Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Gómez Castellanos, J.R.; Gadda, G.; Fraaije, M.W.; Mattevi, A. Same substrate, many reactions: Oxygen activation in flavoenzymes. Chem. Rev. 2018, 118, 1742–1769. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef]
- Torra, J.; Lafaye, C.; Signor, L.; Aumonier, S.; Flors, C.; Shu, X.; Nonell, S.; Gotthard, G.; Royant, A. Tailing miniSOG: Structural bases of the complex photophysics of a favin-binding singlet oxygen photosensitizing protein. Sci. Rep. 2019, 9, 2428. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Lafaye, C.; Aumonier, S.; Torra, J.; Signor, L.; von Stetten, D.; Noirclerc-Savoye, M.; Shu, X.; Ruiz-González, R.; Gotthard, G.; Royant, A.; et al. Ribofavin-binding proteins for singlet oxygen production. Photochem. Photobiol. Sci. 2022, 21, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- Colloc’h, N.; Gabison, L.; Monard, G.; Altarsha, M.; Chiadmi, M.; Marassio, G.; Sopkova-de Oliveira Santos, J.; El Hajji, M.; Castro, B.; Abraini, J.H.; et al. Oxygen pressurized X-ray crystallography: Probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism. Biophys. J. 2008, 95, 2415–2422. [Google Scholar] [CrossRef] [PubMed]
- Matthews, A.; Saleem-Batcha, R.; Sanders, J.N.; Stull, F.; Houk, K.N.; Teufel, R. Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases. Nat. Chem. Biol. 2020, 16, 556–563. [Google Scholar] [CrossRef]
- Auhim, H.S.; Grigorenko, B.L.; Harris, T.K.; Aksakal, O.E.; Polyakov, I.V.; Berry, C.; Gomes, G.d.P.; Alabugin, I.V.; Rizkallah, P.J.; Nemukhin, A.V.; et al. Stalling chromophore synthesis of the fluorescent protein venus reveals the molecular basis of the final oxidation step. Chem. Sci. 2021, 12, 7735–7745. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.; Kim, K.; King, P.; Seibert, M.; Schulten, K. Finding Gas Diffusion Pathways in Proteins: Application to O2 and H2 Transport in CpI [FeFe]-Hydrogenase and the Role of Packing Defects. Structure 2005, 13, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Carpentier, P.; Bourgeois, D.; Field, M. Diffusion Pathways of Oxygen Species in the Phototoxic Fluorescent Protein KillerRed. Photochem. Photobiol. Sci. 2010, 9, 1342–1350. [Google Scholar] [CrossRef]
- Chapagain, P.P.; Regmi, C.K.; Castillo, W. Fluorescent Protein Barrel Fluctuations and Oxygen Diffusion Pathways in mCherry. J. Chem. Phys. 2011, 135, 235101. [Google Scholar] [CrossRef]
- Regmi, C.K.; Bhandari, Y.R.; Gerstman, B.S.; Chapagain, P.P. Exploring the Diffusion of Molecular Oxygen in the Red Fluorescent Protein mCherry Using Explicit Oxygen Molecular Dynamics Simulations. J. Phys. Chem. B 2013, 117, 2247–2253. [Google Scholar] [CrossRef]
- Polyakov, I.V.; Domratcheva, T.M.; Kulakova, A.M.; Nemukhin, A.V.; Grigorenko, B.L. Computational Modeling of the Interaction of Molecular Oxygen with the Flavin-Dependent Enzyme RutA. Supercomput. Front. Innov. 2022, 9, 46–55. [Google Scholar] [CrossRef]
- Polyakov, I.V.; Nemukhin, A.V.; Domratcheva, T.M.; Kulakova, A.M.; Grigorenko, B.L. Quantum-based modeling of protein-ligand interaction: The complex of RutA with uracil and molecular oxygen. Mol. Inform. 2022, 41, 2200175. [Google Scholar] [CrossRef] [PubMed]
- Grigorenko, B.L.; Nemukhin, A.V.; Polyakov, I.V.; Khrenova, M.G.; Krylov, A.I. A light-induced reaction with oxygen leads to chromophore decomposition and irreversible photobleaching in GFP-type proteins. J. Phys. Chem. B 2015, 119, 5444–5452. [Google Scholar] [CrossRef]
- Grigorenko, B.L.; Krylov, A.I.; Nemukhin, A.V. Molecular Modeling Clarifies the Mechanism of Chromophore Maturation in the Green Fluorescent Protein. J. Am. Chem. Soc. 2017, 139, 10239–10249. [Google Scholar] [CrossRef]
- Grigorenko, B.L.; Domratcheva, T.M.; Nemukhin, A.V. QM/MM Modeling of the Flavin Functionalization in the RutA Monooxygenase. Molecules 2023, 28, 2405. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Aleksandrov, A. A Molecular Mechanics Model for Flavins. J. Comp. Chem. 2019, 40, 2834–2842. [Google Scholar] [CrossRef]
- Wang, S.; Hou, K.; Heinz, H. Accurate and Compatible Force Fields for Molecular Oxygen, Nitrogen, and Hydrogen to Simulate Gases, Electrolytes, and Heterogeneous Interfaces. J. Chem. Theory Comput. 2021, 17, 5198–5213. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Chai, J.-D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Melo, M.C.R.; Bernardi, R.C.; Rudack, T.; Scheurer, M.; Riplinger, C.; Phillips, J.C.; Maia, J.D.C.; Rocha, G.B.; Ribeiro, J.V.; Stone, J.E.; et al. NAMD Goes Quantum: An Integrative Suite for Hybrid Simulations. Nat. Methods 2018, 15, 351–354. [Google Scholar] [CrossRef]
- Seritan, S.; Bannwarth, C.; Fales, B.S.; Hohenstein, E.G.; Isborn, C.M.; Kokkila-Schumacher, S.I.L.; Li, X.; Liu, F.; Luehr, N.; Snyder, J.W., Jr.; et al. Terachem: A graphical processing unit-accelerated electronic structure package for large-scale ab initio molecular dynamics. WIREs Comput. Mol. Sci. 2021, 11, e1494. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef]

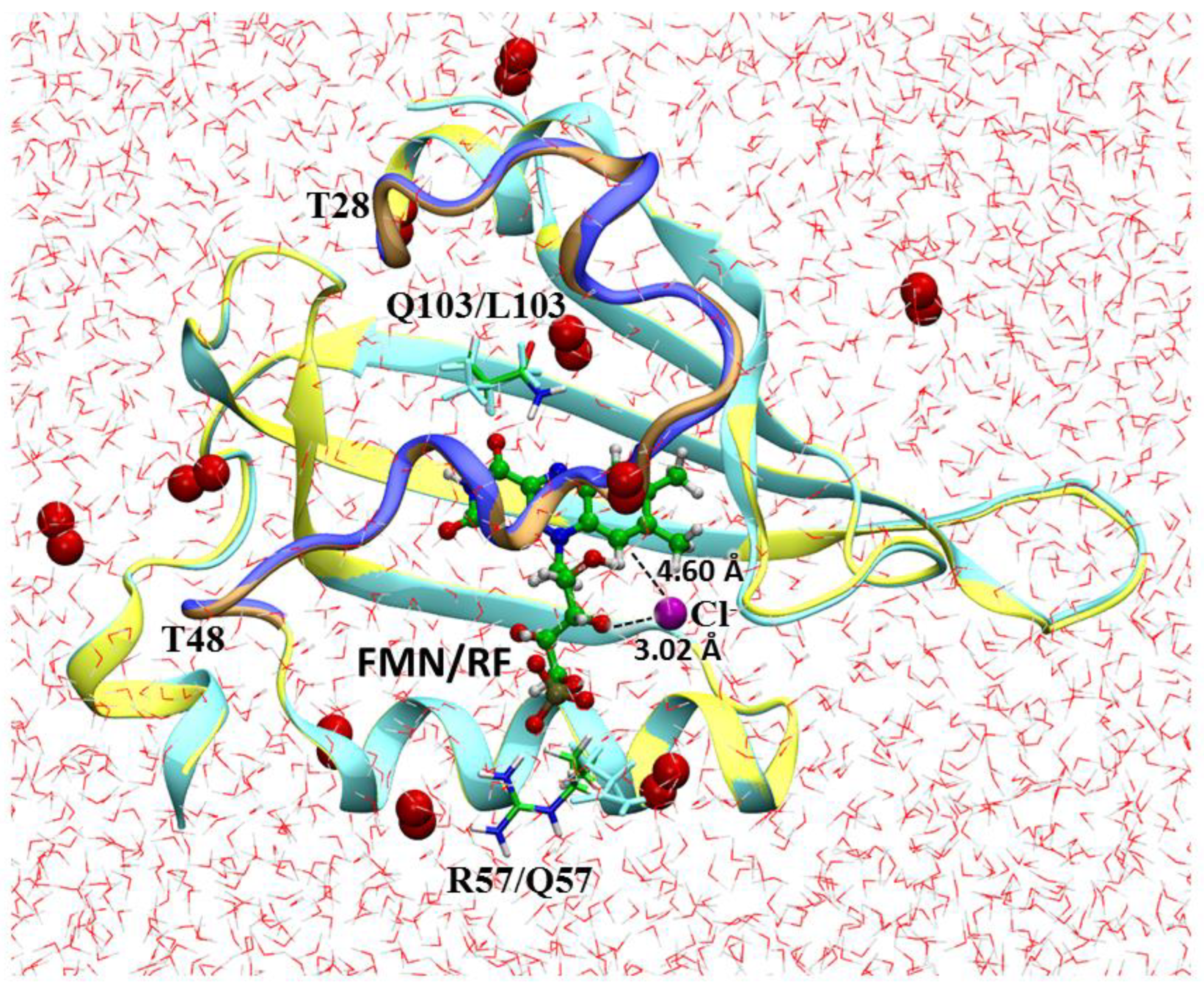
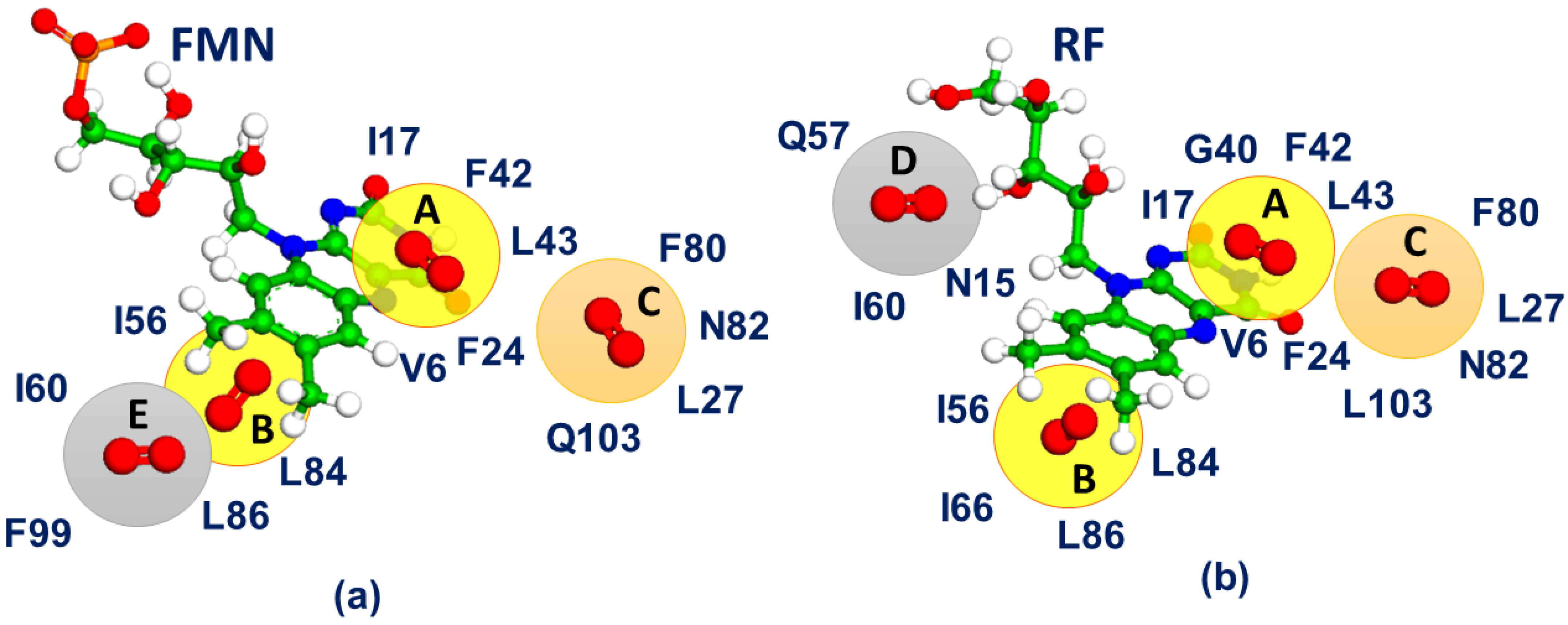

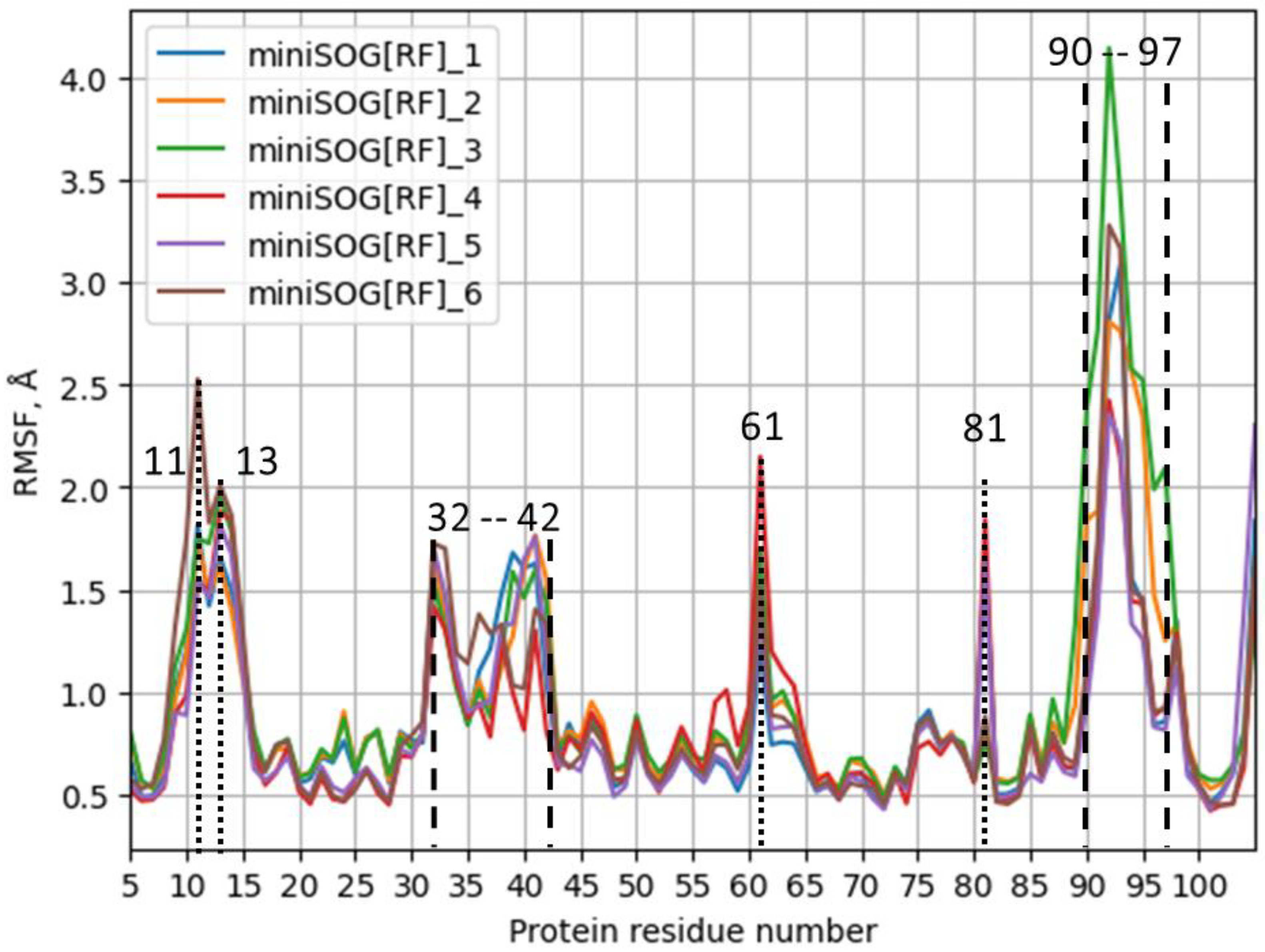
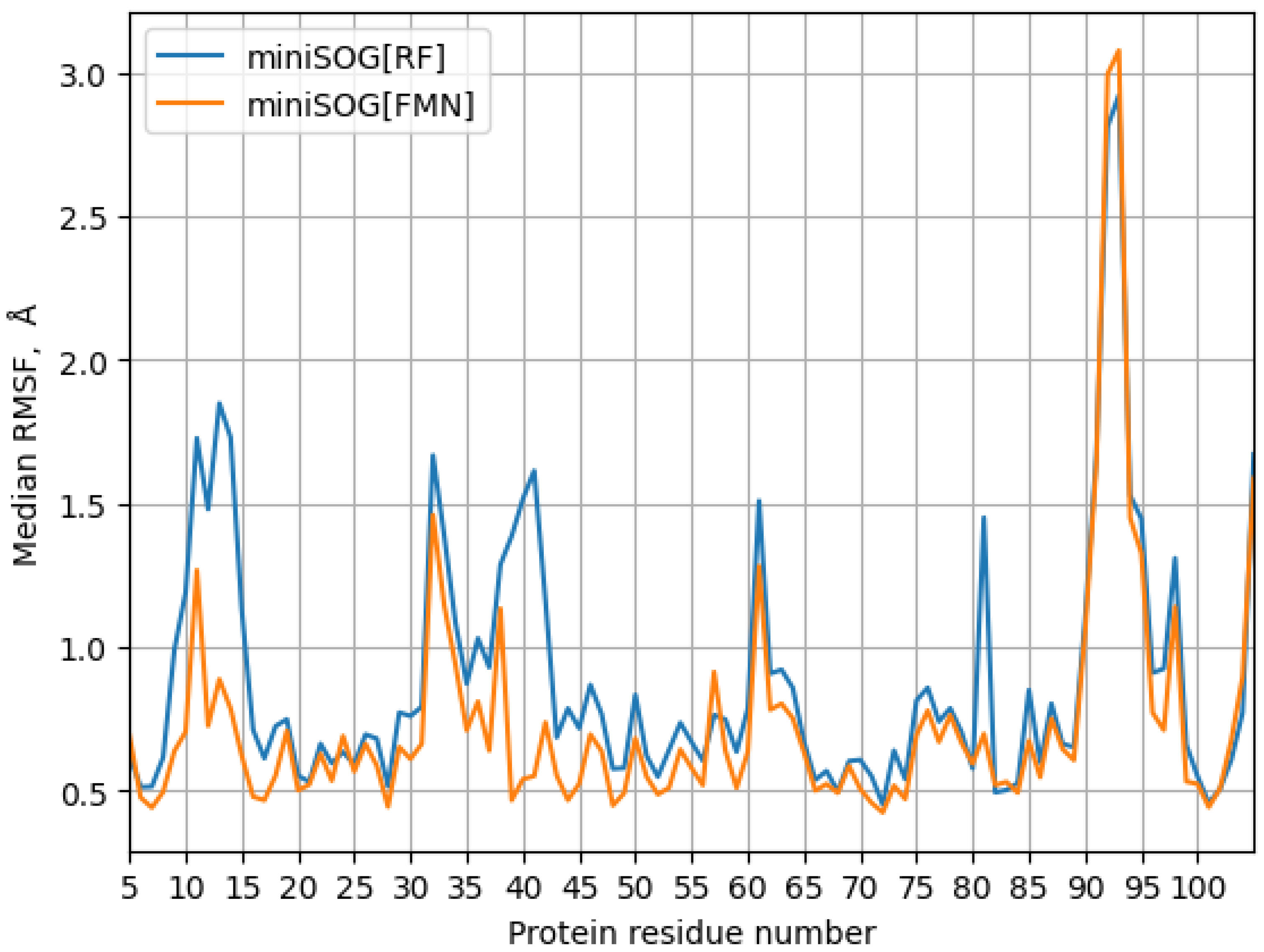
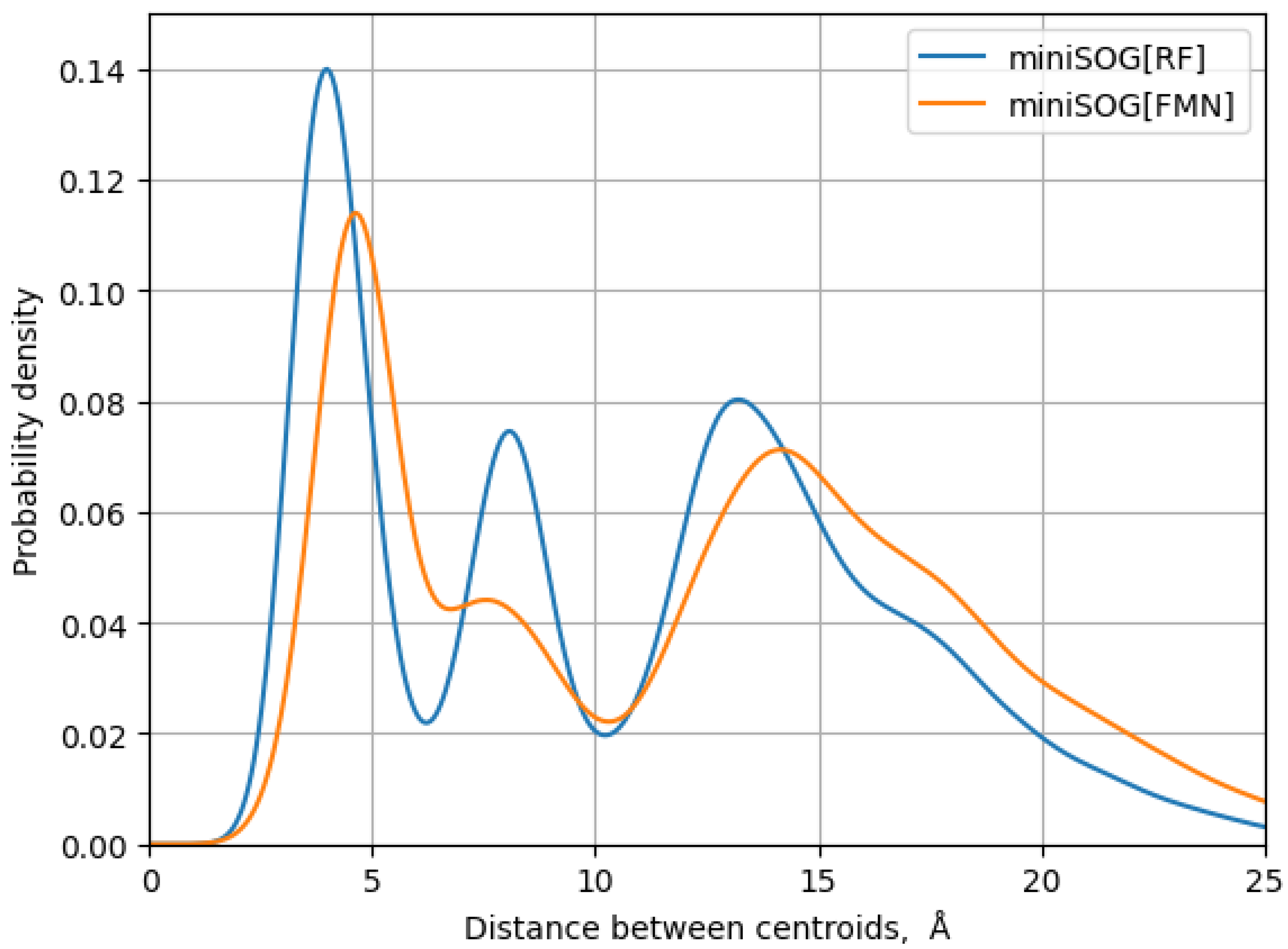
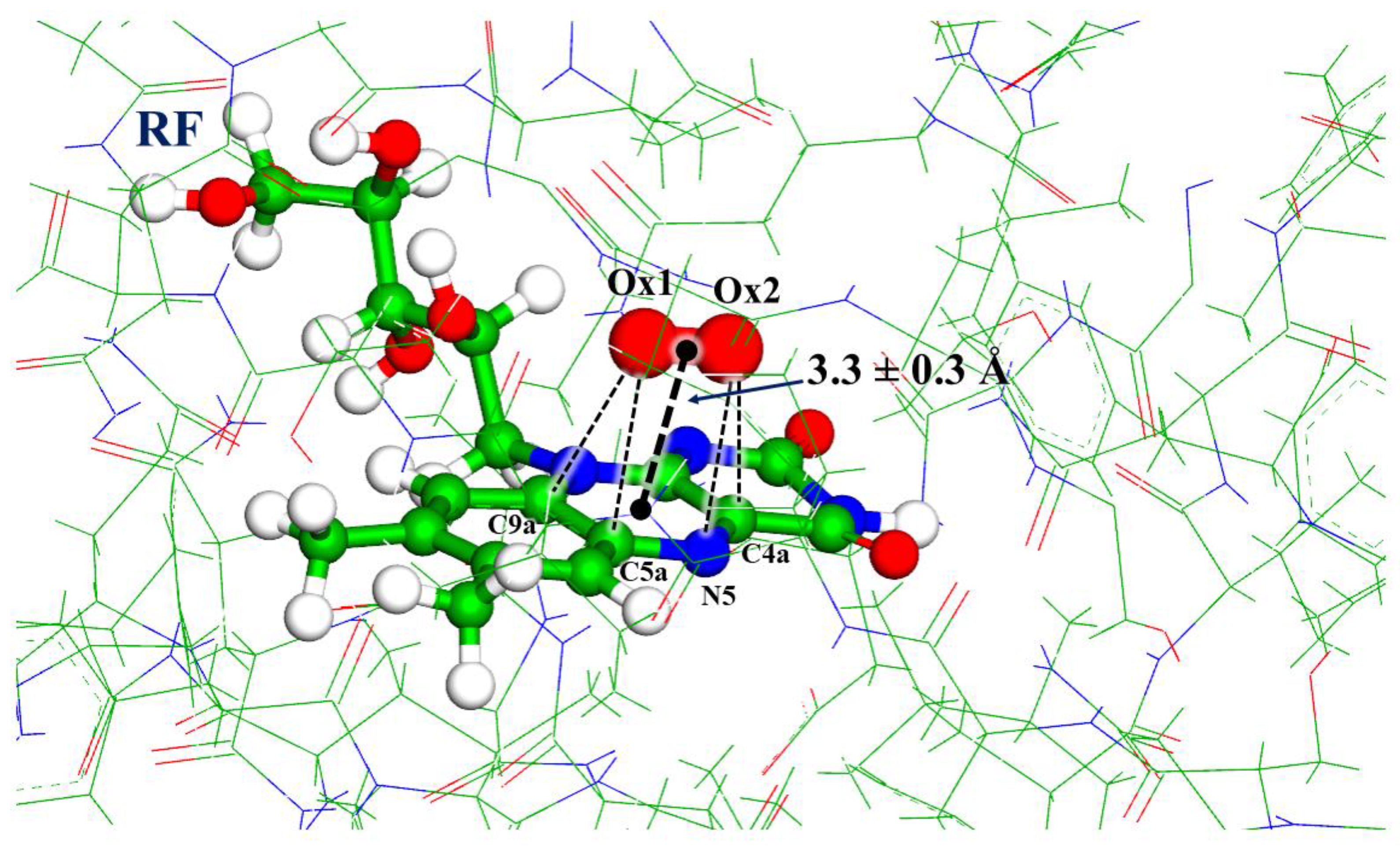
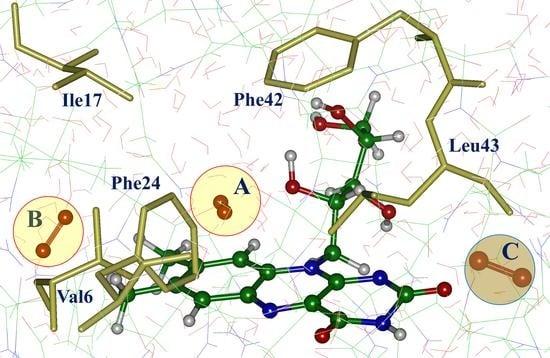
| Model System | BB 5–105 | BB 28–48 | 28–48 noH | ISO |
|---|---|---|---|---|
| miniSOG[FMN] | 0.8 ± 0.2 | 0.7 ± 0.2 | 1.4 ± 0.3 | 0.9 ± 0.2 |
| miniSOG[RF] | 1.1 ± 0.2 | 1.1 ± 0.3 | 2.0 ± 0.3 | 1.0 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polyakov, I.; Kulakova, A.; Nemukhin, A. Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen. Biophysica 2023, 3, 252-262. https://doi.org/10.3390/biophysica3020016
Polyakov I, Kulakova A, Nemukhin A. Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen. Biophysica. 2023; 3(2):252-262. https://doi.org/10.3390/biophysica3020016
Chicago/Turabian StylePolyakov, Igor, Anna Kulakova, and Alexander Nemukhin. 2023. "Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen" Biophysica 3, no. 2: 252-262. https://doi.org/10.3390/biophysica3020016
APA StylePolyakov, I., Kulakova, A., & Nemukhin, A. (2023). Computational Modeling of the Interaction of Molecular Oxygen with the miniSOG Protein—A Light Induced Source of Singlet Oxygen. Biophysica, 3(2), 252-262. https://doi.org/10.3390/biophysica3020016