Theoretical Framework and Methodological Approach for Investigating Potential Associations Between Long COVID and Autism Spectrum Disorder Prevalence
Abstract
1. Introduction
Novel Methodological Contributions
- (1)
- Multi-pathway Mendelian Randomization Architecture: Unlike traditional single-instrument approaches, I propose pathway-stratified MR using distinct genetic instruments for viral entry (ACE2), immune response (HLA), and inflammatory resolution (cytokine pathway genes) to isolate specific causal mechanisms while addressing pleiotropy concerns.
- (2)
- Community-Informed AI Development: Rather than applying existing models to autistic populations, I propose co-design protocols where autistic self-advocates participate directly in feature selection, model validation criteria, and interpretation frameworks—addressing the fundamental limitation of AI systems developed without neurodivergent input.
- (3)
- Convergent Biomarker Specificity Framework: I integrate inflammatory signatures with temporal exposure windows and genetic susceptibility profiles to distinguish Long COVID–autism associations from general inflammatory conditions—a specificity challenge not systematically addressed in existing literature.
2. Proposed Experimental Designs for Causal Inference
2.1. Multi-Stage Pleiotropy-Resistant Mendelian Randomization
- Stage One: Pathway-Specific Instrumental Variable Construction
- Stage Two: Comprehensive Pleiotropy Detection and Mitigation Protocols
- Stage Three: Advanced Sensitivity Analysis and Validation Framework
- Population Stratification and Ancestry-Specific Analysis
- Technical Implementation and Quality Control
2.2. Proposed Controlled Animal Model Studies
2.3. Proposed Natural Experiment Designs
2.4. Alternative Environmental Explanations to Investigate
3. Theoretical Biological Mechanisms: Acute vs. Chronic Immune Activation
3.1. Acute Immune Response Characteristics
3.2. Inflammatory Signature Specificity Analysis
- Comparative Inflammatory Profile Characterization
- Machine Learning Discrimination and Biomarker Classification
- Mechanistic Pathway Integration and Biological Plausibility
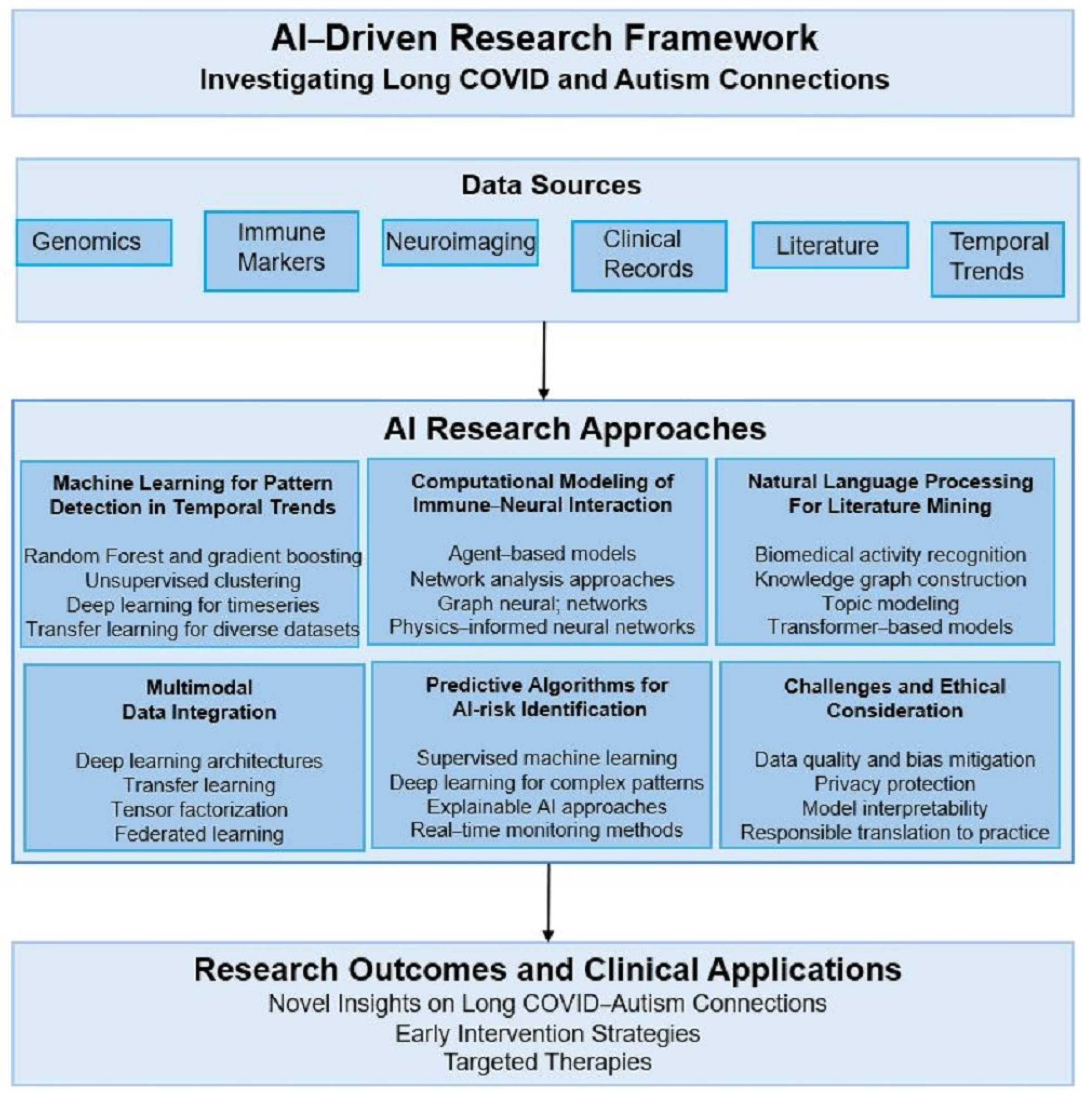
4. AI-Driven Research Implementation Framework
- Multi-Modal Data Integration Architecture
- Integrated AI Workflow Architecture
5. Ethical Framework: Neurodiversity-Affirming Research
5.1. Addressing Pathologization Concerns
5.2. Concrete Community Engagement Implementation
- Four-Stage Co-Design Implementation Protocol
- Stage Three: Ongoing Analysis Oversight and Collaborative Interpretation
- Stage Four: Dissemination Partnership and Knowledge Translation
- Resource Allocation and Institutional Commitment Structures
- Accountability Mechanisms and Quality Assurance
- Practical Implementation Examples and Lessons Learned
5.3. Preventing Research Misapplication
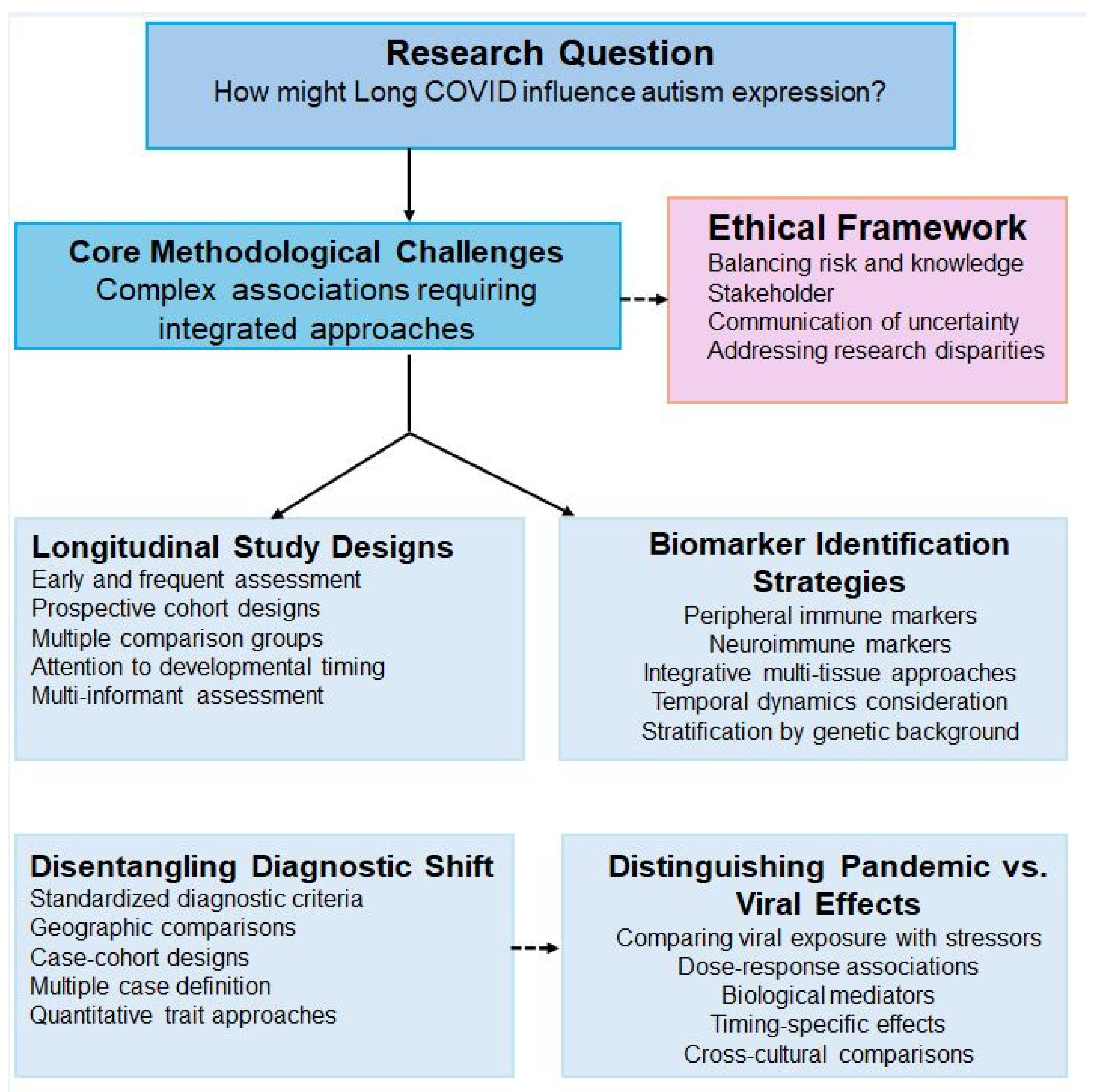
6. Proposed Methodological Framework and Future Directions
6.1. Proposed Longitudinal Study Design Requirements
6.2. Bradford Hill Criteria Application
6.3. Proposed Research Infrastructure and Collaboration
7. Proof-of-Concept Implementation Framework
7.1. Mock Dataset Specifications and Power Analysis
7.2. Expected Performance Metrics and Validation Benchmarks
7.3. Five-Year Implementation and Translation Roadmap
8. Limitations and Methodological Humility
8.1. Current Evidence Constraints
8.2. Ethical Considerations in Causal Claims
8.3. Scientific Rigor and Replication
9. Conclusions
Funding
Conflicts of Interest
References
- Modabbernia, A.; Velthorst, E.; Reichenberg, A. Environmental risk factors for autism: An evidence-based review of systematic reviews and meta-analyses. Mol. Autism 2017, 8, 13. [Google Scholar] [CrossRef]
- Maenner, M.J.; Shaw, K.A.; Bakian, A.V.; Bilder, D.A.; Durkin, M.S.; Esler, A.; Furnier, S.M.; Hallas, L.; Hall-Lande, J.; Hudson, A.; et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2018. MMWR Surveill. Summ. 2021, 70, 1–16. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef]
- Hill, A.B. The environment and disease: Association or causation? Proc. R. Soc. Med. 1965, 58, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.J.; Brugha, T.S.; Erskine, H.E.; Scheurer, R.W.; Vos, T.; Scott, J.G. The epidemiology and global burden of autism spectrum disorders. Psychol. Med. 2015, 45, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Rothman, K.J.; Greenland, S.; Lash, T.L. Modern Epidemiology, 3rd ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2008. [Google Scholar]
- Grimes, D.A.; Schulz, K.F. Bias and causal associations in observational research. Lancet 2002, 359, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Gough, D.; Oliver, S.; Thomas, J. An Introduction to Systematic Reviews, 2nd ed.; SAGE Publications: London, UK, 2017. [Google Scholar]
- Centers for Disease Control and Prevention. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2020. MMWR Surveill. Summ. 2023, 72, 1–14. [Google Scholar]
- Buonsenso, D.; Munblit, D.; De Rose, C.; Sinatti, D.; Ricchiuto, A.; Carfi, A.; Valentini, P. Preliminary evidence of the efficacy of BNT162b2 vaccine on long COVID symptoms in children. Pediatr. Infect. Dis. J. 2023, 42, e22–e28. [Google Scholar]
- Fedak, K.M.; Bernal, A.; Capshaw, Z.A.; Gross, S. Applying the Bradford Hill criteria in the 21st century: How data integration has changed causal inference in molecular epidemiology. Emerg. Themes Epidemiol. 2015, 12, 14. [Google Scholar] [CrossRef]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Geschwind, D.H. Genetics of autism spectrum disorders. Trends Cogn. Sci. 2011, 15, 409–416. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation; Chapman and Hall/CRC: London, UK, 2015. [Google Scholar]
- Angrist, J.D.; Imbens, G.W.; Rubin, D.B. Identification of causal effects using instrumental variables. J. Am. Stat. Assoc. 1996, 91, 444–455. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide association study of severe COVID-19 with respiratory failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [PubMed]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.; et al. Relationship between the ABO blood group and COVID-19 susceptibility. Clin. Infect. Dis. 2021, 73, 328–331. [Google Scholar] [CrossRef]
- Choi, G.B.; Yim, Y.S.; Wong, H.; Kim, S.; Kim, H.; Kim, S.V.; Hoeffer, C.A.; Littman, D.R.; Huh, J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 2016, 351, 933–939. [Google Scholar] [CrossRef]
- Silverman, J.L.; Yang, M.; Lord, C.; Crawley, J.N. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 2010, 11, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Kousathanas, A.; Pairo-Castineira, E.; Rawlik, K.; Stuckey, A.; Odhams, C.A.; Walker, S.; Russell, C.D.; Malinauskas, T.; Wu, Y.; Millar, J.; et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 2022, 607, 97–103. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F. New insights into genetic susceptibility of COVID-19, an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020, 18, 216. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; COVID-19 Team; Aslibekyan, S.; et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021, 53, 801–808. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Estes, M.L.; McAllister, A.K. Maternal immune activation: Implications for neuropsychiatric disorders. Science 2016, 353, 772–777. [Google Scholar] [CrossRef]
- Fernández de Cossío, L.; Guzmán, A.; van der Veldt, S.; Luheshi, G.N. Prenatal infection leads to ASD-like behavior and altered synaptic pruning in the mouse offspring. Brain Behav. Immun. 2017, 63, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Devlin, B.; Roeder, K. Genomic control for association studies. Biometrics 1999, 55, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef]
- Sanderson, E.; Davey Smith, G.; Windmeijer, F.; Bowden, J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int. J. Epidemiol. 2019, 48, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Yarmolinsky, J.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Higgins, J.P.T.; Timpson, N.J.; Dimou, N.; et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA 2021, 326, 1614–1621. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Richmond, R.C.; Davey Smith, G. Mendelian randomization: Concepts and scope. Cold Spring Harb. Perspect. Med. 2022, 12, a040501. [Google Scholar] [CrossRef]
- Gkatzionis, A.; Burgess, S. Contextualizing selection bias in Mendelian randomization: How bad is it likely to be? Int. J. Epidemiol. 2019, 48, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Munafò, M.R.; Davey Smith, G. Robust research needs many lines of evidence. Nature 2018, 553, 399–401. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A. Why most published research findings are false. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed]
- Pashler, H.; Wagenmakers, E.J. Editors’ introduction to the special section on replicability in psychological science: A crisis of confidence? Perspect. Psychol. Sci. 2012, 7, 528–530. [Google Scholar] [CrossRef]
- Open Science Collaboration. Estimating the reproducibility of psychological science. Science 2015, 349, aac4716. [Google Scholar] [CrossRef]
- Nosek, B.A.; Alter, G.; Banks, G.C.; Borsboom, D.; Bowman, S.D.; Breckler, S.J.; Buck, S.; Chambers, C.D.; Chin, G.; Christensen, G.; et al. Promoting an open research culture. Science 2015, 348, 1422–1425. [Google Scholar] [CrossRef]
- Miguel, E.; Camerer, C.; Casey, K.; Cohen, J.; Esterling, K.M.; Gerber, A.; Glennerster, R.; Green, D.P.; Humphreys, M.; Imbens, G.; et al. Promoting transparency in social science research. Science 2014, 343, 30–31. [Google Scholar] [CrossRef] [PubMed]
- Simmons, J.P.; Nelson, L.D.; Simonsohn, U. False-positive psychology: Undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychol. Sci. 2011, 22, 1359–1366. [Google Scholar] [CrossRef]
- Button, K.S.; Ioannidis, J.P.A.; Mokrysz, C.; Nosek, B.A.; Flint, J.; Robinson, E.S.J.; Munafò, M.R. Power failure: Why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013, 14, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences, 2nd ed.; Lawrence Erlbaum Associates: Hillsdale, NJ, USA, 1988. [Google Scholar]
- Lakens, D. Calculating and reporting effect sizes to facilitate cumulative science: A practical primer for t-tests and ANOVAs. Front. Psychol. 2013, 4, 863. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.-S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef]
- Schafer, D.P.; Lehrman, E.K.; Kautzman, A.G.; Koyama, R.; Mardinly, A.R.; Yamasaki, R.; Ransohoff, R.M.; Greenberg, M.E.; Barres, B.A.; Stevens, B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012, 74, 691–705. [Google Scholar] [CrossRef]
- Kazdoba, T.M.; Leach, P.T.; Silverman, J.L.; Crawley, J.N. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis. Res. 2014, 3, 118–133. [Google Scholar] [CrossRef]
- Baker, M.G.; Wilson, N.; Anglemyer, A. Successful elimination of COVID-19 transmission in New Zealand. N. Engl. J. Med. 2020, 383, e56. [Google Scholar] [CrossRef]
- Summers, J.; Cheng, H.-Y.; Lin, H.-H.; Barnard, L.T.; Kvalsvig, A.; Wilson, N.; Baker, M.G. Potential lessons from the Taiwan and New Zealand health responses to the COVID-19 pandemic. Lancet Reg. Health West. Pac. 2020, 4, 100044. [Google Scholar] [CrossRef] [PubMed]
- Woodworth, K.R.; Olsen, E.O.; Neelam, V.; Lewis, E.L.; Galang, R.R.; Oduyebo, T.; Aveni, K.; Yazdy, M.M.; Harvey, E.; Longcore, N.D.; et al. COVID-19 Pregnancy and Infant Linked Outcomes Team (PILOT). Birth and infant outcomes following laboratory-confirmed SARS-CoV-2 infection in pregnancy—SET-NET, 16 jurisdictions, March 29–October 14, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1635–1640. [Google Scholar] [CrossRef]
- Spann, M.N.; Monk, C.; Scheinost, D.; Peterson, B.S. Maternal immune activation during the third trimester is associated with neonatal functional connectivity of the salience network and fetal to toddler behavior. J. Neurosci. 2018, 38, 2877–2886. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Rutter, M.; DiLavore, P.C.; Risi, S.; Gotham, K.; Bishop, S.L. Autism Diagnostic Observation Schedule, Second Edition (ADOS-2) Manual (Part I): Modules 1–4; Western Psychological Services: Torrance, CA, USA, 2012. [Google Scholar]
- Kalkbrenner, A.E.; Windham, G.C.; Serre, M.L.; Akita, Y.; Wang, X.; Hoffman, K.; Thayer, B.P.; Daniels, J.L. Particulate matter exposure, prenatal and postnatal windows of susceptibility, and autism spectrum disorders. Epidemiology 2015, 26, 30–42. [Google Scholar] [CrossRef]
- Grandjean, P.; Landrigan, P.J. Neurobehavioural effects of developmental toxicity. Lancet Neurol. 2014, 13, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.D.; Ebisu, K. Changes in U.S. air pollution during the COVID-19 pandemic. Sci. Total Environ. 2020, 739, 139864. [Google Scholar] [CrossRef]
- Volk, H.E.; Lurmann, F.; Penfold, B.; Hertz-Picciotto, I.; McConnell, R. Traffic-related air pollution, particulate matter, and autism. JAMA Psychiatry 2013, 70, 71–77. [Google Scholar] [CrossRef]
- Becerra, T.A.; Wilhelm, M.; Olsen, J.; Cockburn, M.; Ritz, B. Ambient air pollution and autism spectrum disorders in children. Environ. Health Perspect. 2013, 121, 380–386. [Google Scholar] [CrossRef]
- Entringer, S.; Buss, C.; Wadhwa, P.D. Prenatal stress, development, health and disease risk: A psychobiological perspective. Psychoneuroendocrinology 2015, 62, 366–375. [Google Scholar] [CrossRef]
- Christian, L.M. Psychoneuroimmunology in pregnancy: Immune pathways linking stress with maternal health, adverse birth outcomes, and fetal development. Neurosci. Biobehav. Rev. 2012, 36, 350–361. [Google Scholar] [CrossRef]
- D’Anna-Hernandez, K.L.; Ross, R.G.; Natvig, C.L.; Laudenslager, M.L. Hair cortisol levels as a retrospective marker of hypothalamic-pituitary axis activity throughout pregnancy. Psychoneuroendocrinology 2011, 36, 1312–1318. [Google Scholar]
- Mann, J.R.; McDermott, S.; Bao, H.; Hardin, J.; Gregg, A. Pre-eclampsia, birth weight, and autism spectrum disorders. J. Autism Dev. Disord. 2010, 40, 548–554. [Google Scholar] [CrossRef]
- Downing, C.; Johnson, T.E.; Larson, C.; Leakey, T.I.; Siegfried, R.N.; Rafferty, T.M.; Cooney, C.A. Subtle decreases in DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure: Effects of a methyl-supplemented diet. Alcohol 2011, 45, 681–688. [Google Scholar] [CrossRef]
- Monk, C.; Georgieff, M.K.; Osterholm, E.A. Research review: Maternal prenatal distress and poor nutrition—Mutually influencing risk factors affecting infant neurocognitive development. J. Child Psychol. Psychiatry 2013, 54, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19, a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Li, J.; Garbett, K.; Mirnics, K.; Patterson, P.H. Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci. 2007, 27, 10695–10702. [Google Scholar] [CrossRef]
- Wu, W.L.; Hsiao, E.Y.; Yan, Z.; Mazmanian, S.K.; Patterson, P.H. The placental interleukin-6 signaling controls fetal brain development and microglial development. J. Neuroinflamm. 2017, 14, 9. [Google Scholar]
- Sweeney, M.D.; Sagare, A.P.; Zlokovic, B.V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018, 14, 133–150. [Google Scholar] [CrossRef]
- Varatharaj, A.; Thomas, N.; Ellul, M.A.; Davies, N.W.S.; Pollak, T.A.; Tenorio, E.L.; Sultan, M.; Easton, A.; Breen, G.; Zandi, M.; et al. Neurological and neuropsychiatric complications of COVID-19 in 153 patients: A UK-wide surveillance study. Lancet Psychiatry 2020, 7, 875–882. [Google Scholar] [CrossRef]
- Kanberg, N.; Ashton, N.J.; Andersson, L.-M.; Yilmaz, A.; Lindh, M.; Nilsson, S.; Price, R.W.; Blennow, K.; Zetterberg, H.; Gisslén, M. Neurochemical evidence of astrocytic and neuronal injury commonly found in COVID-19. Neurology 2020, 95, e1754–e1759. [Google Scholar] [CrossRef]
- Ellul, M.A.; Benjamin, L.; Singh, B.; Lant, S.; Michael, B.D.; Easton, A.; Kneen, R.; Defres, S.; Sejvar, J.; Solomon, T. Neurological associations of COVID-19. Lancet Neurol. 2020, 19, 767–783. [Google Scholar] [CrossRef]
- Paterson, R.W.; Brown, R.L.; Benjamin, L.; Nortley, R.; Wiethoff, S.; Bharucha, T.; Jayaseelan, D.L.; Kumar, G.; Raftopoulos, R.E.; Zambreanu, L.; et al. Autism: Maternally derived antibodies specific for fetal brain proteins. Neurotoxicology 2008, 29, 226–231. [Google Scholar]
- Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Pessah, I.; Van de Water, J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav. Immun. 2011, 25, 40–45. [Google Scholar] [CrossRef]
- Masi, A.; Quintana, D.S.; Glozier, N.; Lloyd, A.R.; Hickie, I.B.; Guastella, A.J. Cytokine aberrations in autism spectrum disorder: A systematic review and meta-analysis. Mol. Psychiatry 2015, 20, 440–446. [Google Scholar] [CrossRef]
- Onore, C.; Careaga, M.; Ashwood, P. The role of immune dysfunction in the pathophysiology of autism spectrum disorders. Brain Behav. Immun. 2012, 26, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Careaga, M.; Van de Water, J.; Ashwood, P. Immune dysfunction in autism: A pathway to treatment. Neurotherapeutics 2010, 7, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Chez, M.G.; Dowling, T.; Patel, P.B.; Khanna, P.; Kominsky, M. Elevation of tumor necrosis factor-alpha in cerebrospinal fluid of autistic children. Pediatr. Neurol. 2007, 36, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Vargas, D.L.; Nascimbene, C.; Krishnan, C.; Zimmerman, A.W.; Pardo, C.A. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol. 2005, 57, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Singer, H.S.; Morris, C.M.; Williams, P.N.; Yoon, D.Y.; Hong, J.J.; Zimmerman, A.W. Antibodies against fetal brain in sera of mothers with autistic children. J. Neuroimmunol. 2006, 181, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Braunschweig, D.; Ashwood, P.; Krakowiak, P.; Hertz-Picciotto, I.; Hansen, R.; Croen, L.A.; Pessah, I.N.; Van de Water, J. Autism: Maternally derived antibodies specific for fetal brain proteins. Neurotoxicology 2008, 29, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Croen, L.A.; Braunschweig, D.; Haapanen, L.; Yoshida, C.K.; Fireman, B.; Grether, J.K.; Kharrazi, M.; Hansen, R.L.; Ashwood, P.; Van de Water, J. Maternal mid-pregnancy autoantibodies to fetal brain protein: The early markers for autism study. Biol. Psychiatry 2008, 64, 583–588. [Google Scholar] [CrossRef]
- Brimberg, L.; Mader, S.; Jeganathan, V.; Berlin, R.; Coleman, T.R.; Gregersen, P.K.; Huerta, P.T.; Volpe, B.T.; Diamond, B. Caspr2-reactive antibody cloned from a mother of an ASD child mediates an ASD-like phenotype in mice. Mol. Psychiatry 2016, 21, 1663–1671. [Google Scholar] [CrossRef]
- Lord, C.; Risi, S.; Lambrecht, L.; Cook, E.H.; Leventhal, B.L.; DiLavore, P.C.; Pickles, A.; Rutter, M. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000, 30, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Rutter, M.; Le Couteur, A. Autism Diagnostic Interview-Revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994, 24, 659–685. [Google Scholar] [CrossRef]
- Dawson, G.; Campbell, K.; Hashemi, J.; Lippmann, S.J.; Smith, V.; Carpenter, K.; Egger, H.; Espinosa, S.; Vermeer, S.; Baker, J.; et al. Atypical postural control can be detected via computer vision analysis in toddlers with autism spectrum disorder. Sci. Rep. 2018, 8, 17008. [Google Scholar] [CrossRef]
- Kessler, D.; Angstadt, M.; Sripada, C.S. Growth charting of brain connectivity networks and the identification of attention impairment in youth. JAMA Psychiatry 2016, 73, 481–489. [Google Scholar] [CrossRef]
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front. Neuroinform 2019, 13, 70. [Google Scholar] [CrossRef]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. Neuroimage Clin. 2018, 17, 16–23. [Google Scholar] [CrossRef]
- Levy, S.; Duda, M.; Haber, N.; Wall, D.P. Sparsifying machine learning models identify stable subsets of predictive features for behavioral detection of autism. Mol. Autism 2017, 8, 65. [Google Scholar] [CrossRef]
- Kong, X.; Gong, S.; Su, L.; Howard, N.; Kong, Y. Automatic detection of acromegaly from facial photographs using machine learning methods. EBioMedicine 2018, 27, 94–102. [Google Scholar] [CrossRef]
- Camm-Crosbie, L.; Bradley, L.; Shaw, R.; Baron-Cohen, S.; Cassidy, S. ‘People like me don’t get support’: Autistic adults’ experiences of support and treatment for mental health difficulties, self-injury and suicidality. Autism 2019, 23, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.M. Neurodiversity, quality of life, and autistic adults: Shifting research and professional focuses onto real-life challenges. Disabil. Stud. Q. 2010, 30. [Google Scholar] [CrossRef]
- Vivanti, G. Ask the editor: What is the most appropriate way to talk about individuals with a diagnosis of autism? J. Autism Dev. Disord. 2020, 50, 691–693. [Google Scholar] [CrossRef]
- Washington, P.; Leblanc, E.; Dunlap, K.; Penev, Y.; Kline, A.; Paskov, K.; Sun, M.W.; Chrisman, B.; Stockham, N.; Varma, M.; et al. Precision telemedicine through crowdsourced machine learning: Testing variability of crowd workers for video-based autism feature recognition. J. Pers. Med. 2020, 10, 86. [Google Scholar] [CrossRef]
- Bone, D.; Bishop, S.L.; Black, M.P.; Goodwin, M.S.; Lord, C.; Narayanan, S.S. Use of machine learning to improve autism screening and diagnostic instruments: Effectiveness, efficiency, and multi-instrument fusion. J. Child Psychol. Psychiatry 2016, 57, 927–937. [Google Scholar] [CrossRef]
- Abbas, H.; Garberson, F.; Glover, E.; Wall, D.P. Machine learning for early detection of autism (and other conditions) using a parental questionnaire and home video screening. J. Autism Dev. Disord. 2018, 48, 3431–3441. [Google Scholar]
- Duda, M.; Ma, R.; Haber, N.; Wall, D.P. Use of machine learning for behavioral distinction of autism and ADHD. Transl. Psychiatry 2016, 6, e732. [Google Scholar] [CrossRef]
- Fusaroli, R.; Lambrechts, A.; Bang, D.; Bowler, D.M.; Gaigg, S.B. “Is voice a marker for autism spectrum disorder? A systematic review and meta-analysis”. J. Autism Dev. Disord. 2017, 47, 2962–2982. [Google Scholar] [CrossRef]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659–667. [Google Scholar] [CrossRef]
- Abraham, A.; Milham, M.P.; Di Martino, A.; Craddock, R.C.; Samaras, D.; Thirion, B.; Varoquaux, G. Deriving reproducible biomarkers from multi-site resting-state data: An Autism Spectrum Disorder example. Neuroimage 2017, 147, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.A.; Zielinski, B.A.; Fletcher, P.T.; Alexander, A.L.; Lange, N.; Bigler, E.D.; Lainhart, J.E.; Anderson, J.S. Multisite functional connectivity MRI classification of autism: ABIDE results. Front. Hum. Neurosci. 2013, 7, 599. [Google Scholar] [CrossRef] [PubMed]
- Rasool, A.; Aslam, S.; Xu, Y.; Wang, Y.; Pan, Y.; Chen, W. Deep neurocomputational fusion for ASD diagnosis using multi-domain EEG analysis. Neurocomputing 2025, 641, 130353. [Google Scholar] [CrossRef]
- Kam, T.E.; Suk, H.I.; Lee, S.W. Multiple functional networks modeling for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2017, 38, 5804–5821. [Google Scholar] [CrossRef]
- Campbell, K.; Carpenter, K.L.; Hashemi, J.; Espinosa, S.; Marsan, S.; Borg, J.S.; Chang, Z.; Qiu, Q.; Vermeer, S.; Adler, E.; et al. Computer vision analysis captures atypical attention in toddlers with autism. Autism 2019, 23, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Wall, D.P.; Dally, R.; Luyster, R.; Jung, J.Y.; DeLuca, T.F. Use of artificial intelligence to shorten the behavioral diagnosis of autism. PLoS ONE 2012, 7, e43855. [Google Scholar] [CrossRef]
- Kenny, L.; Hattersley, C.; Molins, B.; Buckley, C.; Povey, C.; Pellicano, E. Which terms should be used to describe autism? Perspectives from the UK autism community. Autism 2016, 20, 442–462. [Google Scholar] [CrossRef]
- Kapp, S.K.; Gillespie-Lynch, K.; Sherman, L.E.; Hutman, T. Deficit, difference, or both? Autism and neurodiversity. Dev. Psychol. 2013, 49, 59–71. [Google Scholar] [CrossRef]
- Singer, J. Neurodiversity: The Birth of an Idea; Neurodiversity Press: Lexington, KY, USA, 2016. [Google Scholar]
- Silberman, S. NeuroTribes: The Legacy of Autism and the Future of Neurodiversity; Avery: New York, NY, USA, 2015. [Google Scholar]
- Pellicano, E.; Stears, M. Bridging autism, science and society: Moving toward an ethically informed approach to autism research. Autism Res. 2011, 4, 271–282. [Google Scholar] [CrossRef]
- Fletcher-Watson, S.; Adams, J.; Brook, K.; Charman, T.; Crane, L.; Cusack, J.; Leekam, S.; Milton, D.; Parr, J.R.; Pellicano, E. Making the future together: Shaping autism research through meaningful participation. Autism 2019, 23, 943–953. [Google Scholar] [CrossRef]
- Nicolaidis, C.; Raymaker, D.; McDonald, K.; Dern, S.; Ashkenazy, E.; Boisclair, C.; Robertson, S.; Baggs, A. Collaboration strategies in nontraditional community-based participatory research partnerships: Lessons from an academic-community partnership with autistic self-advocates. Prog. Community Health Partnersh. 2011, 5, 143–150. [Google Scholar] [CrossRef]
- Crane, L.; Pellicano, E.; Remington, A. Editorial: Toward a more inclusive approach to autism research. Autism 2019, 23, 841–843. [Google Scholar]
- Milton, D.E.M. On the ontological status of autism: The ‘double empathy problem’. Disabil. Soc. 2012, 27, 883–887. [Google Scholar] [CrossRef]
- Chown, N.; Robinson, J.; Beardon, L.; Downing, J. Improving research about us, with us: A draft framework for inclusive autism research. Disabil. Soc. 2017, 32, 720–734. [Google Scholar] [CrossRef]
- Charlton, J.I. Autistic Self Advocacy Network. In Nothing About Us, Without Us: Disability Oppression and Empowerment; University of California Press: Berkeley, CA, USA, 2001. [Google Scholar]
- Raymaker, D.M.; McDonald, K.E.; Ashkenazy, E.; Gerrity, M.; Baggs, A.M.; Kripke, C.; Hourston, S.; Nicolaidis, C. Barriers to healthcare: Instrument development and comparison between autistic adults and adults with and without other disabilities. Autism 2017, 21, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Hernán, M.A. Methods of public health research--strengthening causal inference from observational data. N. Engl. J. Med. 2021, 385, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Robins, J.M.; Hernán, M.A.; Brumback, B. Marginal structural models and causal inference in epidemiology. Epidemiology 2000, 11, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Höfler, M. The Bradford Hill considerations on causality: A counterfactual perspective. Emerg. Themes Epidemiol. 2005, 2, 11. [Google Scholar] [CrossRef]
- Victora, C.G.; Habicht, J.P.; Bryce, J. Evidence-based public health: Moving beyond randomized trials. Am. J. Public Health 2004, 94, 400–405. [Google Scholar] [CrossRef]
- Rothman, K.J. Causes. Am. J. Epidemiol. 1976, 104, 587–592. [Google Scholar] [CrossRef]
- VanderWeele, T.J. Principles of confounder selection. Eur. J. Epidemiol. 2019, 34, 211–219. [Google Scholar] [CrossRef]
- Glymour, M.M.; Weuve, J.; Berkman, L.F.; Kawachi, I.; Robins, J.M. When is baseline adjustment useful in analyses of change? An example with education and cognitive change. Am. J. Epidemiol. 2005, 162, 267–278. [Google Scholar] [CrossRef]
- Pearce, N. Analysis of matched case-control studies. BMJ 2016, 352, i969. [Google Scholar] [CrossRef]
- Rothman, K.J.; Greenland, S. Causation and causal inference in epidemiology. Am. J. Public Health 2005, 95 (Suppl. 1), S144–S150. [Google Scholar] [CrossRef]
- Egger, M.; Davey-Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Ioannidis, J.P.; Trikalinos, T.A. The appropriateness of asymmetry tests for publication bias in meta-analyses: A large survey. CMAJ 2007, 176, 1091–1096. [Google Scholar] [CrossRef]
- Colquhoun, D. An investigation of the false discovery rate and the misinterpretation of p-values. R. Soc. Open Sci. 2014, 1, 140216. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Statistics notes: Absence of evidence is not evidence of absence. BMJ 1995, 311, 485. [Google Scholar] [CrossRef] [PubMed]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P.; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. Lancet 2007, 370, 1453–1457. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rudroff, T. Theoretical Framework and Methodological Approach for Investigating Potential Associations Between Long COVID and Autism Spectrum Disorder Prevalence. NeuroSci 2025, 6, 80. https://doi.org/10.3390/neurosci6030080
Rudroff T. Theoretical Framework and Methodological Approach for Investigating Potential Associations Between Long COVID and Autism Spectrum Disorder Prevalence. NeuroSci. 2025; 6(3):80. https://doi.org/10.3390/neurosci6030080
Chicago/Turabian StyleRudroff, Thorsten. 2025. "Theoretical Framework and Methodological Approach for Investigating Potential Associations Between Long COVID and Autism Spectrum Disorder Prevalence" NeuroSci 6, no. 3: 80. https://doi.org/10.3390/neurosci6030080
APA StyleRudroff, T. (2025). Theoretical Framework and Methodological Approach for Investigating Potential Associations Between Long COVID and Autism Spectrum Disorder Prevalence. NeuroSci, 6(3), 80. https://doi.org/10.3390/neurosci6030080






