Probing Structural Dynamics of Membrane Proteins Using Electron Paramagnetic Resonance Spectroscopic Techniques
Abstract
1. Membrane Proteins
2. Challenges and Recent Improvements Using Biophysical Techniques for Studying Membrane Proteins
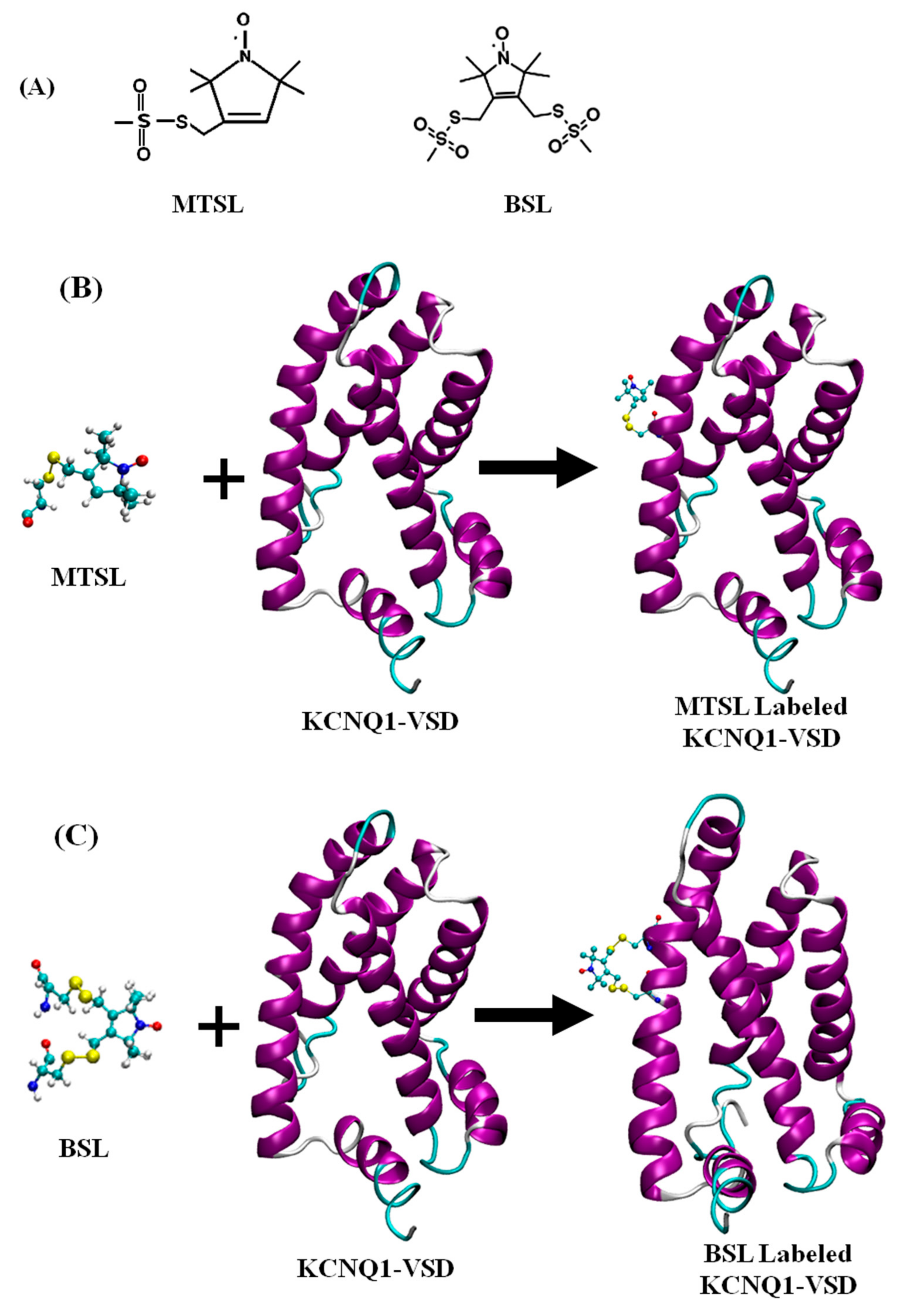
3. Site Directed Spin Labeling (SDSL) Approaches for EPR Spectroscopy
4. Application of EPR Coupled with SDSL for Investigation of Membrane Proteins
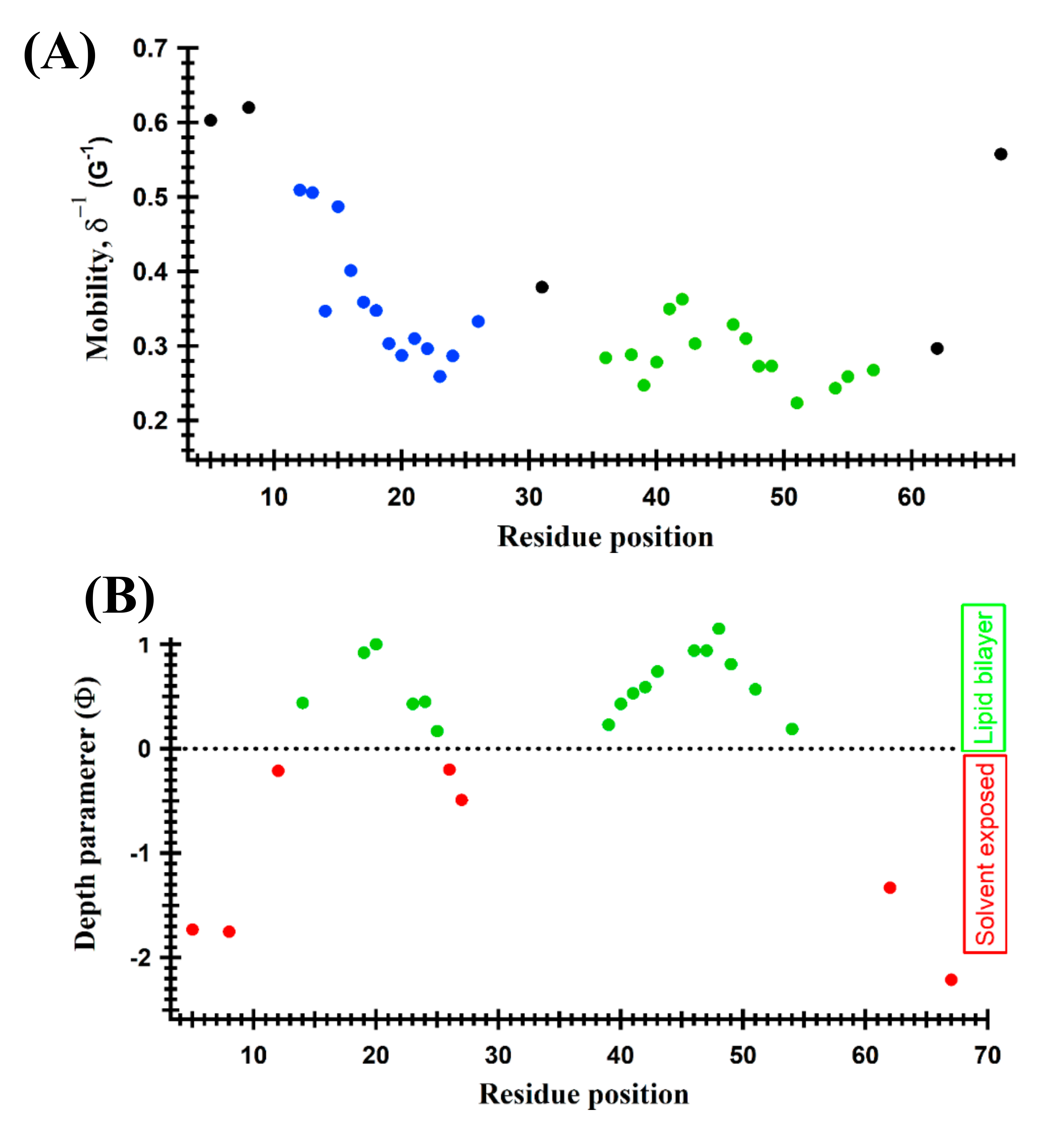
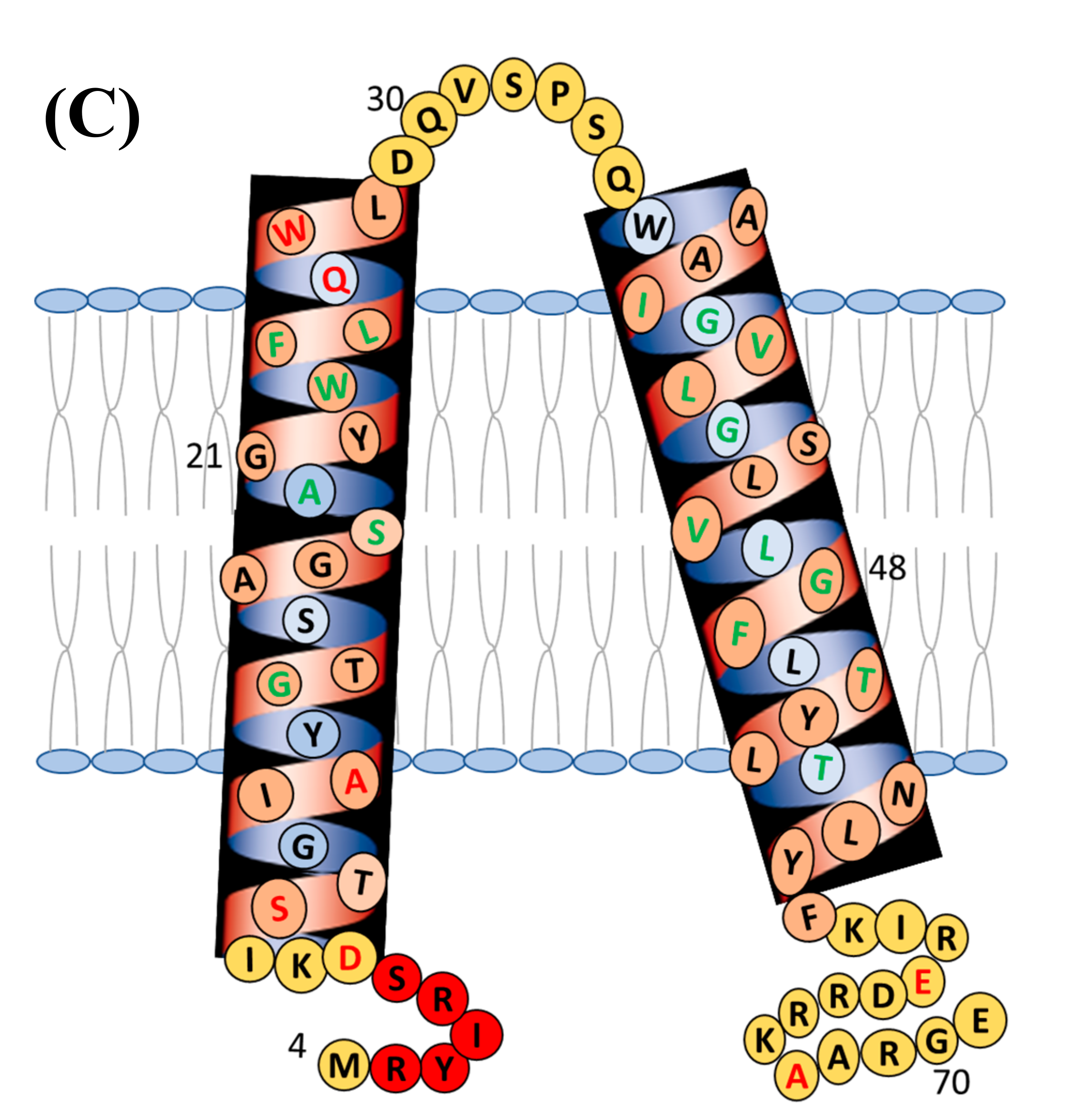
4.1. Structural Topology and Dynamic Properties of Membrane Proteins
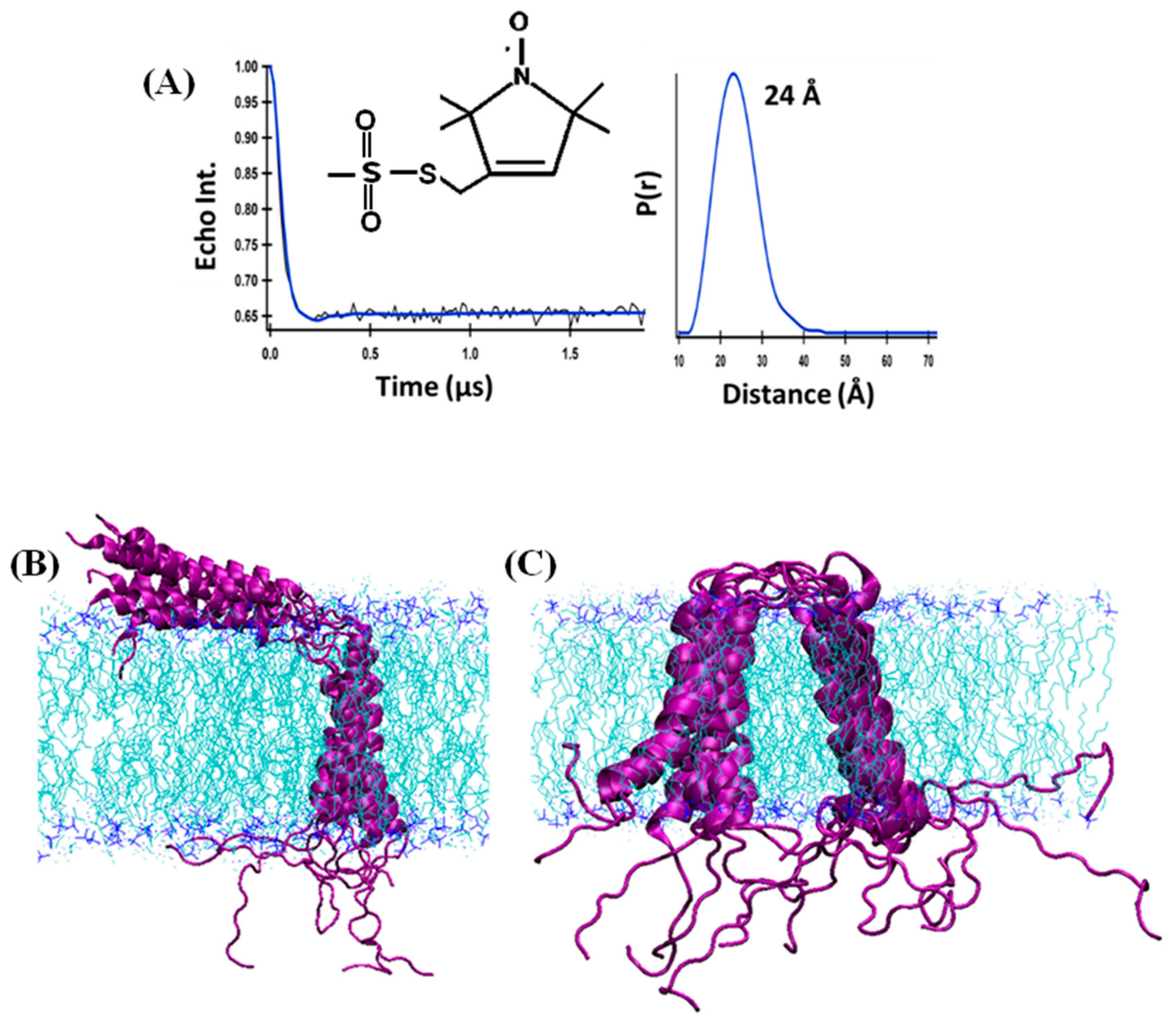
4.2. Distance Measurement on Membrane Proteins Using Dual SDSL EPR Spectroscopy
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Congreve, M.; Marshall, F. The impact of GPCR structures on pharmacology and structure-based drug design. Br. J. Pharmacol. 2010, 159, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Structural biology: The gatekeepers revealed. Nature 2010, 465, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Elrod, D.W. Prediction of membrane protein types and subcellular locations. Proteins Struct. Funct. Genet. 1999, 34, 137–153. [Google Scholar] [CrossRef]
- Engel, A.; Gaub, H.E. Structure and Mechanics of Membrane Proteins. Annu. Rev. Biochem. 2008, 77, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Klug, C.S.; Feix, J.B. Methods and Applications of Site-Directed Spin Labeling EPR Spectroscopy. Methods Cell Biol. 2008, 84, 617–658. [Google Scholar] [PubMed]
- Sahu, I.D.; McCarrick, R.M.; Lorigan, G.A. Use of Electron Paramagnetic Resonance to Solve Biochemical Problems. Biochemistry 2013, 52, 5967–5984. [Google Scholar] [CrossRef] [PubMed]
- Das, B.B.; Park, S.H.; Opella, S.J. Membrane protein structure from rotational diffusion. Biochim. Biophys. Acta 2015, 1848, 229–245. [Google Scholar] [CrossRef]
- Kang, H.J.; Lee, C.; Drew, D. Breaking the barriers in membrane protein crystallography. Int. J. Biochem. Cell Biol. 2013, 45, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Hemminga, M.A.; Berliner, L.J. ESR Spectroscopy in Membrane Biophysics; Springer: New York, NY, USA, 2007. [Google Scholar]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef]
- Sahu, I.D.; McCarrick, R.M.; Troxel, K.R.; Zhang, R.; Smith, H.J.; Dunagan, M.M.; Swartz, M.S.; Rajan, P.V.; Kroncke, B.M.; Sanders, C.R.; et al. DEER EPR Measurements for Membrane Protein Structures via Bifunctional Spin Labels and Lipodisq Nanoparticles. Biochemistry 2013, 52, 6627–6632. [Google Scholar] [CrossRef][Green Version]
- Harding, B.D.; Dixit, G.; Burridge, K.M.; Sahu, I.D.; Dabney-Smith, C.; Edelmann, R.E.; Konkolewicz, D.; Lorigan, G.A. Charac-terizing the structure of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for membrane protein spectroscopic studies. Chem. Phys. Lipids 2019, 218, 65–72. [Google Scholar] [CrossRef]
- Burridge, K.M.; Harding, B.D.; Sahu, I.D.; Kearns, M.M.; Stowe, R.B.; Dolan, M.T.; Edelmann, R.E.; Dabney-Smith, C.; Page, R.C.; Konkolewicz, D.; et al. Simple Derivatization of RAFT-Synthesized Styrene–Maleic Anhydride Copolymers for Lipid Disk Formulations. Biomacromolecules 2020, 21, 1274–1284. [Google Scholar] [CrossRef]
- Sahu, I.D.; Dixit, G.; Reynolds, W.D.; Kaplevatsky, R.; Harding, B.D.; Jaycox, C.K.; McCarrick, R.M.; Lorigan, G.A. Characteriza-tion of the Human KCNQ1 Voltage Sensing Domain (VSD) in Lipodisq Nanoparticles for Electron Paramagnetic Resonance (EPR) Spectroscopic Studies of Membrane Proteins. J. Phys. Chem. B 2020, 124, 2331–2342. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.F.; Clark, E.E.; Sahu, I.D.; Zhang, R.; Frantz, N.D.; Al-Abdul-Wahid, M.S.; Dabney-Smith, C.; Konkolewicz, D.; Lorigan, G.A. Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim. Biophys. Acta 2016, 1858, 2931–2939. [Google Scholar] [CrossRef] [PubMed]
- Orwick-Rydmark, M.; Lovett, J.E.; Graziadei, A.; Lindholm, L.; Hicks, M.R.; Watts, A. Detergent-Free Incorporation of a Sev-en-Transmembrane Receptor Protein into Nanosized Bilayer Lipodisq Particles for Functional and Biophysical Studies. Nano Lett. 2012, 12, 4687–4692. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Sligar, S.G. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010, 584, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Bayburt, T.H.; Sligar, S.G. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003, 12, 2476–2481. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed self-assembly of monodisperse phospholipid bilayer nano-discs with controlled size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef] [PubMed]
- Orwick, M.C.; Judge, P.J.; Procek, J.; Lindholm, L.; Graziadei, A.; Engel, A.; Gröbner, G.; Watts, A. Detergent-Free Formation and Physicochemical Characterization of Nanosized Lipid-Polymer Complexes: Lipodisq. Angew. Chem. Int. Ed. 2012, 51, 4653–4657. [Google Scholar] [CrossRef]
- Jamshad, M.; Lin, Y.-P.; Knowles, T.J.; Parslow, R.A.; Harris, C.; Wheatley, M.; Poyner, D.R.; Bill, R.M.; Thomas, O.R.; Overduin, M.; et al. Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans. 2011, 39, 813–818. [Google Scholar] [CrossRef]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.-P.; Dafforn, T.; Overduin, M. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef]
- Rajesh, S.; Knowles, T.; Overduin, M. Production of membrane proteins without cells or detergents. New Biotechnol. 2011, 28, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Long, A.R.; O’Brien, C.C.; Malhotra, K.; Schwall, C.T.; Albert, A.D.; Watts, A.; Alder, N.N. A detergent-free strategy for the re-constitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol. 2013, 13, 41. [Google Scholar] [CrossRef]
- Jamshad, M.; Grimard, V.; Idini, I.; Knowles, T.J.; Dowle, M.R.; Schofield, N.; Sridhar, P.; Lin, Y.; Finka, R.; Wheatley, M.; et al. Structural analysis of a nano-particle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015, 8, 774–789. [Google Scholar] [CrossRef]
- Lund, A.; Andersson, P.; Eriksson, J.; Hallin, J.; Johansson, T.; Jonsson, R.; Lofgren, H.; Paulin, C.; Tell, A. Automatic fitting pro-cedures for EPR spectra of disordered systems: Matrix diagonalization and perturbation methods applied to fluorocarbon radicals. Spectrochim. Acta Part A 2008, 69, 1294–1300. [Google Scholar] [CrossRef][Green Version]
- Zhang, R.; Sahu, I.D.; Liu, L.; Osatuke, A.; Comer, R.G.; Dabney-Smith, C.; Lorigan, G.A. Characterizing the structure of lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim. Biophys. Acta Biomembr. 2015, 1848, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Upshur, M.A.; Saotome, K.; Sahu, I.D.; McCarrick, R.M.; Feix, J.B.; Lorigan, G.A.; Howard, K.P. Cholesterol-Dependent Conformational Exchange of the C-Terminal Domain of the Influenza A M2 Protein. Biochemistry 2015, 54, 7157–7167. [Google Scholar] [CrossRef]
- Scheidelaar, S.; Koorengevel, M.C.; Pardo, J.D.; Meeldijk, J.D.; Breukink, E.; Killian, J.A. Molecular Model for the Solubilization of Membranes into Nanodisks by Styrene Maleic Acid Copolymers. Biophys. J. 2015, 108, 279–290. [Google Scholar] [CrossRef]
- Dörr, J.M.; Scheidelaar, S.; Koorengevel, M.C.; Dominguez, J.J.; Schäfer, M.; Van Walree, C.A.; Killian, J.A. The styrene–maleic acid copolymer: A versatile tool in membrane research. Eur. Biophys. J. EBJ 2016, 45, 3–21. [Google Scholar] [CrossRef]
- Dörr, J.M.; Koorengevel, M.C.; Schäfer, M.; Prokofyev, A.V.; Scheidelaar, S.; Van Der Cruijsen, E.A.W.; Dafforn, T.R.; Baldus, M.; Killian, J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; Kroncke, B.M.; Zhang, R.; Dunagan, M.M.; Smith, H.J.; Craig, A.; McCarrick, R.M.; Sanders, C.R.; Lorigan, G.A. Structural Investigation of the Transmembrane Domain of KCNE1 in Proteoliposomes. Biochemistry 2014, 53, 6392–6401. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD-Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. Software news and updates—CHARNIM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Liang, B.; Tamm, L.K. NMR as a Tool to Investigate Membrane Protein Structure, Dynamics and Function. Nat. Struct. Mol. Biol. 2016, 23, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Bordag, N.; Keller, S. α-Helical transmembrane peptides: A “Divide and Conquer” approach to membrane proteins. Chem. Phys. Lipids 2010, 163, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Moraes, L.G.; Fázio, M.A.; Vieira, R.F.; Nakaie, C.R.; Miranda, M.T.M.; Schreier, S.; Daffre, S.; Miranda, A. Conformational and functional studies of gomesin analogues by CD, EPR and fluorescence spectroscopies. Biochim. Biophys. Acta Biomembr. 2007, 1768, 52–58. [Google Scholar] [CrossRef]
- Baker, M. Making membrane proteins for structures: A trillion tiny tweaks. Nat. Methods 2010, 7, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Stevens, T.J.; Samso, M. Membrane proteins: The ‘Wild West’ of structural biology. Trends Biochem. Sci. 2003, 28, 137–144. [Google Scholar] [CrossRef]
- Huang, C.; Mohanty, S. Challenging the Limit: NMR Assignment of a 31 kDa Helical Membrane Protein. J. Am. Chem. Soc. 2010, 132, 3662–3663. [Google Scholar] [CrossRef]
- Sahu, I.D.; Lorigan, G.A. Electron Paramagnetic Resonance as a Tool for Studying Membrane Proteins. Biomolecules 2020, 10, 763. [Google Scholar] [CrossRef]
- Columbus, L.; Hubbell, W.L. A new spin on protein dynamics. Trends Biochem. Sci. 2002, 27, 288–295. [Google Scholar] [CrossRef]
- Acharya, K.R.; Lloyd, M.D. The advantages and limitations of protein crystal structures. Trends Pharmacol. Sci. 2005, 26, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR studies of structure and function of biological macromolecules (Nobel Lecture). J. Biomol. NMR 2003, 27, 13–39. [Google Scholar] [CrossRef] [PubMed]
- Schiemann, O.; Prisner, T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007, 40, 1–53. [Google Scholar] [CrossRef]
- Lecoq, L.; Fogeron, M.-L.; Meier, B.H.; Nassal, M.; Böckmann, A. Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies. Viruses 2020, 12, 1069. [Google Scholar] [CrossRef] [PubMed]
- Midgett, C.R.; Madden, D.R. Breaking the bottleneck: Eukaryotic membrane protein expression for high-resolution structural studies. J. Struct. Biol. 2007, 160, 265–274. [Google Scholar] [CrossRef]
- Bahar, I.; Lezon, T.R.; Bakan, A.; Shrivastava, I.H. Normal Mode Analysis of Biomolecular Structures: Functional Mechanisms of Membrane Proteins. Chem. Rev. 2010, 110, 1463–1497. [Google Scholar] [CrossRef]
- Callaway, E. The protein-imaging technique taking over structural biology. Nature 2020, 578, 201. [Google Scholar] [CrossRef]
- Autzen, H.E.; Julius, D.; Cheng, Y.F. Membrane mimetic systems in CryoEM: Keeping membrane proteins in their native envi-ronment. Curr. Opin. Struct. Biol. 2019, 58, 259–268. [Google Scholar] [CrossRef]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.M.G.E.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Huynh, D.T.; Yeates, T.O. A 3.8 angstrom resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nat. Commun. 2019, 10, 1–7. [Google Scholar]
- Fanucci, G.E.; Cafiso, D.S. Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 2006, 16, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; Gross, A.; Langen, R.; Lietzow, M.A. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 1998, 8, 649–656. [Google Scholar] [CrossRef]
- Qin, P.Z.; Dieckmann, T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 2004, 14, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Berliner, L.J. From spin-labeled proteins to in vivo EPR applications. Eur. Biophys. J. 2010, 39, 579–588. [Google Scholar] [CrossRef]
- Speicher, D.W. Characterization of protein primary structure. Dev. Biol. Stand. 1998, 96, 27–28. [Google Scholar]
- Klare, J.P.; Steinhoff, H.-J. Spin labeling EPR. Photosynth. Res. 2009, 102, 377–390. [Google Scholar] [CrossRef]
- Hubbell, W.L.; Mchaourab, H.S.; Altenbach, C.; Lietzow, M.A. Watching proteins move using site-directed spin labeling. Structure 1996, 4, 779–783. [Google Scholar] [CrossRef]
- Hubbell, W.L.; López, C.J.; Altenbach, C.; Yang, Z. Technological advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 2013, 23, 725–733. [Google Scholar] [CrossRef]
- Claxton, D.P.; Kazmier, K.; Mishra, S.; McHaourab, H.S. Navigating Membrane Protein Structure, Dynamics, and Energy Landscapes Using Spin Labeling and EPR Spectroscopy. Methods Enzymol. 2015, 564, 349–387. [Google Scholar]
- Weil, J.A.; Bolton, J.R. Electron Paramagnetic Resonance: Elementary Theory and Practical Applications; Wiley-Interscience: Hoboken, NJ, USA, 2007. [Google Scholar]
- Goldfarb, D.; Stoll, S. EPR Spectroscopy: Fundamentals and Methods; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 2018. [Google Scholar]
- Roessler, M.M.; Salvadori, E. Principles and applications of EPR spectroscopy in the chemical sciences. Chem. Soc. Rev. 2018, 47, 2534–2553. [Google Scholar] [CrossRef] [PubMed]
- Bordignon, E.; Steinhoff, H.-J. Membrane Protein Structure and Dynamics Studied by Site-Directed Spin-Labeling ESR. In ESR Spectroscopy in Membrane Biophysics; Springer: Boston, MA, USA, 2007; Volume 27, pp. 129–164. [Google Scholar]
- Altenbach, C.; Flitsch, S.L.; Khorana, H.G.; Hubbell, W.L. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry 1989, 28, 7806–7812. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, C.; Froncisz, W.; Hyde, J.; Hubbell, W. Conformation of spin-labeled melittin at membrane surfaces investigated by pulse saturation recovery and continuous wave power saturation electron paramagnetic resonance. Biophys. J. 1989, 56, 1183–1191. [Google Scholar] [CrossRef]
- Altenbach, C.; Marti, T.; Khorana, H.G.; Hubbell, W.L. Transmembrane protein structure: Spin labeling of bacteriorhodopsin mutants. Science 1990, 248, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Steinhoff, H.-J. Multi-Frequency EPR Spectroscopy Studies of the Structure and Conformational Changes of Site-Directed Spin Labelled Membrane Proteins. In Supramolecular Structure and Function 8; Springer: Boston, MA, USA, 2005; Volume 8, pp. 157–177. [Google Scholar]
- Cornish, V.W.; Benson, D.R.; Altenbach, C.A.; Hideg, K.; Hubbell, W.L.; Schultz, P.G. Site-specific incorporation of biophysical probes into proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 2910–2914. [Google Scholar] [CrossRef]
- Roser, P.; Schmidt, M.J.; Drescher, M.; Summerer, D. Site-directed spin labeling of proteins for distance measurements in vitro and in cells. Org. Biomol. Chem. 2016, 14, 5468–5476. [Google Scholar] [CrossRef]
- Haugland, M.M.; Lovett, J.E.; Anderson, E.A. Advances in the synthesis of nitroxide radicals for use in biomolecule spin labelling. Chem. Soc. Rev. 2018, 47, 668–680. [Google Scholar] [CrossRef]
- Karthikeyan, G.; Bonucci, A.; Casano, G.; Gerbaud, G.; Abel, S.; Thomé, V.; Kodjabachian, L.; Magalon, A.; Guigliarelli, B.; Belle, V.; et al. A Bioresistant Nitroxide Spin Label for In-Cell EPR Spectroscopy: In Vitro and In Oocytes Protein Structural Dynamics Studies. Angew. Chem. Int. Ed. 2018, 57, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Bleicken, S.; Assafa, T.E.; Zhang, H.; Elsner, C.; Ritsch, I.; Pink, M.; Rajca, S.; Jeschke, G.; Rajca, A. Bordignon, E. gem-Diethyl Pyrroline Nitroxide Spin Labels: Synthesis, EPR Characterization, Rotamer Libraries and Biocompatibility. Chemistryopen 2019, 8, 1057–1065. [Google Scholar] [CrossRef]
- Sahu, I.D.; Craig, A.F.; Dunagum, M.M.; McCarrick, R.M.; Lorigan, G.A. Characterization of bifunctional spin labels for inves-tigating the structural and dynamic properties of membrane proteins using EPR spectroscopy. J. Phys. Chem. B 2017, 121, 9185–9195. [Google Scholar] [CrossRef]
- McCaffrey, J.E.; James, Z.M.; Svensson, B.; Binder, B.P.; Thomas, D.D. A bifunctional spin label reports the structural topology of phospholamban in magnetically-aligned bicelles. J. Magn. Reson. 2016, 262, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wanderling, S.; Sompornpisut, P.; Perozo, E.; Somponspisut, P. Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nat. Struct. Mol. Biol. 2014, 21, 160–166. [Google Scholar] [CrossRef]
- Lösel, R.M.; Philipp, R.; Kálai, T.; Hideg, K.; Trommer, W.E. Synthesis and Application of Novel Bifunctional Spin Labels. Bioconjug. Chem. 1999, 10, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Haugland, M.M.; Anderson, E.A.; Lovett, J.E. Tuning the properties of nitroxide spin labels for use in electron paramagnetic resonance spectroscopy through chemical modification of the nitroxide framework. In Electron Paramagnetic Resonance; Chechik, V., Murphy, D.M., Eds.; Royal Society of Chemistry (RSC): London, UK, 2017; pp. 1–34. [Google Scholar]
- Sahu, I.D.; Craig, A.F.; Dunagan, M.M.; Troxel, K.R.; Zhang, R.; Meiberg, A.G.; Harmon, C.N.; McCarrick, R.M.; Kroncke, B.M.; Sanders, C.R.; et al. Probing Structural Dynamics and Topology of the KCNE1 Membrane Protein in Lipid Bilayers via Site-Directed Spin Labeling and Electron Paramagnetic Resonance Spectroscopy. Biochemistry 2015, 54, 6402–6412. [Google Scholar] [CrossRef]
- Basak, S.; Chatterjee, S.; Chakrapani, S. Site directed spin labeling and EPR spectroscopic studies of pntameric ligand-gated ion channels. JOVE J. Vis. Exp. 2016, 113, 54127. [Google Scholar]
- Sahu, I.D.; Lorigan, G.A. Biophysical EPR Studies Applied to Membrane Proteins. J. Phys. Chem. Biophys. 2015, 5, 188. [Google Scholar] [CrossRef]
- Jeschke, G.; Bender, A.; Schweikardt, T.; Panek, G.; Decker, H.; Paulsen, H. Localization of the N-terminal Domain in Light-harvesting Chlorophyll a/b Protein by EPR Measurements. J. Biol. Chem. 2005, 280, 18623–18630. [Google Scholar] [CrossRef]
- Mchaourab, H.S.; Perozo, E. Determination of Protein Folds and Conformational Dynamics Using Spin-Labeling EPR Spec-troscopy. In Biological Magnetic Resonance; Berliner, L., Eaton, G., Eaton, S., Eds.; Springer: New York, NY, USA, 2002; pp. 185–247. [Google Scholar]
- Perozo, E.; Cortes, D.M.; Cuello, L.G. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 1998, 5, 459–469. [Google Scholar] [CrossRef]
- Vasquez, V.; Sotomayor, M.; Cortes, D.M.; Roux, B.; Schulten, K.; Perozo, E. Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol. 2008, 378, 55–70. [Google Scholar] [CrossRef]
- Hustedt, E.J.; Beth, A.H. Nitroxide spin-spin interactions: Applications to protein structure and dynamics. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 129–153. [Google Scholar] [CrossRef]
- Brown, L.J.; Hare, J.E. Electron Paramagnetic Resonance: Site-Directed Spin Labeling; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Wunnicke, D.; Hänelt, I. The Synergetic Effects of Combining Structural Biology and EPR Spectroscopy on Membrane Proteins. Crystals 2017, 7, 117. [Google Scholar] [CrossRef]
- Hustedt, E.; Smirnov, A.; Laub, C.; Cobb, C.; Beth, A. Molecular distances from dipolar coupled spin-labels: The global analysis of multifrequency continuous wave electron paramagnetic resonance data. Biophys. J. 1997, 72, 1861–1877. [Google Scholar] [CrossRef]
- Ghimire, H.; Hustedt, E.J.; Sahu, I.D.; Inbaraj, J.J.; McCarrick, R.; Mayo, D.J.; Benedikt, M.R.; Lee, R.T.; Grosser, S.M.; Lorigan, G.A. Distance Measurements on a Dual-Labeled TOAC AChR M2δ Peptide in Mechanically Aligned DMPC Bilayers via Dipolar Broadening CW-EPR Spectroscopy. J. Phys. Chem. B 2012, 116, 3866–3873. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, E.J.; Stein, R.A.; Sethaphong, L.; Brandon, S.; Zhou, Z.; DeSensi, S.C. Dipolar Coupling between Nitroxide Spin Labels: The Development and Application of a Tether-in-a-Cone Model. Biophys. J. 2006, 90, 340–356. [Google Scholar] [CrossRef]
- Banham, J.E.; Baker, C.M.; Ceola, S.; Day, I.J.; Grant, G.H.; Groenen, E.J.; Rodgers, C.T.; Jeschke, G.; Timmel, C.R. Distance measurements in the borderline region of applicability of CW EPR and DEER: A model study on a homologous series of spin-labelled peptides. J. Magn. Reson. 2008, 191, 202–218. [Google Scholar] [CrossRef] [PubMed]
- Rabenstein, M.D.; Shin, Y.K. Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. USA 1995, 92, 8239–8243. [Google Scholar] [CrossRef] [PubMed]
- Czogalla, A.; Pieciul, A.; Jezierski, A.; Sikorski, A.F. Attaching a spin to a protein—Site-directed spin labeling in structural biology. Acta Biochim. Pol. 2007, 54, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Mandal, T.; Hustedt, E.J.; Song, L.; Oh, K.J. CW EPR and DEER Methods to Determine BCL-2 Family Protein Structure and Interactions: Application of Site-Directed Spin Labeling to BAK Apoptotic Pores. In BCL-2 Family Proteins; Methods in Molecular Biology; Gavathiotis, E., Ed.; Humana Press: New York, NY, USA, 2019; Volume 1877, pp. 257–303. [Google Scholar]
- Jeschke, G.; Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007, 9, 1895–1910. [Google Scholar] [CrossRef]
- Schweiger, A.; Jeschke, G. Principles of Pulse Electron Paramagnetic Resonance; Oxford University Press: New York, NY, USA, 2001. [Google Scholar]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000, 142, 331–340. [Google Scholar] [CrossRef]
- Jeschke, G.; Chechik, V.; Ionita, P.; Godt, A.; Zimmermann, H.; Banham, J.; Timmel, C.R.; Hilger, D.; Jung, H. Deer Analysis 2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006, 30, 473–498. [Google Scholar] [CrossRef]
- Ibáñez, L.F.; Jeschke, G.; Stoll, S. DeerLab: A comprehensive software package for analyzing dipolar electron paramagnetic resonance spectroscopy data. Magn. Reson. 2020, 1, 209–224. [Google Scholar] [CrossRef]
- Worswick, S.G.; Spencer, J.A.; Jeschke, G.; Kuprov, I. Deep neural network processing of DEER data. Sci. Adv. 2018, 4, eaat5218. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; Lorigan, G.A. Site-Directed Spin Labeling EPR for Studying Membrane Proteins. BioMed Res. Int. 2018, 2018, 3248289. [Google Scholar] [CrossRef]
- Sahu, I.D.; Lorigan, G.A. EPR Techniques, Spin Labeling and Spin Trapping. In Encyclopedia of Analytical Science; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; pp. 315–327. [Google Scholar]
- Borbat, P.P.; McHaourab, H.S.; Freed, J.H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: Study of T4 lysozyme. J. Am. Chem. Soc. 2002, 124, 5304–5314. [Google Scholar] [CrossRef]
- Jeschke, G. DEER Distance Measurements on Proteins. Annu. Rev. Phys. Chem. 2012, 63, 419–446. [Google Scholar] [CrossRef] [PubMed]
- Milov, A.D.; Tsvetkov, Y.D.; Formaggio, F.; Crisma, M.; Toniolo, C.; Raap, J. Self-assembling properties of membrane-modifying peptides studied by PELDOR and CW-ESR spectroscopies. J. Am. Chem. Soc. 2000, 122, 3843–3848. [Google Scholar] [CrossRef]
- Hilger, D.; Jung, H.; Padan, E.; Wegener, C.; Vogel, K.P.; Steinhoff, H.J.; Jeschke, G. Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E. coli. Biophys. J. 2005, 89, 1328–1338. [Google Scholar] [CrossRef]
- Ahammad, T.A.; Drew, D.L.; Sahu, I.D.; Khan, R.H.; Butcher, B.J.; Serafin, R.A.; Galende, A.P.; McCarrick, R.M.; Lorigan, G.A. Conformational Differences are Observed for the Active and Inactive Forms of Pinholin S21 using DEER Spectroscopy. Phys. Chem. B 2020, 124, 11396–11405. [Google Scholar] [CrossRef]
- Bordignon, E.; Bleicken, S. New limits of sensitivity of site-directed spin labeling electron paramagnetic resonance for membrane proteins. Biochim. Biophys. Acta Biomembr. 2018, 1860, 841–853. [Google Scholar] [CrossRef]
- Feintuch, A.; Otting, G.; Goldfarb, D. Gd3+ Spin Labeling for Measuring Distances in Biomacromolecules: Why and How? In Methods in Enzymology; Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part A; Elsevier: Amsterdam, The Netherlands, 2015; Volume 563, pp. 415–457. [Google Scholar]
- Jassoy, J.J.; Berndhäuser, A.; Duthie, F.; Kühn, S.P.; Hagelueken, G.; Schiemann, O. Versatile Trityl Spin Labels for Nanometer Distance Measurements on Biomolecules In Vitro and within Cells. Angew. Chem. Int. Ed. 2017, 56, 177–181. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Ji, M.; Cunningham, T.F.; Saxena, S. Cu2+ as an ESR Probe of Protein Structure and Function. In Methods in Enzymology; Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part A; Elsevier: Amsterdam, The Netherlands, 2015; Volume 563, pp. 459–481. [Google Scholar]
- Joseph, B.; Sikora, A.; Cafiso, D.S. Ligand Induced Conformational Changes of a Membrane Transporter in E. coli Cells Observed with DEER/PELDOR. J. Am. Chem. Soc. 2016, 138, 1844–1847. [Google Scholar] [CrossRef]
- Yardeni, E.H.; Bahrenberg, T.; Stein, R.A.; Mishra, S.; Zomot, E.; Graham, B.; Tuck, K.L.; Huber, T.; Bibi, E.; McHaourab, H.S.; et al. Probing the solution structure of the E. coli multidrug transporter MdfA using DEER distance measurements with nitroxide and Gd(III) spin labels. Sci. Rep. 2019, 9, 12528. [Google Scholar] [CrossRef]
- Joseph, B.; Tormyshev, V.M.; Rogozhnikova, O.Y.; Akhmetzyanov, D.; Bagryanskaya, E.G.; Prisner, T.F. Selective High-Resolution Detection of Membrane Protein-Ligand Interaction in Native Membranes Using Trityl-Nitroxide PELDOR. Angew. Chem. Int. Ed. 2016, 55, 11538–11542. [Google Scholar] [CrossRef]
- Altenbach, C.; Yang, K.; Farrens, D.L.; Farahbakhsh, Z.T.; Khorana, H.G.; Hubbell, W.L. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: A site-directed spin-labeling study. Biochemistry 1996, 35, 12470–12478. [Google Scholar] [CrossRef] [PubMed]
- Hubbell, W.L.; Altenbach, C. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Curr. Opin. Struct. Biol. 1994, 4, 566–573. [Google Scholar] [CrossRef]
- Fajer, P.G. Site directed spin labelling and pulsed dipolar electron paramagnetic resoonance (double electron-electron resonance) of force activation in muscle. J. Phys. Condens. Matter 2005, 17, S1459–S1469. [Google Scholar] [CrossRef]
- Tait, C.E.; Stoll, S. Coherent pump pulses in Double Electron Electron Resonance spectroscopy. Phys. Chem. Chem. Phys. 2016, 18, 18470–18485. [Google Scholar] [CrossRef]
- Klug, C.S.; Feix, J.B. SDSL: A survey of biological applications. Biol. Magn. Reson. 2004, 24, 269–308. [Google Scholar]
- Stoll, S.; Schweiger, A. Easyspin: Simulating cw ESR spectra. Biol. Magn. Reson. 2007, 27, 299–321. [Google Scholar]
- Soria, M.A.; Cervantes, S.A.; Bajakian, T.H.; Siemer, A.B. The Functional Amyloid Orb2A Binds to Lipid Membranes. Biophys. J. 2017, 113, 37–47. [Google Scholar] [CrossRef]
- Victor, K.G.; Cafiso, D.S. Location and Dynamics of Basic Peptides at the Membrane Interface: Electron Paramagnetic Resonance Spectroscopy of Tetramethyl-Piperidine-N-Oxyl-4-Amino-4-Carboxylic Acid-Labeled Peptides. Biophys. J. 2001, 81, 2241–2250. [Google Scholar] [CrossRef][Green Version]
- Yu, L.; Wang, W.; Ling, S.; Liu, S.; Xiao, L.; Xin, Y.; Lai, C.; Xiong, Y.; Zhang, L.; Tian, C. CW-EPR studies revealed different motional properties and oligomeric states of the integrin beta(1a) transmembrane domain in detergent micelles or liposomes. Sci. Rep. 2015, 5, 7848. [Google Scholar] [CrossRef] [PubMed]
- Altenbach, C.; Greenhalgh, D.A.; Khorana, H.G.; Hubbell, W.L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 1994, 91, 1667–1671. [Google Scholar] [CrossRef]
- Cortes, D.M.; Cuello, L.G.; Perozo, E. Molecular architecture of full-length KcsA—Role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 2001, 117, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Voss, J.; He, M.M.; Hubbell, W.L.; Kaback, H.R. Site-directed spin labeling demonstrates that transmembrane domain XII in the lactose permease of Escherichia coli is an alpha-helix. Biochemistry 1996, 35, 12915–12918. [Google Scholar] [CrossRef]
- Song, Y.; Hustedt, E.J.; Brandon, S.; Sanders, C.R. Competition Between Homodimerization and Cholesterol Binding to the C99 Domain of the Amyloid Precursor Protein. Biochemistry 2013, 52, 5051–5064. [Google Scholar] [CrossRef]
- Perozo, E.; Hubbell, W.L. Transmembrane voltage control in liposomes- the use of bacteriorhodopsin as a light-driven current source. Biophys. J. 1993, 64, A222. [Google Scholar]
- Mokdad, A.; Herrick, D.Z.; Kahn, A.K.; Andrews, E.; Kim, M.; Cafiso, D.S. Ligand-Induced Structural Changes in the Escherichia coli Ferric Citrate Transporter Reveal Modes for Regulating Protein-Protein Interactions. J. Mol. Biol. 2012, 423, 818–830. [Google Scholar] [CrossRef][Green Version]
- Aziz, A.; Hess, J.F.; Budamagunta, M.S.; Voss, J.C.; FitzGerald, P.G. Site-directed Spin Labeling and Electron Paramagnetic Resonance Determination of Vimentin Head Domain Structure. J. Biol. Chem. 2010, 285, 15278–15285. [Google Scholar] [CrossRef]
- Dong, J.; Yang, G.; Mchaourab, H.S. Structural Basis of Energy Transduction in the Transport Cycle of MsbA. Science 2005, 308, 1023–1028. [Google Scholar] [CrossRef]
- Malmberg, N.J.; Falke, J.J. Use of EPR power saturation toanalyze the membrane-docking geometries of peripheral proteins: A applications to C2 domains. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 71–90. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, Y.G.; Thorgeirsson, T.E.; Shin, Y.-K. Topology of an Amphiphilic Mitochondrial Signal Sequence in the Membrane-Inserted State: A Spin Labeling Study. Biochemistry 1994, 33, 14221–14226. [Google Scholar] [CrossRef] [PubMed]
- Klug, C.S.; Su, W.Y.; Feix, J.B. Mapping of the residues involved in a proposed beta-strand located in the ferric enterobactin receptor FepA using site-directed spin-labeling. Biochemistry 1997, 36, 13027–13033. [Google Scholar] [CrossRef]
- Carter, J.D.; Mathias, J.D.; Gomez, E.F.; Ran, Y.; Xu, F.; Galiano, L.; Tran, N.Q.; D’Amore, P.W.; Wright, C.S.; Chakravorty, D.K.; et al. Characterizing Solution Surface Loop Conformational Flexibility of the GM2 Activator Protein. J. Phys. Chem. B 2014, 118, 10607–10617. [Google Scholar] [CrossRef]
- Ahammad, T.; Drew, D.L.; Khan, R.H.; Sahu, I.D.; Faul, E.; Li, T.; Lorigan, G.A. Structural Dynamics and Topology of the Inactive Form of S21 Holin in a Lipid Bilayer Using Continuous-Wave Electron Paramagnetic Resonance Spectroscopy. J. Phys. Chem. B 2020, 124, 5370–5379. [Google Scholar] [CrossRef]
- Ahammad, T.; Drew, D.L., Jr.; Sahu, I.D.; Serafin, R.A.; Clowes, K.R.; Lorigan, G.A. Continuous Wave Electron Paramagnetic Resonance Spectroscopy Reveals the Structural Topology and Dynamic Properties of Active Pinholin S2168 in a Lipid Bilayer. J. Phys. Chem. B 2019, 123, 8048–8056. [Google Scholar] [CrossRef] [PubMed]
- Dixit, G.; Sahu, I.D.; Reynolds, W.D.; Wadsworth, T.M.; Harding, B.D.; Jaycox, C.K.; Dabney-Smith, C.; Sanders, C.R.; Lorigan, G.A. Probing the Dynamics and Structural Topology of the Reconstituted Human KCNQ1 Voltage Sensor Domain (Q1-VSD) in Lipid Bilayers Using Electron Paramagnetic Resonance Spectroscopy. Biochemistry 2019, 58, 965–973. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Ghosh, B.; Hanson, S.M.; Raspanti, J.M.; Grant, V.A.; Diarra, G.M.; Schuh, A.M.; Satyshur, K.A.; Klug, C.S.; Czajkowski, C. Site-Directed Spin Labeling Reveals Pentameric Ligand-Gated Ion Channel Gating Motions. PLoS Biol. 2013, 11, e1001714. [Google Scholar] [CrossRef]
- Pang, T.; Savva, C.G.; Fleming, K.G.; Struck, D.K.; Young, R. Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. USA 2009, 106, 18966–18971. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.; Park, T.; Young, R. Mutational analysis of the S21pinholin. Mol. Microbiol. 2010, 76, 68–77. [Google Scholar] [CrossRef]
- Mosslehy, W.; Voskoboynikova, N.; Colbasevici, A.; Ricke, A.; Klose, D.; Klare, J.P.; Mulkidjanian, A.Y.; Steinhoff, H.J. Conformational Dynamics of Sensory Rhodopsin II in Nanolipoprotein and Styrene-Maleic Acid Lipid Particles. Photochem. Photobiol. 2019, 95, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.-W.; Zheng, T.-Y.; Kao, C.-J.; Horng, J.-C. Determination of Interspin Distance Distributions by cw-ESR Is a Single Linear Inverse Problem. Biophys. J. 2009, 97, 930–936. [Google Scholar] [CrossRef][Green Version]
- Meyer, A.; Dechert, S.; Dey, S.; Höbartner, C.; Bennati, M. Measurement of Angstrom to Nanometer Molecular Distances with 19 F Nuclear Spins by EPR/ENDOR Spectroscopy. Angew. Chem. Int. Ed. 2020, 59, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Czogalla, A.; Jaszewski, A.R.; Diakowski, W.; Bok, E.; Jezierski, A.; Sikorski, A.F. Structural insight into an ankyrin-sensitive lipid-binding site of erythroid beta-spectrin. Mol. Membr. Biol. 2007, 24, 215–224. [Google Scholar] [CrossRef]
- Essen, L.O.; Siegert, R.; Lehmann, W.D.; Oesterhelt, D. Lipid patches in membrane protein oligomers: Crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. USA 1998, 95, 11673–11678. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Wang, W.; Yu, L.; Peng, J.; Cai, X.; Xiong, Y.; Hayati, Z.; Zhang, L.; Zhang, Z.; Song, L.; et al. Structure of an E. coli integral membrane sulfurtransferase and its structural transition upon SCN− binding defined by EPR-based hybrid method. Sci. Rep. 2016, 6, 20025. [Google Scholar] [CrossRef]
- Sahu, I.D.; Hustedt, E.J.; Ghimire, H.; Inbaraj, J.J.; McCarrick, R.M.; Lorigan, G.A. CW dipolar broadening EPR spectroscopy and mechanically aligned bilayers used to measure distance and relative orientation between two TOAC spin labels on an antimicrobial peptide. J. Magn. Reson. 2014, 249, 72–79. [Google Scholar] [CrossRef]
- Hanelt, I.; Wunnicke, D.; Muller-Trimbusch, M.; Vor der Bruggen, M.; Kraus, I.; Bakker, E.P.; Steinhoff, H.J. Membrane Region M2C2 in Subunit KtrB of the K+ Uptake System KtrAB from Vibrio alginolyticus Forms a Flexible Gate Controlling K+ Flux—An electron paramagnetic resonance study. J. Biol. Chem. 2010, 285, 28210–28219. [Google Scholar] [CrossRef]
- Steinhoff, H.-J. Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin labeling reveal protein-protein and protein-oligonucleotide interaction. Biol. Chem. 2004, 385, 913–920. [Google Scholar] [CrossRef]
- Wegener, A.-A.; Klare, J.P.; Engelhard, M.; Steinhoff, H.-J. Structural insights into the early steps of receptor–transducer signal transfer in archaeal phototaxis. EMBO J. 2001, 20, 5312–5319. [Google Scholar] [CrossRef]
- Scarpelli, F.; Drescher, M.; Rutters-Meijneke, T.; Holt, A.; Rijkers, D.T.S.; Killian, J.A.; Huber, M. Aggregation of Transmembrane Peptides Studied by Spin-Label EPR. J. Phys. Chem. B 2009, 113, 12257–12264. [Google Scholar] [CrossRef][Green Version]
- Yu, L.; Wang, W.; Ling, S.; He, Y.; Xiao, L.; Wu, K.; Zhang, L.; Tian, C. Distance measurement between two flexible sites in proteins in high viscosity medium at physiological temperature using continuous wave EPR. Protein Cell 2014, 5, 334–337. [Google Scholar] [CrossRef][Green Version]
- El Mkami, H.; Ward, R.; Bowman, A.; Owen-Hughes, T.; Norman, D.G. The spatial effect of protein deuteration on nitroxide spin-label relaxation: Implications for EPR distance measurement. J. Magn. Reson. 2014, 248, 36–41. [Google Scholar] [CrossRef]
- Schmidt, T.; Walti, M.A.; Baber, J.L.; Hustedt, E.J.; Clore, G.M. Long Distance Measurements up to 160 angstrom in the GroEL Tetradecamer Using Q-Band DEER EPR Spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 15905–15909. [Google Scholar] [CrossRef]
- Mchaourab, H.S.; Steed, P.R.; Kazmier, K. Toward the Fourth Dimension of Membrane Protein Structure: Insight into Dynamics from Spin-Labeling EPR Spectroscopy. Structure 2011, 19, 1549–1561. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Mchaourab, H.S. Increased Sensitivity and Extended Range of Distance Measurements in Spin-Labeled Membrane Proteins: Q-Band Double Electron-Electron Resonance and Nanoscale Bilayers. Biophys. J. 2010, 98, L18–L20. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Bortolus, M.; Mchaourab, H.S. Conformational Cycle of the ABC Transporter MsbA in Liposomes: Detailed Analysis Using Double Electron–Electron Resonance Spectroscopy. J. Mol. Biol. 2009, 393, 586–597. [Google Scholar] [CrossRef]
- Endeward, B.; Butterwick, J.A.; MacKinnon, R.; Prisner, T.F. Pulsed Electron−Electron Double-Resonance Determination of Spin-Label Distances and Orientations on the Tetrameric Potassium Ion Channel KcsA. J. Am. Chem. Soc. 2009, 131, 15246–15250. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Ramlall, T.F.; Borbat, P.P.; Freed, J.H.; Eliezer, D. Membrane-Bound α-Synuclein Forms an Extended Helix: Long-Distance Pulsed ESR Measurements Using Vesicles, Bicelles, and Rodlike Micelles. J. Am. Chem. Soc. 2008, 130, 12856–12857. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ellena, J.F.; Kim, A.M.; Cafiso, D.S. Substrate-Dependent Unfolding of the Energy Coupling Motif of a Membrane Transport Protein Determined by Double Electron-Electron Resonance. Biochemistry 2006, 45, 10847–10854. [Google Scholar] [CrossRef][Green Version]
- Polyhach, Y.; Bordignon, E.; Tschaggelar, R.; Gandra, S.; Godt, A.; Jeschke, G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 2012, 14, 10762–10773. [Google Scholar] [CrossRef]
- Cunningham, T.F.; Putterman, M.R.; Desai, A.; Horne, W.S.; Saxena, S. The Double-Histidine Cu2+-Binding Motif: A Highly Rigid, Site-Specific Spin Probe for Electron Spin Resonance Distance Measurements. Angew. Chem. Int. Ed. 2015, 54, 6330–6334. [Google Scholar] [CrossRef]
- Li, C.-C.; Hung, C.-L.; Yeh, P.-S.; Li, C.-E.; Chiang, Y.-W. Doubly spin-labeled nanodiscs to improve structural determination of membrane proteins by ESR. RSC Adv. 2019, 9, 9014–9021. [Google Scholar] [CrossRef]
- Jao, C.C.; Hegde, B.G.; Chen, J.; Haworth, I.S.; Langen, R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. USA 2008, 105, 19666–19671. [Google Scholar] [CrossRef]
- Milikisiyants, S.; Wang, S.; Munro, R.A.; Donohue, M.; Ward, M.E.; Bolton, D.; Brown, L.S.; Smirnova, T.I.; Ladizhansky, V.; Smirnov, A.I. Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints. J. Mol. Biol. 2017, 429, 1903–1920. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Han, W.; Fiorin, G.; Islam, S.M.; Schulten, K.; Roux, B. Structural Refinement of Proteins by Restrained Molecular Dynamics Simulations with Non-interacting Molecular Fragments. PLoS Comput. Biol. 2015, 11, e1004368. [Google Scholar] [CrossRef] [PubMed]
- Vicente, E.F.; Sahu, I.D.; Costa-Filho, A.J.; Cilli, E.M.; Lorigan, G.A. Conformational changes of the HsDHODH N-terminal Microdomain via DEER Spectroscopy. J. Phys. Chem. B 2015, 119, 8693–8697. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Borbat, P.P.; Norman, H.D.; Freed, J.H. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Sci. Rep. 2015, 5, 11757. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R. Nanoscale lipid membrane mimetics in spin-labeling and electron paramagnetic resonance spectroscopy studies of protein structure and function. Nanotechnol. Rev. 2017, 6, 75–92. [Google Scholar] [CrossRef]
- Dixit, M.; Kim, S.; Matthews, G.F.; Erreger, K.; Galli, A.; Cobb, C.E.; Hustedt, E.J.; Beth, A.H. Structural Arrangement of the Intracellular Ca2+ Binding Domains of the Cardiac Na+/Ca2+ Exchanger (NCX1.1): Effects of Ca2+ binding. J. Biol. Chem. 2013, 288, 4194–4207. [Google Scholar] [CrossRef] [PubMed]
- Hilger, D.; Polyhach, Y.; Jung, H.; Jeschke, G. Backbone Structure of Transmembrane Domain IX of the Na+/Proline Transporter PutP of Escherichia coli. Biophys. J. 2009, 96, 217–225. [Google Scholar] [CrossRef]
- Kroncke, B.M.; Van Horn, W.D.; Smith, J.; Kang, C.; Welch, R.C.; Song, Y.; Nannemann, D.P.; Taylor, K.C.; Sisco, N.J.; George, A.L.; et al. Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci. Adv. 2016, 2, e1501228. [Google Scholar] [CrossRef]
- Barrett, P.J.; Song, Y.; Van Horn, W.D.; Hustedt, E.J.; Schafer, J.M.; Hadziselimovic, A.; Beel, A.J.; Sanders, C.R. The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol. Science 2012, 336, 1168–1171. [Google Scholar] [CrossRef] [PubMed]
- Mullen, A.; Hall, J.; Diegel, J.; Hassan, I.; Fey, A.; Macmillan, F. Membrane transporters studied by EPR spectroscopy: Structure determination and elucidation of functional dynamics. Biochem. Soc. Trans. 2016, 44, 905–915. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Basak, S.; Schmandt, N.; Gicheru, Y.; Chakrapani, S. Crystal structure and dynamics of a lipid induced potential desensitized-state of a pentameric ligand-gated channel. Elife 2017, 6, e23886. [Google Scholar] [CrossRef]
- Herneisen, A.L.; Sahu, I.D.; McCarrick, R.M.; Feix, J.B.; Lorigan, G.A.; Howard, K.P. A Budding-Defective M2 Mutant Exhibits Reduced Membrane Interaction, Insensitivity to Cholesterol, and Perturbed Interdomain Coupling. Biochemistry 2017, 56, 5955–5963. [Google Scholar] [CrossRef]
- Kumar, P.; Van Son, M.; Zheng, T.; Valdink, D.; Raap, J.; Kros, A.; Huber, M. Coiled-coil formation of the membrane-fusion K/E peptides viewed by electron paramagnetic resonance. PLoS ONE 2018, 13, e0191197. [Google Scholar] [CrossRef] [PubMed]
- Louis, J.M.; Baber, J.L.; Ghirlando, R.; Aniana, A.; Bax, A.; Roche, J. Insights into the Conformation of the Membrane Proximal Regions Critical to the Trimerization of the HIV-1 gp41 Ectodomain Bound to Dodecyl Phosphocholine Micelles. PLoS ONE 2016, 11, e0160597. [Google Scholar] [CrossRef] [PubMed]
- Puljung, M.C.; DeBerg, H.A.; Zagotta, W.N.; Stoll, S. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc. Natl. Acad. Sci. USA 2014, 111, 9816–9821. [Google Scholar] [CrossRef] [PubMed]
- Martens, C.; Stein, R.A.; Masureel, M.; Roth, A.; Mishra, S.; Dawaliby, R.; Konijnenberg, A.A.; Sobott, A.K.F.; Govaerts, C.; McHaourab, H.S. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol. 2016, 23, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Riederer, E.A.; Focke, P.J.; Georgieva, E.R.; Akyuz, N.; Matulef, K.; Borbat, P.P.; Freed, J.H.; Blanchard, S.C.; Boudker, O.; Valiyaveetil, F.I. A Facile approach forte in vitro assembly of multimeric membrane transport proteins. Elife 2018, 7, e36478. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Borbat, P.P.; Ginter, C.S.; Freed, J.H.; Boudker, O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 2013, 20, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Nyenhuis, D.A.; Nilaweera, T.D.; Cafiso, D.S. Native Cell Environment Constrains Loop Structure in the Escherichia coli Cobalamin Transporter BtuB. Biophys. J. 2020, 119, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D. Conformational Dynamics of the Extracellular Loop of BtuB in Whole Cells. Biophys. J. 2020, 119, 1470–1471. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, I.D.; Lorigan, G.A. Probing Structural Dynamics of Membrane Proteins Using Electron Paramagnetic Resonance Spectroscopic Techniques. Biophysica 2021, 1, 106-125. https://doi.org/10.3390/biophysica1020009
Sahu ID, Lorigan GA. Probing Structural Dynamics of Membrane Proteins Using Electron Paramagnetic Resonance Spectroscopic Techniques. Biophysica. 2021; 1(2):106-125. https://doi.org/10.3390/biophysica1020009
Chicago/Turabian StyleSahu, Indra D., and Gary A. Lorigan. 2021. "Probing Structural Dynamics of Membrane Proteins Using Electron Paramagnetic Resonance Spectroscopic Techniques" Biophysica 1, no. 2: 106-125. https://doi.org/10.3390/biophysica1020009
APA StyleSahu, I. D., & Lorigan, G. A. (2021). Probing Structural Dynamics of Membrane Proteins Using Electron Paramagnetic Resonance Spectroscopic Techniques. Biophysica, 1(2), 106-125. https://doi.org/10.3390/biophysica1020009




