Analysis of Enzyme Conformation Dynamics Using Single-Molecule Förster Resonance Energy Transfer (smFRET)
Abstract
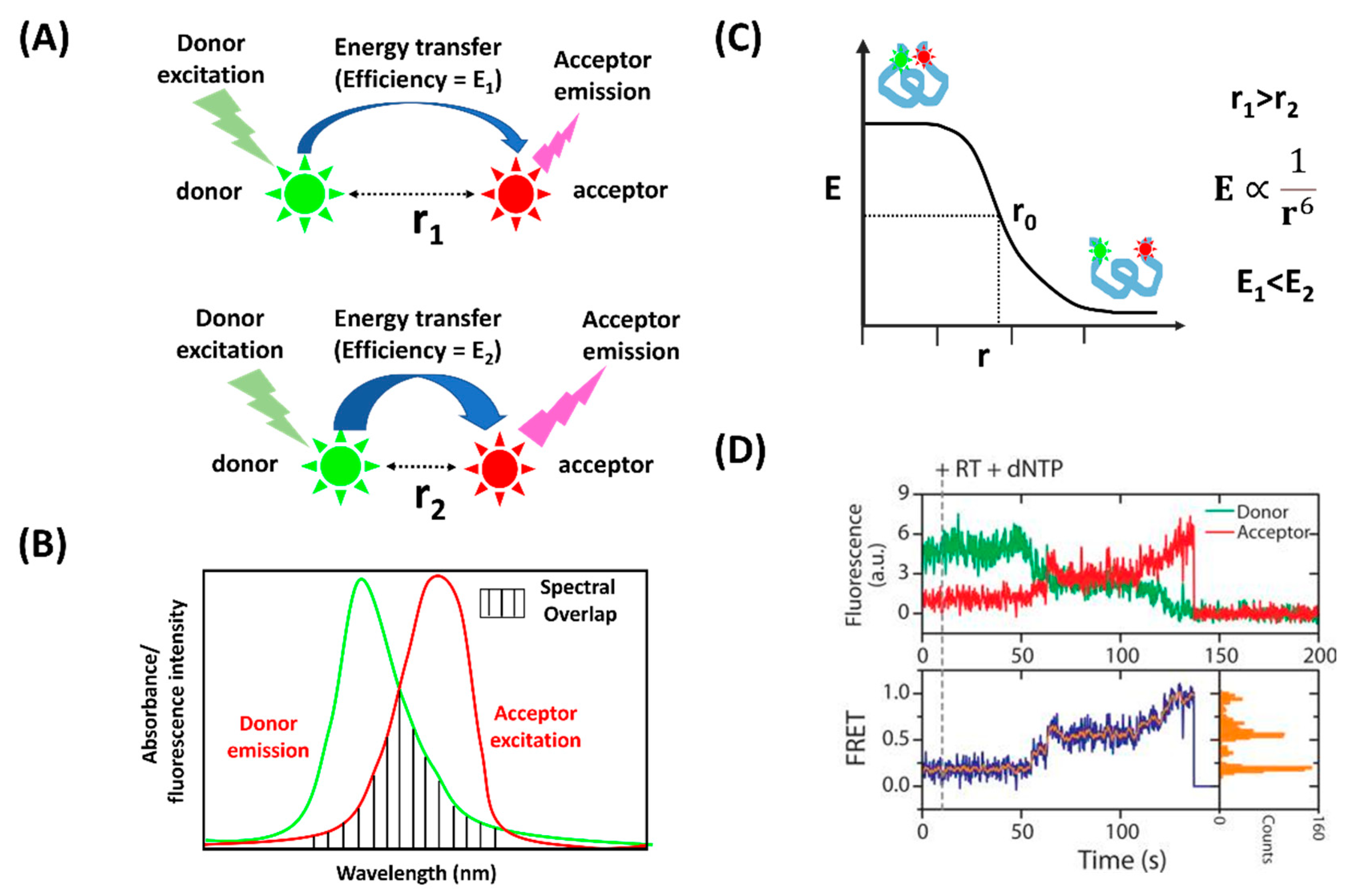
:1. Introduction
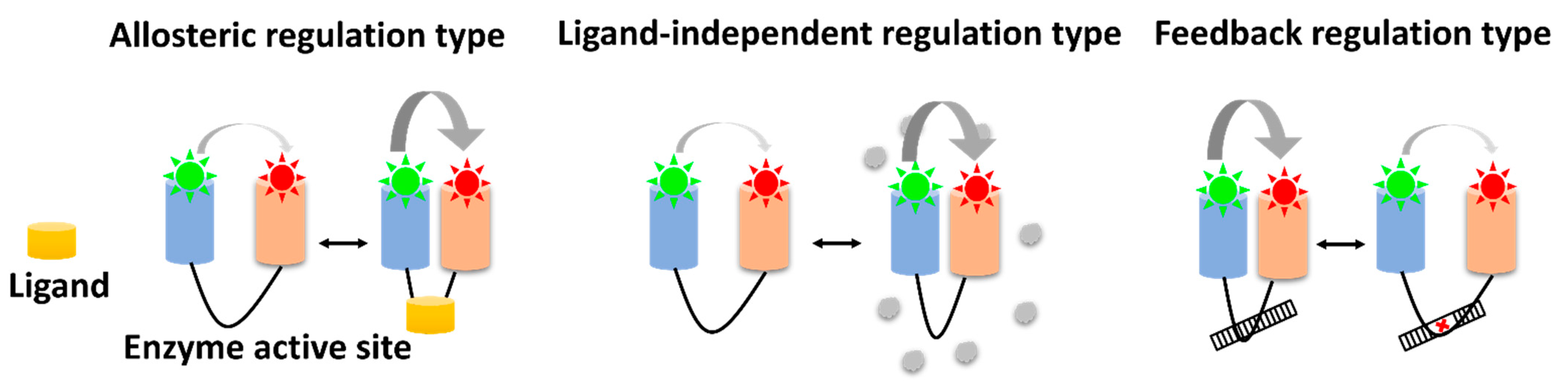
2. Enzyme Conformational Dynamics
3. Allosteric Regulation Type
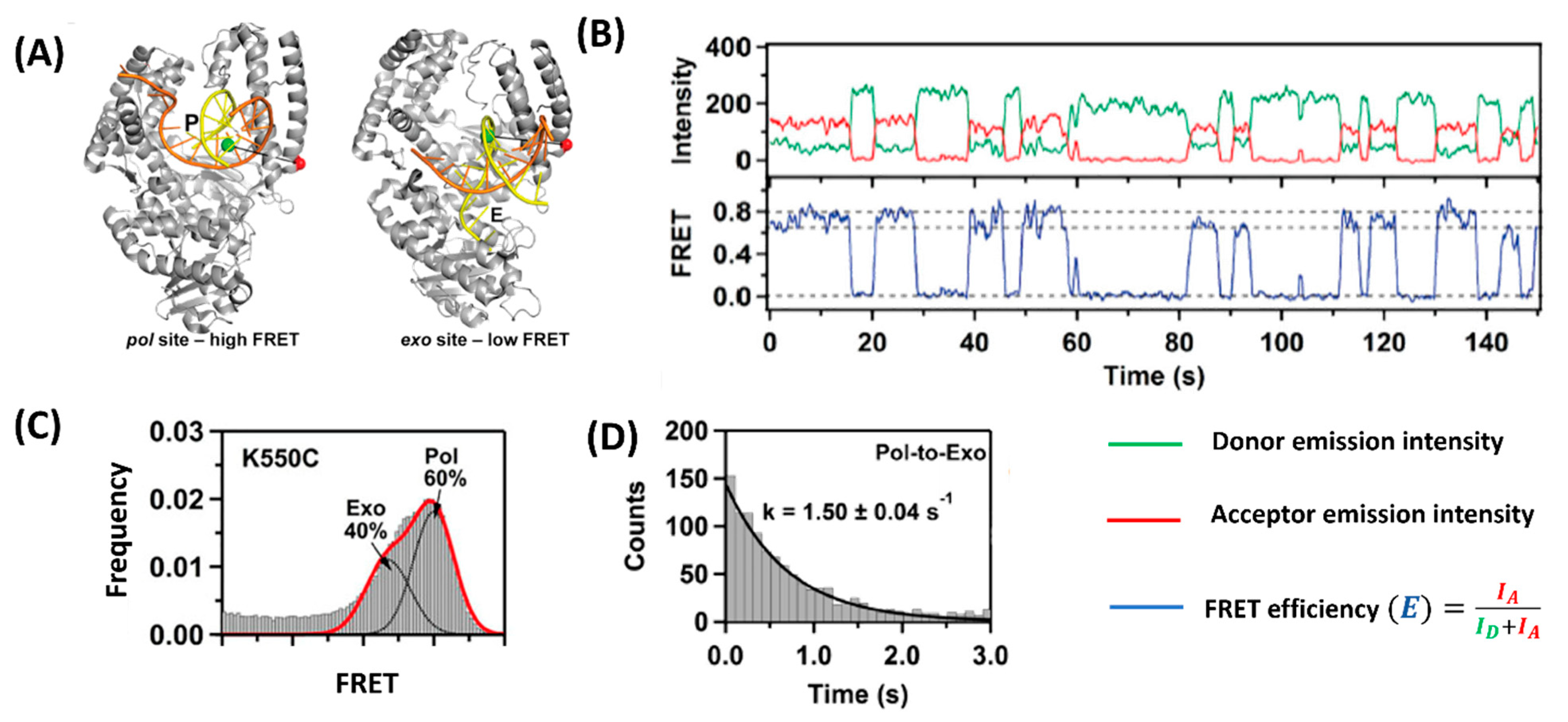
4. Ligand-Independent Regulation Type
5. Feedback Regulation Type
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joo, C.; Balci, H.; Ishitsuka, Y.; Buranachai, C.; Ha, T. Advances in Single-Molecule Fluorescence Methods for Molecular Biology. Annu. Rev. Biochem. 2008, 77, 51–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, E.; Barth, A.; Hendrix, J.; Ambrose, B.; Birkedal, V.; Blanchard, S.C.; Börner, R.; Sung Chung, H.; Cordes, T.; Craggs, T.D.; et al. FRET-based dynamic structural biology: Challenges, perspectives and an appeal for open-science practices. eLife 2021, 10, e60416. [Google Scholar] [CrossRef] [PubMed]
- Lerner, E.; Cordes, T.; Ingargiola, A.; Alhadid, Y.; Chung, S.; Michalet, X.; Weiss, S. Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 2018, 359, eaan1133. [Google Scholar] [CrossRef] [Green Version]
- Sasmal, D.K.; Pulido, L.E.; Kasal, S.; Huang, J. Single-molecule fluorescence resonance energy transfer in molecular biology. Nanoscale 2016, 8, 19928–19944. [Google Scholar] [CrossRef] [Green Version]
- Orevi, T.; Lerner, E.; Rahamim, G.; Amir, D.; Haas, E. Ensemble and single-molecule detected time-resolved FRET methods in studies of protein conformations and dynamics. Methods Mol. Biol. 2014, 1076, 113–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wallrabe, H.; Seo, S.-A.; Periasamy, A. FRET microscopy in 2010: The legacy of Theodor Förster on the 100th anniversary of his birth. ChemPhysChem Eur. J. Chem. Phys. Phys. Chem. 2011, 12, 462–474. [Google Scholar] [CrossRef] [Green Version]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef] [Green Version]
- Selvin, P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef]
- Hellenkamp, B.; Schmid, S.; Doroshenko, O.; Opanasyuk, O.; Kühnemuth, R.; Rezaei Adariani, S.; Ambrose, B.; Aznauryan, M.; Barth, A.; Birkedal, V.; et al. Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study. Nat. Methods 2018, 15, 669–676. [Google Scholar] [CrossRef]
- Kalinin, S.; Peulen, T.; Sindbert, S.; Rothwell, P.J.; Berger, S.; Restle, T.; Goody, R.S.; Gohlke, H.; Seidel, C.A.M. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods 2012, 9, 1218–1225. [Google Scholar] [CrossRef]
- Dimura, M.; Peulen, T.O.; Hanke, C.A.; Prakash, A.; Gohlke, H.; Seidel, C.A.M. Quantitative FRET studies and integrative modeling unravel the structure and dynamics of biomolecular systems. Curr. Opin. Struct. Biol. 2016, 40, 163–185. [Google Scholar] [CrossRef] [PubMed]
- Larsen, K.P.; Choi, J.; Prabhakar, A.; Puglisi, E.V.; Puglisi, J.D. Relating Structure and Dynamics in RNA Biology. Cold Spring Harb. Perspect. Biol. 2019, 11, a032474. [Google Scholar] [CrossRef] [Green Version]
- Lipman, E.A.; Schuler, B.; Bakajin, O.; Eaton, W.A. Single-molecule measurement of protein folding kinetics. Science 2003, 301, 1233–1235. [Google Scholar] [CrossRef] [PubMed]
- Mazal, H.; Haran, G. Single-molecule FRET methods to study the dynamics of proteins at work. Curr. Opin. Biomed. Eng. 2019, 12, 8–17. [Google Scholar] [CrossRef]
- Michalet, X.; Weiss, S.; Jäger, M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem. Rev. 2006, 106, 1785–1813. [Google Scholar] [CrossRef]
- Lee, J.; Crickard, J.B.; Reese, J.C.; Lee, T.-H. Single-molecule FRET method to investigate the dynamics of transcription elongation through the nucleosome by RNA polymerase II. Methods 2019, 159–160, 51–58. [Google Scholar] [CrossRef]
- Liu, S.; Abbondanzieri, E.A.; Rausch, J.W.; Le Grice, S.F.J.; Zhuang, X. Slide into action: Dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science 2008, 322, 1092–1097. [Google Scholar] [CrossRef]
- Deniz, A.A.; Dahan, M.; Grunwell, J.R.; Ha, T.; Faulhaber, A.E.; Chemla, D.S.; Weiss, S.; Schultz, P.G. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Förster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA 1999, 96, 3670–3675. [Google Scholar] [CrossRef] [Green Version]
- Deniz, A.A.; Laurence, T.A.; Beligere, G.S.; Dahan, M.; Martin, A.B.; Chemla, D.S.; Dawson, P.E.; Schultz, P.G.; Weiss, S. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 5179–5184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Talaga, D.S.; Lau, W.L.; Lu, H.S.M.; DeGrado, W.F.; Hochstrasser, R.M. Folding dynamics of single GCN-4 peptides by fluorescence resonant energy transfer confocal microscopy. Chem. Phys. 1999, 247, 69–83. [Google Scholar] [CrossRef]
- Ha, T.; Zhuang, X.; Kim, H.D.; Orr, J.W.; Williamson, J.R.; Chu, S. Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl. Acad. Sci. USA 1999, 96, 9077–9082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, T.; Enderle, T.; Ogletree, D.F.; Chemla, D.S.; Selvin, P.R.; Weiss, S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA 1996, 93, 6264–6268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, T.; Ting, A.Y.; Liang, J.; Caldwell, W.B.; Deniz, A.A.; Chemla, D.S.; Schultz, P.G.; Weiss, S. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. USA 1999, 96, 893–898. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Hu, D.; Vorpagel, E.; Lu, H.P. Probing Single-Molecule T4 Lysozyme Conformational Dynamics by Intramolecular Fluorescence Energy Transfer. J. Phys. Chem. B 2003, 107, 7947. [Google Scholar] [CrossRef]
- Lu, H.P. Probing single-molecule protein conformational dynamics. Acc. Chem. Res. 2005, 38, 557–565. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, H.P. Bunching effect in single-molecule T4 lysozyme nonequilibrium conformational dynamics under enzymatic reactions. J. Phys. Chem. B 2010, 114, 6669–6674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, J.A.; Duderstadt, K.; Watkins, L.P.; Bhattacharyya, S.; Brokaw, J.; Chu, J.W.; Yang, H. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc. Natl. Acad. Sci. USA 2007, 104, 18055–18060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Atas, E.; Lindqvist, L.; Sonenberg, N.; Pelletier, J.; Meller, A. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res. 2012, 40, 6199–6207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, B.M.; Loper, J.; Najarro, K.; Stone, M.D. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA 2012, 18, 653–660. [Google Scholar] [CrossRef] [Green Version]
- Jansson, L.I.; Stone, M.D. Single-Molecule Analysis of Reverse Transcriptase Enzymes. Cold Spring Harb Perspect. Biol. 2019, 11, a032458. [Google Scholar] [CrossRef]
- Gabba, M.; Poblete, S.; Rosenkranz, T.; Katranidis, A.; Kempe, D.; Züchner, T.; Winkler, R.G.; Gompper, G.; Fitter, J. Conformational state distributions and catalytically relevant dynamics of a hinge-bending enzyme studied by single-molecule FRET and a coarse-grained simulation. Biophys. J. 2014, 107, 1913–1923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branigan, E.; Carlos Penedo, J.; Hay, R.T. Ubiquitin transfer by a RING E3 ligase occurs from a closed E2~ubiquitin conformation. Nat. Commun. 2020, 11, 2846. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, H.; Dienerowitz, F.; Dienerowitz, M. Single-molecule FRET combined with electrokinetic trapping reveals real-time enzyme kinetics of individual F-ATP synthases. Nanoscale 2022, 14, 2327–2336. [Google Scholar] [CrossRef]
- Zhuang, X.; Kim, H.; Pereira, M.J.; Babcock, H.P.; Walter, N.G.; Chu, S. Correlating structural dynamics and function in single ribozyme molecules. Science 2002, 296, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.; Wilson, T.J.; Nahas, M.K.; Clegg, R.M.; Lilley, D.M.; Ha, T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. USA 2003, 100, 9308–9313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Li, Y.; Mukherjee, S.; Wu, Y.; Yan, H.; Lu, H.P. Probing single-molecule enzyme active-site conformational state intermittent coherence. J. Am. Chem. Soc. 2011, 133, 14389–14395. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Lu, M.; Lu, H.P. Single-molecule photon stamping FRET spectroscopy study of enzymatic conformational dynamics. Phys. Chem. Chem. Phys. 2013, 15, 770–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; He, Y.; Lu, H.P. Manipulating and probing enzymatic conformational fluctuations and enzyme-substrate interactions by single-molecule FRET-magnetic tweezers microscopy. Phys. Chem. Chem. Phys. 2014, 16, 13052–13058. [Google Scholar] [CrossRef]
- Rutkauskas, D.; Petkelyte, M.; Naujalis, P.; Sasnauskas, G.; Tamulaitis, G.; Zaremba, M.; Siksnys, V. Restriction enzyme Ecl18kI-induced DNA looping dynamics by single-molecule FRET. J. Phys. Chem. B 2014, 118, 8575–8582. [Google Scholar] [CrossRef] [PubMed]
- Tutkus, M.; Marciulionis, T.; Sasnauskas, G.; Rutkauskas, D. DNA-Endonuclease Complex Dynamics by Simultaneous FRET and Fluorophore Intensity in Evanescent Field. Biophys. J. 2017, 112, 850–858. [Google Scholar] [CrossRef] [Green Version]
- Tutkus, M.; Sasnauskas, G.; Rutkauskas, D. Probing the dynamics of restriction endonuclease NgoMIV-DNA interaction by single-molecule FRET. Biopolymers 2017, 107, e23075. [Google Scholar] [CrossRef] [PubMed]
- Götz, C.; Hinze, G.; Gellert, A.; Maus, H.; von Hammerstein, F.; Hammerschmidt, S.J.; Lauth, L.M.; Hellmich, U.A.; Schirmeister, T.; Basché, T. Conformational Dynamics of the Dengue Virus Protease Revealed by Fluorescence Correlation and Single-Molecule FRET Studies. J. Phys. Chem. B 2021, 125, 6837–6846. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.; Eleftheriadis, N.; Karathanou, K.; Smit, J.H.; Portaliou, A.G.; Chatzi, K.E.; Karamanou, S.; Bondar, A.N.; Gouridis, G.; Economou, A. A nexus of intrinsic dynamics underlies translocase priming. Structure 2021, 29, 846–858.e7. [Google Scholar] [CrossRef]
- Berezhna, S.Y.; Gill, J.P.; Lamichhane, R.; Millar, D.P. Single-molecule Förster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I. J. Am. Chem. Soc. 2012, 134, 11261–11268. [Google Scholar] [CrossRef] [Green Version]
- Millar, D.P. Conformational Dynamics of DNA Polymerases Revealed at the Single-Molecule Level. Front. Mol. Biosci. 2022, 9, 826593. [Google Scholar] [CrossRef] [PubMed]
- Karam, P.; Powdrill, M.H.; Liu, H.W.; Vasquez, C.; Mah, W.; Bernatchez, J.; Götte, M.; Cosa, G. Dynamics of hepatitis C virus (HCV) RNA-dependent RNA polymerase NS5B in complex with RNA. J. Biol. Chem. 2014, 289, 14399–14411. [Google Scholar] [CrossRef] [Green Version]
- Mazumder, A.; Wang, A.; Uhm, H.; Ebright, R.H.; Kapanidis, A.N. RNA polymerase clamp conformational dynamics: Long-lived states and modulation by crowding, cations, and nonspecific DNA binding. Nucleic Acids Res. 2021, 49, 2790–2802. [Google Scholar] [CrossRef]
- Osuka, S.; Isomura, K.; Kajimoto, S.; Komori, T.; Nishimasu, H.; Shima, T.; Nureki, O.; Uemura, S. Real-time observation of flexible domain movements in CRISPR-Cas9. EMBO J. 2018, 37, e96941. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Peng, S.; Sun, R.; Lin, J.; Wang, N.; Chen, C. The Conformational Dynamics of Cas9 Governing DNA Cleavage Are Revealed by Single-Molecule FRET. Cell Rep. 2018, 22, 372–382. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Cui, Y.; Zhang, Y.; Zhang, Y.; Liang, M.; Chen, H.; Lan, J.; Song, G.; Lou, J. The initiation, propagation and dynamics of CRISPR-SpyCas9 R-loop complex. Nucleic Acids Res. 2018, 46, 350–361. [Google Scholar] [CrossRef] [Green Version]
- Cheung, A.C.M.; Cramer, P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 2011, 471, 249–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kettenberger, H.; Armache, K.-J.; Cramer, P. Complete RNA Polymerase II Elongation Complex Structure and Its Interactions with NTP and TFIIS. Mol. Cell 2004, 16, 955–965. [Google Scholar] [CrossRef] [Green Version]
- Komissarova, N.; Kashlev, M. RNA Polymerase Switches between Inactivated and Activated States By Translocating Back and Forth along the DNA and the RNA. J. Biol. Chem. 1997, 272, 15329–15338. [Google Scholar] [CrossRef] [Green Version]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [Green Version]
- Nishimasu, H.; Cong, L.; Yan, W.X.; Ran, F.A.; Zetsche, B.; Li, Y.; Kurabayashi, A.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Staphylococcus aureus Cas9. Cell 2015, 162, 1113–1126. [Google Scholar] [CrossRef] [Green Version]
- Szczelkun, M.D.; Tikhomirova, M.S.; Sinkunas, T.; Gasiunas, G.; Karvelis, T.; Pschera, P.; Siksnys, V.; Seidel, R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. USA 2014, 111, 9798–9803. [Google Scholar] [CrossRef] [Green Version]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Ban, T.; Zhu, J.K.; Melcher, K.; Xu, H.E. Structural mechanisms of RNA recognition: Sequence-specific and non-specific RNA-binding proteins and the Cas9-RNA-DNA complex. Cell. Mol. Life Sci. 2015, 72, 1045–1058. [Google Scholar] [CrossRef]
- Jiang, F.; Zhou, K.; Ma, L.; Gressel, S.; Doudna, J.A. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015, 348, 1477–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josephs, E.A.; Kocak, D.D.; Fitzgibbon, C.J.; McMenemy, J.; Gersbach, C.A.; Marszalek, P.E. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 2015, 43, 8924–8941. [Google Scholar] [CrossRef] [Green Version]
- Hohng, S.; Joo, C.; Ha, T. Single-molecule three-color FRET. Biophys. J. 2004, 87, 1328–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Lee, S.; Ragunathan, K.; Joo, C.; Ha, T.; Hohng, S. Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 2010, 49, 9922–9925. [Google Scholar] [CrossRef] [PubMed]
- Joo, C.; Ha, T. Imaging and identifying impurities in single-molecule FRET studies. Cold Spring Harbar Protoc. 2012, 2012, 1109–1112. [Google Scholar] [CrossRef]
- Ha, T.; Tinnefeld, P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 2012, 63, 595–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Kunzelmann, S.; Webb, M.R.; Ha, T. Detecting intramolecular conformational dynamics of single molecules in short distance range with subnanometer sensitivity. Nano Lett. 2011, 11, 5482–5488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohlbein, J.; Craggs, T.D.; Cordes, T. Alternating-laser excitation: Single-molecule FRET and beyond. Chem. Soc. Rev. 2014, 43, 1156–1171. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.; Louis, J.M.; Gopich, I.V.; Chung, H.S. Three-Color Single-Molecule FRET and Fluorescence Lifetime Analysis of Fast Protein Folding. J. Phys. Chem. B 2018, 122, 11702–11720. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Park, J.; Dahmen, K.A.; Chemla, Y.R.; Ha, T. A comparative study of multivariate and univariate hidden Markov modelings in time-binned single-molecule FRET data analysis. J. Phys. Chem. B 2010, 114, 5386–5403. [Google Scholar] [CrossRef]
- McKinney, S.A.; Joo, C.; Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006, 91, 1941–1951. [Google Scholar] [CrossRef] [Green Version]
- Gordon, M.P.; Ha, T.; Selvin, P.R. Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. USA 2004, 101, 6462–6465. [Google Scholar] [CrossRef] [Green Version]
- Barth, A.; Voith von Voithenberg, L.; Lamb, D.C. Quantitative Single-Molecule Three-Color Förster Resonance Energy Transfer by Photon Distribution Analysis. J. Phys. Chem. B 2019, 123, 6901–6916. [Google Scholar] [CrossRef]
- König, S.L.; Hadzic, M.; Fiorini, E.; Börner, R.; Kowerko, D.; Blanckenhorn, W.U.; Sigel, R.K. BOBA FRET: Bootstrap-based analysis of single-molecule FRET data. PLoS ONE 2013, 8, e84157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirchi, M.; Tsukanov, R.; Khamis, R.; Tomov, T.E.; Berger, Y.; Khara, D.C.; Volkov, H.; Haran, G.; Nir, E. Photon-by-Photon Hidden Markov Model Analysis for Microsecond Single-Molecule FRET Kinetics. J. Phys. Chem. B 2016, 120, 13065–13075. [Google Scholar] [CrossRef]
- Wilson, H.; Wang, Q. ABEL-FRET: Tether-free single-molecule FRET with hydrodynamic profiling. Nat. Methods 2021, 18, 816–820. [Google Scholar] [CrossRef]
- Kaur, A.; Ellison, M.; Dhakal, S. MASH-FRET: A Simplified Approach for Single-Molecule Multiplexing Using FRET. Anal. Chem. 2021, 93, 8856–8863. [Google Scholar] [CrossRef] [PubMed]
- Hirata, E.; Kiyokawa, E. Future Perspective of Single-Molecule FRET Biosensors and Intravital FRET Microscopy. Biophys. J. 2016, 111, 1103–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Type | Enzyme | References |
|---|---|---|
| Allosteric regulation | Staphylococcal nuclease (SNase) | [23] |
| T4-lysozyme | [24,25,26] | |
| Adenylate kinase (AK) | [27] | |
| RNA helicase (eIF4A) | [28] | |
| Telomerase ribonucleoprotein (RNP) | [29,30] | |
| Phosphoglycerate kinase (PGK) | [31] | |
| RING E3 ligase | [32] | |
| ATP synthase | [33] | |
| Ligand-independent regulation type | Hairpin ribozyme | [34,35] |
| 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK) | [36,37,38] | |
| Restriction endonucleases (Ecl18kI and NgoMIV) | [39,40,41] | |
| Dengue virus protease (DENV-PR) | [42] | |
| Sec preprotein translocase | [43] | |
| Feedback regulation type | RNA Polymerases I and II | [44,45,46,47] |
| CRISPR-associated protein 9 (Cas9) | [48,49,50] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huynh, M.; Sengupta, B. Analysis of Enzyme Conformation Dynamics Using Single-Molecule Förster Resonance Energy Transfer (smFRET). Biophysica 2022, 2, 123-134. https://doi.org/10.3390/biophysica2020014
Huynh M, Sengupta B. Analysis of Enzyme Conformation Dynamics Using Single-Molecule Förster Resonance Energy Transfer (smFRET). Biophysica. 2022; 2(2):123-134. https://doi.org/10.3390/biophysica2020014
Chicago/Turabian StyleHuynh, Mai, and Bhaswati Sengupta. 2022. "Analysis of Enzyme Conformation Dynamics Using Single-Molecule Förster Resonance Energy Transfer (smFRET)" Biophysica 2, no. 2: 123-134. https://doi.org/10.3390/biophysica2020014
APA StyleHuynh, M., & Sengupta, B. (2022). Analysis of Enzyme Conformation Dynamics Using Single-Molecule Förster Resonance Energy Transfer (smFRET). Biophysica, 2(2), 123-134. https://doi.org/10.3390/biophysica2020014






