1. Introduction
It is well known that RNA is a string of nitrogenous bases containing the sequence of instructions for constructing amino acids, as well as other interesting information. The organization suggests a strictly deterministic procedure. Once having established the beginning of a sequence, the nucleotides are considered three by three, thus forming a series of
codons. Each codon is therefore identified by a triplet of symbols, taken from the four available, i.e.,
Uracil (U),
Cytosine (C),
Adenine (A), and
Guanine (G). There are 64 possible combinations. Each of them allows us to identify a single amino acid according to the famous conversion chart (see
Table 1), obtained thanks to the pioneering efforts of M. Nirenberg and coworkers [
1].
The result is extraordinary, but not perfectly convincing from the theoretical viewpoint. In fact, there are only 21 amino acids, so there are several codons that lead to the same result. The table therefore displays some redundancy (called
codon degeneracy) and there does not seem to exist a clear law that allows the combinations to be thinned out in a logical way. In particular, there are eight amino acids having four or more codon representations (
Ser,
Leu,
Pro,
Arg,
Thr,
Val,
Ala,
Gly), and 12 amino acids having one to three codon representations (
Phe, Leu, Tyr, Cys, Trp, His, Gin, Ile, Asn, Lys, Asp, Glu). Finally, there are four special codons. One of them corresponds to the amino acid
Met, that determines the beginning of a protein. Each of the last three codons represent the command
stop that marks instead the end of a protein. Many attempts have been made in the past to try to better formalize the table (see, e.g., [
2]). Given that nature makes its resources available in a very parsimonious way, the aim is to understand if there are hidden relationships that justify the existence of 64 combinations when in the end only a third would be necessary.
The idea we propose here is to interpret the genetic code in a dynamic way. RNA is not trivially a score to be read and performed by other, more or less complex, molecules. Instead, it is itself the entire musical instrument, which decrypts and, at the same time, plays appropriate chords associated with each codon. The trick is to assume that the message carried by a codon can be repeated periodically so as to become a sort of sound. Different amino acids will correspond to different sounds, but there will be less sounds than codons. The idea presented here is simple but could, if tested with scientific rigor, prove to be a breakthrough in the study of the genetic code and how its information is conveyed.
The paper is organized as follows. In
Section 2, we provide a little mathematical introduction to the problem. The first part requires a basic knowledge of combinatorics. In the the second part, we propose a set of periodic functions which will later be combined to explicitly construct what we will define as sounds. The results of this procedures will be reported in
Section 3, while in
Section 4 we we will give the reasons that underlie our choices. The conclusions are presented in
Section 5.
2. Methods
To start, we want to calculate the number of periodic sequences that are obtained by indefinitely repeating a triplet of symbols, taken from a vocabulary of only four symbols. The calculation is straightforward.
Suppose to have the following list of four elements: . We can consider all possible triplets that can be formed using the above characters. We know that all combinations amount to 64. We then take the periodic signal generated by the repetition of a given triplet for an indefinite number of times. For example, if we choose , the corresponding “sound” is . The question is: how many different sequences can be built? First of all, there exist four triplets , , , , that generate trivial constant monochromatic sequences. We are left with another 60 interesting cases. Let us now observe that the sequence originating for example from coincides with those generated by or . Indeed, the above three combinations give rise to the same sound, therefore they are redundant. This means that it is possible to group within a single class the three triplets that produce a specific sound. Dividing 60 by 3, we obtain 20 classes, i.e., 20 different possible sounds. Incidentally, with the exception of Met, 20 is also the number of amino acids. The coincidence is somewhat surprising. However, we have not finished yet. There are eight sounds where the symbols are all different. It may not be a just coincidence that exactly eight amino acids admit representations with four or six redundant codons. There are 12 sequences where a given symbol appears twice, such as for example: . Do they correspond to the other 12 amino acids? Finally, there are four monochromatic sounds. Do they correspond to the start and stop signals?
We now show how to mathematically generate
different sounds starting from certain assigned frequencies. Let us say right away that we will not be able to fully solve the problem of rewriting with a minimal number of entries the conversion chart of
Table 1. In practice, we will not be able to associate in automatic way the generic symbols
with the classic ones representing the nitrogenous bases, i.e.,: U, C, A, G. Indeed, it will soon be evident that a straight relation does not hold. We think that, physically, a set of three ordered codons constitutes a unique oscillating electric circuit which elaborates three resonating frequencies (described by some of the symbols
) which, in turn, produce a characteristic sound. We do not yet know exactly how these circuits work. We provide some ideas in
Section 4. The purpose of this paper is mainly to interpret the genetic code encrypted in the DNA in an active way, which is realized through the emission of electromagnetic messages at a distance. For the reasons that will be better expressed in
Section 4, we believe that DNA is not only a data container but also transmits its data into the environment. At the moment, this conjecture allows us to understand why the symbols U, C, A, G produce redundant combinations, while this is no longer verified if we use symbols
. The connection between the two sets of symbols is yet to be found. Note that we are not just sidestepping the problem with semantic speculation, we are offering a new way to understand how DNA works.
Let us put our ideas into mathematical language. We denote by
the generic triplet of values obtained by choosing randomly (with possible repetition) three members from a given set
of numerical values. According to what has been said in
Section 1, we want the triplet
to be equivalent to
and
(in fact, they all generate the same sound). Instead,
will not be equivalent in general to
, corresponding to another sound in our construction. Successively, we set
and define the functions of the variable
x:
The above are periodic functions with period equal to
. The first three (
1), (
2), (
3) constitute a classic trigonometric Lagrangian basis with respect to the nodes
. The elements of the basis actually attain a value of 1 at one of the nodes and 0 at the remaining two. Function (
4), on the other hand, vanishes at all nodes. Thus, for given values of
, the function (
5) suitably combines the previous functions. We observe the following facts.
For a fixed
a, we have the relations,
:
The second relation above says that, when the elements of the triplet are the same (
), the corresponding function is purely periodic, having a frequency equal to
.
Other properties to be pointed out are:
We finally have:
This last property says that an even permutation of the indices produces the same wave, up to a phase change. With this, we want to express, for instance, that
,
,
(triplets subject to an even number of permutations) will generate the same sound. The same must be true for triplets
,
,
(still linked by even permutations). However, we do not want the first set of triplets to produce the same sound as the second set, although they use the same symbols, since the two sets are linked by odd permutations. To this end, an appropriate choice of parameters
must be implemented.
3. Results
We propose the following values for our parameters:
Such a choice is arbitrary, meaning that other choices can work in a similar way. This is true, except for some critical choices that we are not going to list here. According to the second equation in (
6), when all indices are equal to
a we obtain a monochromatic wave of frequency
. We avoid the trivial graphical representation in this case. Otherwise, using (
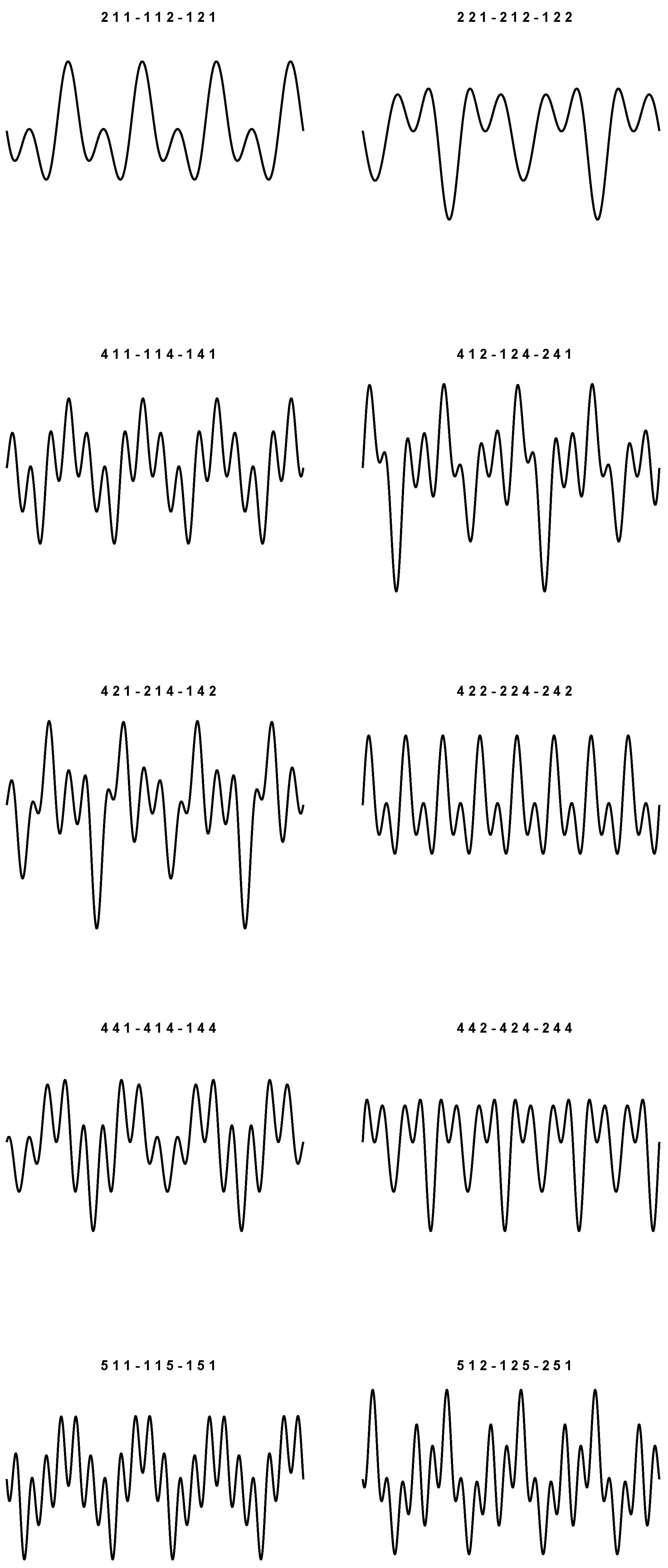
9), we obtain 20 sounds which are plotted in
Figure 1 and
Figure 2. In particular, things have been arranged so that the periodic plots are all different. The values used to simulate each wave are listed at the top of the figures.
As prefigured, for each plot, the corresponding triplets are the same up to even permutations. In the context of these representations, we do not want to state that the graphs shown are actually the real ones. We just want to argue that it is in fact possible to build 20 different plots, and that there are no other possible displacements (besides the four pure frequencies), even if we have three-by-three permutations of four symbols in play.
As we said, the idea is to associate each sound with an amino acid, and vice versa. In this way there is no more redundancy. Other periodic signals can be obtained with different parameter choices. However, the essence of the concept is the same. The problem remains of how a sound can be generated from a single codon, respecting the rules of the conversion
Table 1. Some hints about this are provided in the paragraphs to follow.
In the framework of differential models, a simple oscillator is obtained through the solution of the system:
that has its natural frequency equal to
. An alternative writing of (
10) is:
with
. Here, the matrix
M has imaginary eigenvalues
.
We then propose the following generalization:
where
has now six components. To better understand what we have in mind, let us refer to
Figure 3.
Each nitrogenous base of a given codon can be viewed as an oscillatory electric circuit having its own resonance frequency (, , , if we refer to the figure). Altogether, they constitute a new global circuit, where links are expressed with the help of some coupling constants , . These constants vary depending on the special type of bases involved. For example, the constant coupling Cytosine and Adenine is expected to be different from that coupling Cytosine and Guanine. This produces a characteristic time behavior of the corresponding solution . To know the main frequencies involved, the eigenvalues of the matrix M relative to the specific codon should be computed. The codon is not just the union of the uncoupled bases, but contains information on their exact displacement through the knowledge of , . In truth, just three parameters should be enough to determine a specific sound. Thus, there must be a sort of decoding procedure that selects the values , , , allowing for the construction of the final sound. For example, it could be a filter that considers only the three frequencies appearing with greater magnitude in the Fourier expansion of some component of the vector .
For a theoretical analysis, each codon should be replaced by an electrical network based on the conduction properties of the chemical elements involved. Such a circuit has its own resonance frequencies which, in turn, should produce a sound. As will be revealed in the next section, the electromagnetic sound could be naturally emitted as biophoton or run along the phosphate backbone. The design of the electric diagram and the choice of the corresponding parameters (resistors and capacitors, to be evaluated based on the various coupling constants) are still in a primordial stage. The laboratory analysis of DNA photon emission is also in an early stage to allow for reasonable comparisons. For the reasons set out above, it is at the moment difficult to give a more detailed description of the quantities involved in the whole process.
4. Discussion
It has been ascertained under various circumstances that the DNA double helix can be associated with some electric circuit. The results are both experimental and computational. They confirm, for instance, the conductivity of the periodic phosphate backbone [
3,
4], thanks to its one-dimensional periodic structure. Electronic properties of DNA and its nucleotides have been studied, for instance, in [
5,
6,
7,
8]. We observe that the presence of nitrogen can give compounds a tendency to flip from one configuration to another, activating spontaneous oscillations. Concatenations of logic gates using basic molecules as switches have been taken into consideration [
9,
10]. A model of DNA composed of multiple oscillatory RLC circuits is examined in [
11]. These micro circuits are joined by the phosphate backbones that collect the data.
All these results reinforce the idea that DNA should be interpreted in a dynamic way. Therefore, together with the intrinsically static message written in the codons, we also must consider a high probability that this should be assigned a periodic sound, which brings us back to the content of this paper. Obviously, we do not have concrete proof of these conjectures, but common sense tells us that we could be on a good path.
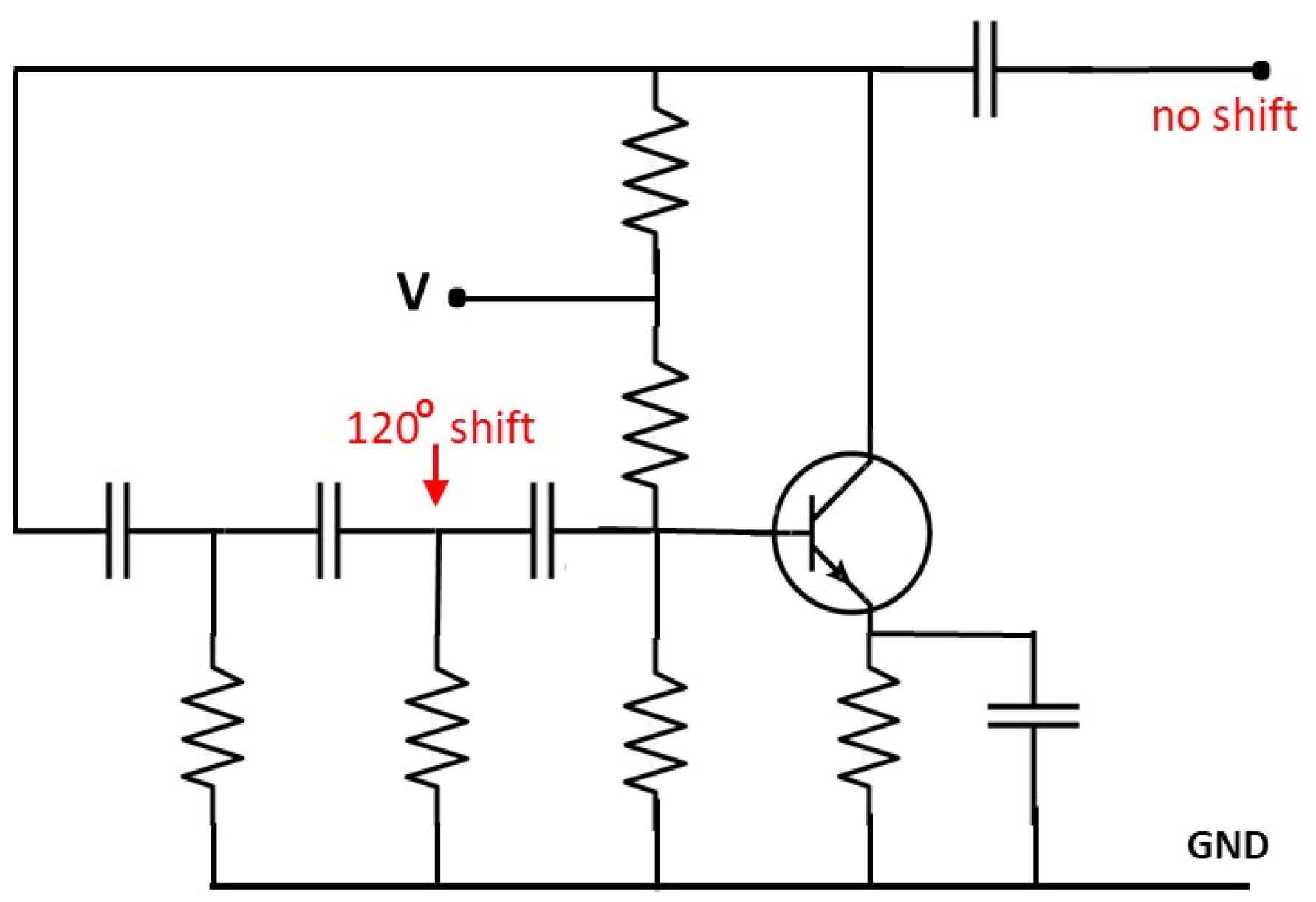
Electric circuits displaying a phase shift equal to
can be easily built (see
Figure 4). Signals such as those studied in the previous section can be generated by collecting the outputs of three devices of this type resonating at different frequencies. Each circuit has its own RC network, and the sum of the frequencies is obtained by imposing suitable shifting parameters so to recover periodic composite waves. This highlights the reciprocal localization properties between nitrogenous bases. The functions in (
1)-(
2)-(
3) are effectively one-third the wavelength out of phase and the sum of the corresponding signals would actually inspire the construction of an appropriate electrical device.
We have no element at this time to continue our investigation. A way should be proposed to assign to each nitrogenous base an electric circuit. The global circuit associated with each codon should then produce a periodic sound (among the 20 + 4 possible). In principle, this is not difficult. However, it is difficult to respect the hidden rules of the codon table. For example, the generic sequence GGX (where X is any of the symbols U, C, A, G) corresponds to Glycine (last box in
Table 1). For some unknown reason, the electric circuit associated with this codon resonates with the couple GG and inhibits the presence of the third component. In terms of the elements of the matrix
M in (
12), this means that
and the coupling constant
associated with
Gli–Gli affects the eigenvalues distribution so that the roles of
and
become secondary. We undertake to study this issue further in a future paper. Meanwhile, let us add some other considerations in favor of a vision in which DNA is an instrument actually played, and not just a static collection of data. The detailed solution of this problem certainly has considerable implications, both from a practical viewpoint and from the concept we have regarding the structure of the genetic information encoded in DNA. New horizons in encryption algorithms suggest the use of RNA dynamic for decoding and coding [
12,
13]. Although these implementations do not currently seem to be closely related to the object of this work, they may serve as a point of reference for future investigations.
The analysis of vibration frequency maps carried out for nucleobases [
14,
15] shows strong DNA activity in producing or absorbing biophotons. Such an intense electromagnetic turmoil regulates the most fundamental biological processes. This aspect strengthens the hypothesis of signals originating from DNA, which remotely guide the main functions of a single cell, but which can extend further, transferring information between groups of cells [
16,
17,
18]. These aspects have also been taken into consideration in [
19] (Section 3.7) and [
20] in a more general context, where the chemical elements themselves arise from an agglomeration of atomic nuclei and electromagnetic manifestations that connect them in a peculiar way characterizing the compound.
The electromagnetic frequencies involved in biological phenomena are in the Terahertz range (
Hertz) [
21]. The wavelengths are therefore of the order of the hundredth or tenth of a millimeter. In terms of applications, such studies are especially important in the medical field, where the sending of intense impulses at frequencies in the THz range can damage or repair organic tissues. The consequences of exposure to high-energy THz radiation are examined in [
22]. An analysis of the emission spectrum of DNA is presented, for example, in [
23], using appropriate instrumentation. Indeed, such experiments usually require very sophisticated equipment [
24]. Through THz waves it is possible to detect changes in DNA and construct precise resonance fingerprints. This allows us to identify cancer cells, for example [
25].
Finally, we must not forget that DNA is a twisted strand that is difficult to access from a physical viewpoint. Moreover, the cell is a system overcrowded with molecules [
26,
27], in which it is practically impossible to create connections except through communications that take place with the exchange of electromagnetic impulses. Of course, it is necessary to assume that suitable receiving stations exist. The RNA polymerase could be one of them, thus eliminating direct contact with DNA, which appears unrealistic in in vivo situations.
We collect the most salient facts here at the bottom:
The genetic information contained in the cell nucleus is extremely difficult to access, at least from a chemical-physical point of view;
Experimental evidence demonstrates that information inside and outside the cells can be propagated through electromagnetic impulses, commonly in the THz range;
The branch known as biophotonics shows that the emission and absorption of photons, at various energy states, is a common phenomenon also at the level of living organisms;
It is therefore not surprising to think that, even in and around DNA, messages are mainly transmitted through traveling electromagnetic waves;
It is not even too bold to think that these messages are not just pulses, but real electromagnetic waves that are reproduced periodically, giving the idea that DNA is more of a transmitting station than a static list of symbols. The studies performed on DNA by various authors are able to confirm that this hypothesis is plausible. It should be noted that, from a measurement point of view, these messages could be very difficult to identify, given that they are of very high frequency and whose shapes can be very complex;
If the above is true, then we have the basis for solving the codon degeneracy problem elegantly. In fact, the redundancy associated with reading the DNA as a mere list of stationary instructions disappears if we switch to the dynamic version instead.