators with Enhanced Proteolytic Stability. Biomacromolecules 2016, 17, 437–445. [Google Scholar] [CrossRef]
- Vincent, J.S.; Levin, I.W. Interaction of ferricytochrome c with cardiolipin multilayers: A resonance Raman study. J. Am. Chem. Soc. 1986, 108, 3551–3554. [Google Scholar] [CrossRef]
- Klaiss-Luna, M.C.; Jemioła-Rzemińska, M.; Strzałka, K.; Manrique-Moreno, M. Understanding the Biophysical Interaction of LTX-315 with Tumoral Model Membranes. Int. J. Mol. Sci. 2022, 24, 581. [Google Scholar] [CrossRef]
- Blume, A.; Huebner, W.; Messner, G. Fourier transform infrared spectroscopy of 13C:O labeled phospholipids hydrogen bonding to carbonyl groups. Biochemistry 1988, 27, 8239–8249. [Google Scholar] [CrossRef]
- Dyck, M.; Kerth, A.; Blume, A.; Lösche, M. Interaction of the Neurotransmitter, Neuropeptide Y, with Phospholipid Membranes: Infrared Spectroscopic Characterization at the Air/Water Interface†. J. Phys. Chem. B 2006, 110, 22152–22159. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).