MiRNA-Mediated Regulation of S100B: A Review
Abstract
1. Introduction
1.1. The S100β Protein

1.2. S100β Is a Promising Therapeutic Target
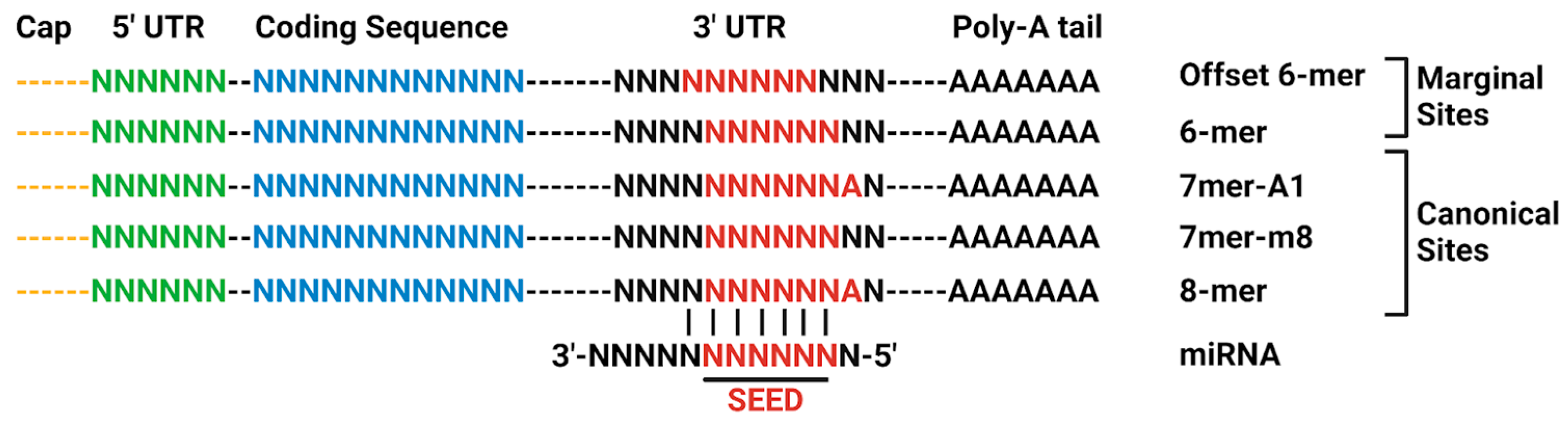
1.3. MicroRNA Regulation of Protein Expression
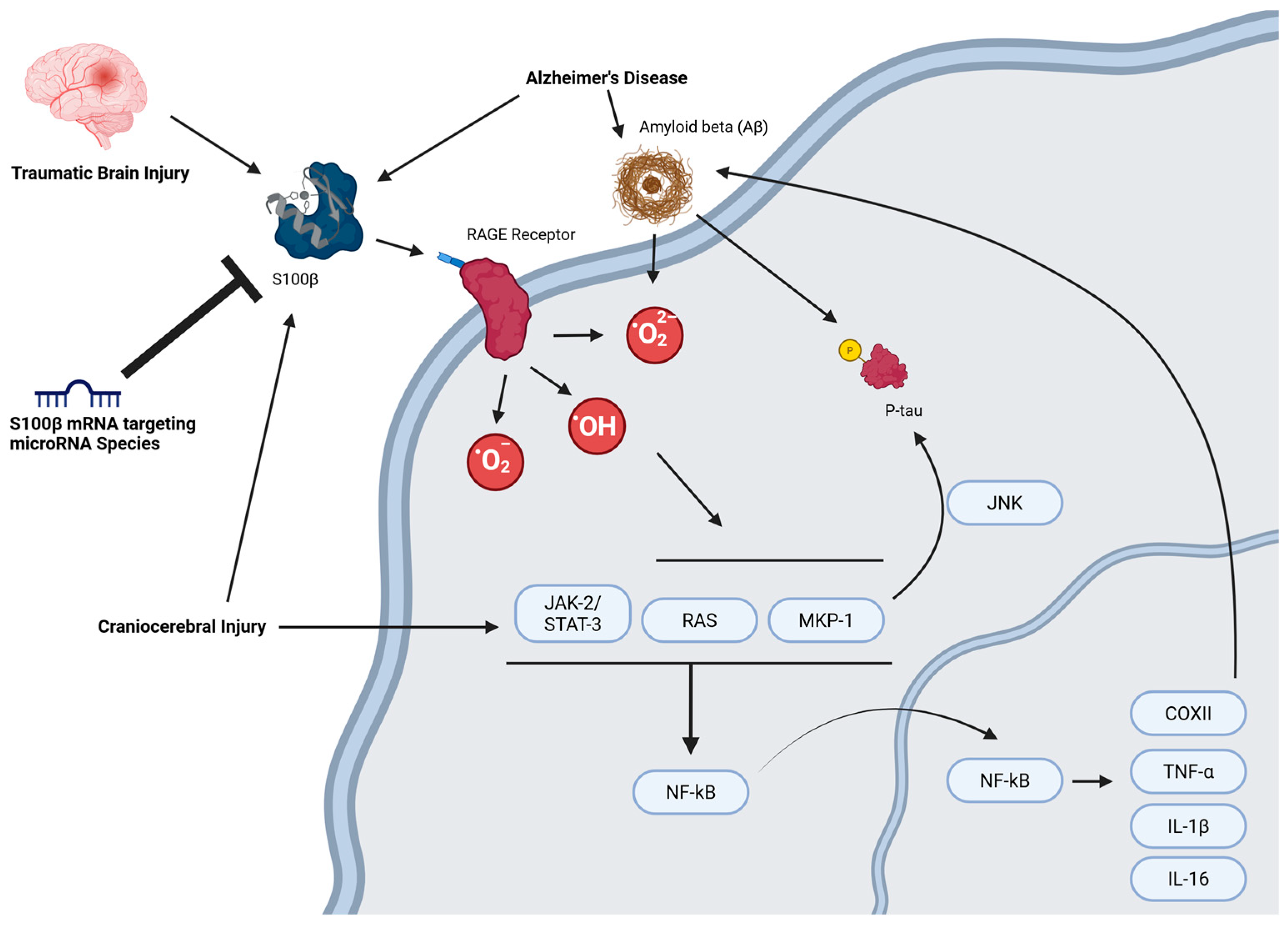
2. The miRNA-S100β Axis in Neurological Disease
MiRNAs That Directly Target S100β mRNA
3. Discussion
4. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5HT | serotonin |
| AD | Alzheimer’s disease |
| AGO2 | argonaute-2 |
| ALS | amyotrophic lateral sclerosis |
| Aβ | Amyloid-β peptide |
| Bcl-2 | B-cell lymphoma 2 |
| bFGF | basic fibroblast growth factor |
| BM | brain metastases |
| Ca | Calcium |
| CapZ | CapZ: capping protein Z; |
| CI | craniocerebral injury |
| circRNA | circular RNA |
| CoxII | cytochrome c oxidase subunit II |
| DKK-1 | Dickopff-1 |
| EpRE | electrophile responsive elements |
| ERK1 | mitogen-activated protein kinase 3 |
| ERK2 | mitogen-activated protein kinase 1 |
| FGFR1 | basic fibroblast growth factor receptor 1 |
| GFAP | glial fibrillary acidic proteins |
| GSK3β | glycogen synthase kinase 3β |
| Hdm | human homolog of murine double minute 2 |
| HIE | hypoxic–ischemic encephalopathy |
| HSP90 | heat shock protein 90 |
| IF | intermediate filament |
| IL-1β | interleukin-1β |
| IL-13 | interleukin-13 |
| IL-4 | interleukin-4 |
| IFN-y | interferon gamma |
| iNOS | inducible nitric oxide synthase |
| IS | ischemic shock |
| JAK2 | Janus kinase 2 |
| JNK | c-Jun N-terminal kinase |
| lncRNA | long noncoding RNA |
| MCAO | middle cerebral artery occlusion |
| MEK | mitogen-activated protein kinase kinases |
| miRNAs, miR- | microRNAs |
| MN | motor neuron |
| mSOD1 | mutant superoxide dismutase 1 |
| NDR | nuclear dbf2-related |
| NF-κB | nuclear factor κB |
| NO | nitric oxide |
| NRF2 | nuclear factor erythroid 2 related factor 2 |
| NSCLC | non-small-cell lung cancer |
| NSE | neuron-specific enolase |
| OGD/R | oxygen and glucose deprivation/reoxygenation |
| p53 | tumor protein p53 |
| PD | Parkinson’s disease |
| PKCα | protein kinase Cα |
| POLII | RNA polymerase II |
| pri-miRNA | primary miRNA |
| p-tau | hyperphosphorylated tau |
| RAGE/AGER | receptor for advanced glycation end product |
| RISC | RNA-induced silencing complex; |
| ROS | reactive oxygen species |
| ROS-GC1 | rod outer segment membrane guanylate cyclase type 1 |
| RSK1 | ribosomal protein S6 kinase α-1 |
| S100β | S100 calcium-binding protein B |
| SOD1 | superoxide dismutase 1 |
| STAT3 | signal transducer and activator of transcription 3 |
| tau | microtubule-associated protein tau |
| TBI | traumatic brain injury |
| TNF-α | tumor necrosis factor α |
| TRAX | translin associated factor X |
| UCH-L1 | ubiquitin carboxy-terminal hydrolase L1 |
| VEGF | vascular endothelial growth factor |
| VS | vinyl sulfone |
| XIST | X-inactive specific transcript |
| Zn | Zinc |
Appendix A
| “High-Quality” Predictions | ||||
| miRNA | Position in the UTR | Seed Match | Context++ Score Percentile | Weighted Context++ Score |
| hsa-miR-330-3p | 127–133 | 7mer-m8 | 97 | −0.22 |
| hsa-miR-1287-5p | 394–401 | 8mer | 99 | −0.90 |
| hsa-miR-6762-5p | 223–230 | 8mer | 99 | −0.65 |
| hsa-miR-4705 | 133–140 | 8mer | 99 | −0.64 |
| hsa-miR-6845-5p | 223–230 | 8mer | 99 | −0.64 |
| hsa-miR-6876-3p | 54–61 | 8mer | 99 | −0.63 |
| hsa-miR-626 | 54–61 | 8mer | 99 | −0.60 |
| hsa-miR-548m | 372–379 | 8mer | 99 | −0.58 |
| hsa-miR-6736-3p | 80–87 | 8mer | 99 | −0.57 |
| hsa-miR-6797-3p | 39–46 | 8mer | 99 | −0.56 |
| hsa-miR-6076 | 39–46 | 8mer | 99 | −0.56 |
| hsa-miR-1182 | 308–315 | 8mer | 99 | −0.56 |
| hsa-miR-769-5p | 261–268 | 8mer | 99 | −0.55 |
| hsa-miR-6791-5p | 227–234 | 8mer | 99 | −0.52 |
| hsa-miR-20b-3p | 333–340 | 8mer | 99 | −0.52 |
| hsa-miR-3160-5p | 47–54 | 8mer | 99 | −0.51 |
| hsa-miR-4292 | 227–234 | 8mer | 99 | −0.51 |
| hsa-miR-1470 | 319–326 | 8mer | 99 | −0.48 |
| hsa-miR-6823-5p | 326–333 | 8mer | 99 | −0.46 |
| hsa-miR-3978 | 155–162 | 8mer | 99 | −0.45 |
| hsa-miR-3941 | 209–216 | 8mer | 99 | −0.37 |
| hsa-miR-7856-5p | 357–364 | 8mer | 99 | −0.28 |
| hsa-miR-196b-5p | 370–376 | 7mer-m8 | 98 | −0.46 |
| hsa-miR-196a-5p | 370–376 | 7mer-m8 | 98 | −0.46 |
| hsa-miR-3190-3p | 365–371 | 7mer-m8 | 98 | −0.44 |
| hsa-miR-4288 | 52–58 | 7mer-m8 | 98 | −0.38 |
| hsa-miR-4650-5p | 203–209 | 7mer-m8 | 98 | −0.38 |
| hsa-miR-1261 | 392–398 | 7mer-m8 | 98 | −0.37 |
| hsa-miR-632 | 52–58 | 7mer-m8 | 98 | −0.30 |
| hsa-miR-8485 | 208–215 | 8mer | 98 | −0.30 |
| hsa-miR-4464 | 15–21 | 7mer-m8 | 98 | −0.29 |
| hsa-miR-4329 | 263–269 | 7mer-m8 | 98 | −0.29 |
| hsa-miR-3135b | 395–401 | 7mer-1A | 98 | −0.29 |
| hsa-miR-6730-5p | 18–24 | 7mer-m8 | 98 | −0.28 |
| hsa-miR-583 | 152–158 | 7mer-m8 | 98 | −0.28 |
| hsa-miR-6849-5p | 367–373 | 7mer-1A | 98 | −0.28 |
| hsa-miR-4748 | 15–21 | 7mer-m8 | 98 | −0.25 |
| hsa-miR-7162-5p | 114–120 | 7mer-m8 | 97 | −0.36 |
| hsa-miR-6811-5p | 177–183 | 7mer-m8 | 97 | −0.36 |
| hsa-miR-593-3p | 31–37 | 7mer-m8 | 97 | −0.34 |
| hsa-miR-1178-5p | 309–315 | 7mer-1A | 97 | −0.34 |
| hsa-miR-516a-3p | 114–120 | 7mer-m8 | 97 | −0.33 |
| hsa-miR-516b-3p | 114–120 | 7mer-m8 | 97 | −0.33 |
| hsa-miR-3681-5p | 367–373 | 7mer-1A | 97 | −0.31 |
| hsa-miR-4430 | 92–98 | 7mer-m8 | 97 | −0.28 |
| hsa-miR-345-5p | 256–262 | 7mer-m8 | 97 | −0.23 |
| “Low-Quality” Predicitions | ||||
| miRNA | Position in the UTR | seed match | context++ score percentile | weighted context++ score |
| hsa-miR-3671 | 403–410 | 8mer | 99 | 0.00 |
| hsa-miR-3133 | 420–427 | 8mer | 99 | 0.00 |
| hsa-miR-3928-3p | 433–440 | 8mer | 99 | 0.00 |
| hsa-miR-1255b-5p | 437–444 | 8mer | 99 | 0.00 |
| hsa-miR-1255a | 437–444 | 8mer | 99 | 0.00 |
| hsa-miR-1247-3p | 505–512 | 8mer | 99 | 0.00 |
| hsa-miR-6765-3p | 525–532 | 8mer | 99 | 0.00 |
| hsa-miR-4534 | 568–575 | 8mer | 99 | 0.00 |
| hsa-miR-8082 | 568–575 | 8mer | 99 | 0.00 |
| hsa-miR-3612 | 658–665 | 8mer | 99 | 0.00 |
| hsa-miR-650 | 658–665 | 8mer | 99 | 0.00 |
| hsa-miR-4443 | 659–666 | 8mer | 99 | 0.00 |
| hsa-miR-371b-3p | 694–701 | 8mer | 99 | 0.00 |
| hsa-miR-607 | 404–411 | 8mer | 99 | 0.00 |
| hsa-miR-6499-3p | 412–418 | 7mer-m8 | 98 | 0.00 |
| hsa-miR-665 | 552–559 | 8mer | 98 | 0.00 |
| hsa-miR-650 | 669–676 | 8mer | 98 | 0.00 |
| hsa-miR-3612 | 669–676 | 8mer | 98 | 0.00 |
| hsa-miR-329-5p | 148–154 | 7mer-m8 | 97 | −0.21 |
| hsa-miR-4420 | 266–272 | 7mer-m8 | 97 | −0.19 |
| hsa-miR-3912-3p | 429–435 | 7mer-m8 | 97 | 0.00 |
| hsa-miR-6132 | 473–480 | 8mer | 97 | 0.00 |
| hsa-miR-6836-5p | 473–480 | 8mer | 97 | 0.00 |
| hsa-miR-6514-5p | 567–573 | 7mer-m8 | 97 | 0.00 |
| hsa-miR-3622a-5p | 667–673 | 7mer-m8 | 97 | 0.00 |
| hsa-miR-3135b | 672–679 | 8mer | 97 | 0.00 |
| hsa-miR-24-3p | 711–718 | 8mer | 97 | 0.00 |
| hsa-miR-340-3p | 33–39 | 7mer-m8 | 96 | −0.48 |
| hsa-miR-4532 | 346–352 | 7mer-m8 | 96 | −0.42 |
| hsa-miR-5587-3p | 347–353 | 7mer-m8 | 96 | −0.40 |
| hsa-miR-4640-5p | 222–228 | 7mer-m8 | 96 | −0.38 |
| hsa-miR-6511b-5p | 177–183 | 7mer-m8 | 96 | −0.31 |
| hsa-miR-1227-5p | 224–230 | 7mer-m8 | 96 | −0.31 |
| hsa-miR-3120-5p | 53–59 | 7mer-m8 | 96 | −0.27 |
| hsa-miR-3652 | 92–98 | 7mer-m8 | 96 | −0.26 |
| hsa-miR-128-2-5p | 224–230 | 7mer-1A | 96 | −0.26 |
| hsa-miR-181b-2-3p | 266–272 | 7mer-m8 | 96 | −0.25 |
| hsa-miR-181b-3p | 266–272 | 7mer-m8 | 96 | −0.25 |
| hsa-miR-187-5p | 102–108 | 7mer-m8 | 96 | −0.24 |
| hsa-miR-34c-3p | 268–274 | 7mer-1A | 96 | −0.22 |
| hsa-miR-4419b | 70–76 | 7mer-1A | 96 | −0.13 |
| hsa-miR-5011-3p | 442–448 | 7mer-m8 | 96 | 0.00 |
| hsa-miR-4772-5p | 517–524 | 8mer | 96 | 0.00 |
| hsa-miR-6827-3p | 33–39 | 7mer-m8 | 95 | −0.45 |
| hsa-miR-128-1-5p | 224–230 | 7mer-1A | 95 | −0.31 |
| hsa-miR-4786-5p | 262–268 | 7mer-1A | 95 | −0.26 |
| hsa-miR-6854-5p | 84–90 | 7mer-m8 | 95 | −0.23 |
| hsa-miR-4700-3p | 23–29 | 7mer-1A | 95 | −0.22 |
| hsa-miR-6787-3p | 82–88 | 7mer-m8 | 95 | −0.20 |
| hsa-miR-1322 | 181–187 | 7mer-m8 | 95 | −0.20 |
| hsa-miR-603 | 209–215 | 7mer-1A | 95 | −0.16 |
| hsa-miR-4652-3p | 336–342 | 7mer-1A | 95 | −0.13 |
| hsa-miR-3929 | 70–76 | 7mer-1A | 95 | −0.12 |
| hsa-miR-4478 | 70–76 | 7mer-1A | 95 | −0.12 |
| hsa-miR-4519 | 413–419 | 7mer-m8 | 95 | 0.00 |
| hsa-miR-4773 | 418–424 | 7mer-m8 | 95 | 0.00 |
| hsa-miR-4717-5p | 612–618 | 7mer-m8 | 95 | 0.00 |
| hsa-miR-4661-3p | 651–657 | 7mer-m8 | 95 | 0.00 |
| hsa-miR-125b-1-3p | 314–320 | 7mer-m8 | 94 | −0.36 |
| hsa-miR-4726-5p | 222–228 | 7mer-m8 | 94 | −0.34 |
| hsa-miR-23a-5p | 324–330 | 7mer-m8 | 94 | −0.26 |
| hsa-miR-129-2-3p | 61–67 | 7mer-m8 | 94 | −0.24 |
| hsa-miR-6852-5p | 226–232 | 7mer-m8 | 94 | −0.22 |
| hsa-miR-6768-5p | 206–212 | 7mer-m8 | 94 | −0.19 |
| hsa-miR-329-3p | 209–215 | 7mer-1A | 94 | −0.16 |
| hsa-miR-4738-5p | 540–546 | 7mer-m8 | 94 | 0.00 |
| hsa-miR-579-5p | 718–724 | 7mer-1A | 94 | 0.00 |
| hsa-miR-6881-5p | 343–349 | 7mer-m8 | 93 | −0.34 |
| hsa-miR-23b-5p | 324–330 | 7mer-m8 | 93 | −0.28 |
| hsa-miR-6829-5p | 288–294 | 7mer-m8 | 93 | −0.27 |
| hsa-miR-602 | 302–308 | 7mer-1A | 93 | −0.27 |
| hsa-miR-129-1-3p | 61–67 | 7mer-m8 | 93 | −0.23 |
| hsa-miR-6892-5p | 356–362 | 7mer-1A | 93 | −0.23 |
| hsa-miR-7151-3p | 24–30 | 7mer-1A | 93 | −0.22 |
| hsa-miR-452-5p | 243–249 | 7mer-1A | 93 | −0.20 |
| hsa-miR-362-3p | 209–215 | 7mer-1A | 93 | −0.13 |
| hsa-miR-490-5p | 440–446 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-4690-5p | 498–504 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-6874-3p | 574–580 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-149-5p | 589–595 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-3943 | 600–606 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-6809-5p | 655–661 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-4443 | 670–676 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-1225-3p | 705–711 | 7mer-m8 | 93 | 0.00 |
| hsa-miR-325-3p | 735–742 | 8mer | 93 | 0.00 |
| hsa-miR-645 | 71–77 | 7mer-m8 | 92 | −0.28 |
| hsa-miR-2114-5p | 230–236 | 7mer-m8 | 92 | −0.21 |
| hsa-miR-3529-3p | 100–106 | 7mer-1A | 92 | −0.16 |
| hsa-miR-1305 | 145–151 | 7mer-1A | 92 | −0.07 |
| hsa-miR-1305 | 406–412 | 7mer-1A | 92 | 0.00 |
| hsa-miR-186-5p | 421–427 | 7mer-1A | 92 | 0.00 |
| hsa-miR-4532 | 506–512 | 7mer-1A | 92 | 0.00 |
| hsa-miR-4663 | 535–541 | 7mer-m8 | 92 | 0.00 |
| hsa-miR-148b-5p | 574–580 | 7mer-m8 | 92 | 0.00 |
| hsa-miR-4737 | 579–585 | 7mer-1A | 92 | 0.00 |
| hsa-miR-1184 | 645–651 | 7mer-m8 | 92 | 0.00 |
| hsa-miR-671-5p | 685–691 | 7mer-m8 | 92 | 0.00 |
| hsa-miR-6803-3p | 721–727 | 7mer-m8 | 92 | 0.00 |
| hsa-miR-892c-3p | 243–249 | 7mer-1A | 91 | −0.17 |
| hsa-miR-4676-3p | 243–249 | 7mer-1A | 91 | −0.17 |
| hsa-miR-4719 | 130–136 | 7mer-1A | 91 | −0.10 |
| hsa-miR-335-3p | 138–144 | 7mer-1A | 91 | −0.06 |
| hsa-miR-155-5p | 398–404 | 7mer-m8 | 91 | 0.00 |
| hsa-miR-3915 | 434–440 | 7mer-m8 | 91 | 0.00 |
| hsa-miR-4653-3p | 566–572 | 7mer-1A | 91 | 0.00 |
| hsa-miR-6840-5p | 702–708 | 7mer-m8 | 91 | 0.00 |
| hsa-miR-4693-5p | 244–250 | 7mer-1A | 90 | −0.20 |
| hsa-miR-4308 | 228–234 | 7mer-1A | 90 | −0.17 |
| hsa-miR-5189-3p | 459–465 | 7mer-m8 | 90 | 0.00 |
| hsa-miR-3937 | 515–521 | 7mer-1A | 90 | 0.00 |
| hsa-miR-3972 | 542–548 | 7mer-m8 | 90 | 0.00 |
| hsa-miR-4502 | 571–577 | 7mer-1A | 90 | 0.00 |
| hsa-miR-5580-5p | 590–596 | 7mer-m8 | 90 | 0.00 |
| hsa-miR-371a-3p | 695–701 | 7mer-1A | 90 | 0.00 |
| hsa-miR-3943 | 704–710 | 7mer-m8 | 90 | 0.00 |
| hsa-miR-6501-3p | 708–714 | 7mer-1A | 90 | 0.00 |
| hsa-miR-4666b | 41–47 | 7mer-1A | 89 | −0.18 |
| hsa-miR-3686 | 335–341 | 7mer-1A | 89 | −0.18 |
| hsa-miR-3137 | 334–340 | 7mer-1A | 89 | −0.17 |
| hsa-miR-5011-3p | 41–47 | 7mer-1A | 89 | −0.14 |
| hsa-miR-1305 | 137–143 | 7mer-1A | 89 | −0.04 |
| hsa-miR-324-5p | 509–515 | 7mer-m8 | 89 | 0.00 |
| hsa-miR-1225-3p | 601–607 | 7mer-m8 | 89 | 0.00 |
| hsa-miR-4743-3p | 336–342 | 7mer-1A | 88 | −0.10 |
| hsa-miR-3622a-3p | 526–532 | 7mer-1A | 88 | 0.00 |
| hsa-miR-2276-3p | 66–72 | 7mer-m8 | 87 | −0.11 |
| hsa-miR-4686 | 446–452 | 7mer-1A | 87 | 0.00 |
| hsa-miR-3918 | 472–478 | 7mer-m8 | 87 | 0.00 |
| hsa-miR-3622b-3p | 526–532 | 7mer-1A | 87 | 0.00 |
| hsa-miR-1202 | 542–548 | 7mer-m8 | 87 | 0.00 |
| hsa-miR-4487 | 592–598 | 7mer-m8 | 87 | 0.00 |
| hsa-miR-3177-3p | 284–290 | 7mer-1A | 86 | −0.28 |
| hsa-miR-20a-3p | 444–450 | 7mer-1A | 86 | 0.00 |
| hsa-miR-3134 | 569–575 | 7mer-1A | 86 | 0.00 |
| hsa-miR-361-3p | 598–604 | 7mer-m8 | 86 | 0.00 |
| hsa-miR-7156-3p | 644–650 | 7mer-m8 | 86 | 0.00 |
| hsa-miR-346 | 52–58 | 7mer-1A | 85 | −0.22 |
| hsa-miR-5095 | 24–30 | 7mer-1A | 85 | −0.15 |
| hsa-miR-3978 | 89–95 | 7mer-m8 | 85 | −0.04 |
| hsa-miR-186-5p | 186–192 | 7mer-m8 | 85 | −0.02 |
| hsa-miR-1249-3p | 480–486 | 7mer-m8 | 85 | 0.00 |
| hsa-miR-663a | 292–298 | 7mer-1A | 84 | −0.30 |
| hsa-miR-642a-5p | 321–327 | 7mer-1A | 84 | −0.10 |
| hsa-miR-1287-5p | 673–679 | 7mer-1A | 84 | 0.00 |
| hsa-miR-887-3p | 732–738 | 7mer-1A | 84 | 0.00 |
| hsa-miR-4477b | 357–363 | 7mer-1A | 83 | −0.14 |
| hsa-miR-374a-3p | 270–276 | 7mer-1A | 83 | −0.10 |
| hsa-miR-466 | 210–216 | 7mer-1A | 83 | −0.09 |
| hsa-miR-1908-5p | 292–298 | 7mer-1A | 82 | −0.27 |
| hsa-miR-3180-5p | 555–561 | 7mer-1A | 82 | 0.00 |
| hsa-miR-4762-3p | 572–578 | 7mer-m8 | 82 | 0.00 |
| hsa-miR-33b-3p | 695–701 | 7mer-1A | 82 | 0.00 |
| hsa-miR-519e-3p | 695–701 | 7mer-1A | 82 | 0.00 |
| hsa-miR-642b-5p | 322–328 | 7mer-m8 | 81 | −0.12 |
| hsa-miR-515-3p | 695–701 | 7mer-1A | 80 | 0.00 |
| hsa-miR-6895-5p | 471–477 | 7mer-m8 | 79 | 0.00 |
| hsa-miR-6515-5p | 660–666 | 7mer-1A | 79 | 0.00 |
| hsa-miR-3618 | 305–311 | 7mer-m8 | 78 | −0.22 |
| hsa-miR-4667-3p | 320–326 | 7mer-1A | 78 | −0.12 |
| hsa-miR-3192-3p | 572–578 | 7mer-1A | 78 | 0.00 |
| hsa-miR-4672 | 210–216 | 7mer-1A | 77 | −0.08 |
| hsa-miR-578 | 45–51 | 7mer-1A | 77 | −0.06 |
| hsa-miR-3140-3p | 560–566 | 7mer-1A | 77 | 0.00 |
| hsa-miR-6787-5p | 292–298 | 7mer-1A | 75 | −0.24 |
| hsa-miR-6744-5p | 438–444 | 7mer-1A | 75 | 0.00 |
| hsa-miR-505-5p | 603–609 | 7mer-1A | 75 | 0.00 |
| hsa-miR-5692a | 401–407 | 7mer-m8 | 73 | 0.00 |
| hsa-miR-875-3p | 90–96 | 7mer-m8 | 72 | −0.07 |
| hsa-miR-6811-5p | 475–481 | 7mer-1A | 71 | 0.00 |
| hsa-miR-6125 | 687–693 | 7mer-1A | 71 | 0.00 |
| hsa-miR-6511b-5p | 475–481 | 7mer-1A | 70 | 0.00 |
| hsa-miR-6828-5p | 501–507 | 7mer-m8 | 69 | 0.00 |
| hsa-miR-383-5p.1 | 518–524 | 7mer-1A | 69 | 0.00 |
| hsa-miR-6722-3p | 474–480 | 7mer-1A | 68 | 0.00 |
| hsa-miR-4299 | 522–528 | 7mer-m8 | 68 | 0.00 |
| hsa-miR-1909-3p | 474–480 | 7mer-1A | 66 | 0.00 |
| hsa-miR-551b-5p | 121–127 | 7mer-m8 | 62 | −0.02 |
| hsa-miR-4290 | 692–698 | 7mer-1A | 58 | 0.00 |
| hsa-miR-3194-5p | 542–548 | 7mer-1A | 54 | 0.00 |
| hsa-miR-1277-5p | 197–203 | 7mer-1A | 52 | −0.02 |
| hsa-miR-4284 | 712–718 | 7mer-1A | 34 | 0.00 |
| hsa-miR-3135b | 605–611 | 7mer-m8 | 32 | 0.00 |
| hsa-miR-6841-3p | 477–483 | 7mer-m8 | 24 | 0.00 |
| hsa-miR-4786-3p | 490–496 | 7mer-m8 | 18 | 0.00 |
References
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef]
- Cancemi, P.; Di Cara, G.; Albanese, N.N.; Costantini, F.; Marabeti, M.R.; Musso, R.; Lupo, C.; Roz, E.; Pucci-Minafra, I. Large-scale proteomic identification of S100 proteins in breast cancer tissues. BMC Cancer 2010, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- Langeh, U.; Singh, S. Targeting S100B Protein as a Surrogate Biomarker and its Role in Various Neurological Disorders. Curr. Neuropharmacol. 2020, 19, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Sorci, G.; Riuzzi, F.; Arcuri, C.; Bianchi, R.; Brozzi, F.; Tubaro, C.; Giambanco, I. S100B’s double life: Intracellular regulator and extracellular signal. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2009, 1793, 1008–1022. [Google Scholar] [CrossRef]
- Leite, M.C.; Galland, F.; Guerra, M.C.; Rodrigues, L.; Taday, J.; Monteforte, P.T.; Hirata, H.; Gottfried, C.; Donato, R.; Smaili, S.; et al. Astroglial S100B Secretion Is Mediated by Ca2+ Mobilization from Endoplasmic Reticulum: A Study Using Forskolin and DMSO as Secretagogues. Int. J. Mol. Sci. 2023, 24, 16576. [Google Scholar] [CrossRef]
- Davey, G.E.; Murmann, P.; Heizmann, C.W. Intracellular Ca2+ and Zn2+ Levels Regulate the Alternative Cell Density-dependent Secretion of S100B in Human Glioblastoma Cells. J. Biol. Chem. 2001, 276, 30819–30826. [Google Scholar] [CrossRef]
- Selistre, N.G.; Rodrigues, L.; Federhen, B.C.; Gayger-Dias, V.; Taday, J.; Wartchow, K.M.; Gonçalves, C.-A. S100B Secretion in Astrocytes, Unlike C6 Glioma Cells, Is Downregulated by Lactate. Metabolites 2023, 14, 7. [Google Scholar] [CrossRef]
- de Souza, D.F.; Leite, M.C.; Quincozes-Santos, A.; Nardin, P.; Tortorelli, L.S.; Rigo, M.M.; Gottfried, C.; Leal, R.B.; Gonçalves, C.-A. S100B secretion is stimulated by IL-1β in glial cultures and hippocampal slices of rats: Likely involvement of MAPK pathway. J. Neuroimmunol. 2009, 206, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.C.; Tortorelli, L.S.; Galland, F.; Da Ré, C.; Negri, E.; Engelke, D.S.; Rodrigues, L.; Leite, M.C.; Gonçalves, C.-A. Lipopolysaccharide modulates astrocytic S100B secretion: A study in cerebrospinal fluid and astrocyte cultures from rats. J. Neuroinflamm. 2011, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, R.; Demel, G.; König, H.-G.; Gross, U.; Prehn, J.; Raabe, A.; Seifert, V.; Kögel, D. Active secretion of S100B from astrocytes during metabolic stress. Neuroscience 2006, 141, 1697–1701. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Ye, M.; Resch, J.M.; Jedrychowski, M.P.; Hu, B.; Lowell, B.B.; Ginty, D.D.; Spiegelman, B.M. Innervation of thermogenic adipose tissue via a calsyntenin 3β–S100b axis. Nature 2019, 569, 229–235. [Google Scholar] [CrossRef]
- Baudier, J.; Mochly-Rosen, D.; Newton, A.; Lee, S.H.; Koshland, D.E.; Cole, R.D. Comparison of S100b protein with calmodulin: Interactions with melittin and microtubule-associated .tau. proteins and inhibition of phosphorylation of .tau. proteins by protein kinase C. Biochemistry 1987, 26, 2886–2893. [Google Scholar] [CrossRef]
- Rothermundt, M.; Peters, M.; Prehn, J.H.; Arolt, V. S100B in brain damage and neurodegeneration. Microsc. Res. Tech. 2003, 60, 614–632. [Google Scholar] [CrossRef]
- Donaldson, C.; Barber, K.R.; Shaw, G.S.; Kay, C.M. Human S100b protein: Formation of a tetramer from synthetic calcium-binding site peptides. Protein Sci. 1995, 4, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Drohat, A.C.; Baldisseri, D.M.; Rustandi, R.R.; Weber, D.J. Solution Structure of Calcium-Bound Rat S100B(ββ) As Determined by Nuclear Magnetic Resonance Spectroscopy. Biochemistry 1998, 37, 2729–2740. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2009, 1793, 993–1007. [Google Scholar] [CrossRef] [PubMed]
- Prez, K.; Fan, L. Structural Basis for S100B Interaction with Its Target Proteins. J. Mol. Genet. Med. 2018, 12, 366. [Google Scholar] [PubMed]
- Wilder, P.T.; Lin, J.; Bair, C.L.; Charpentier, T.H.; Yang, D.; Liriano, M.; Varney, K.M.; Lee, A.; Oppenheim, A.B.; Adhya, S.; et al. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2006, 1763, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, D.B.; Weber, D.J. The Calcium-Dependent Interaction of S100B with Its Protein Targets. Cardiovasc. Psychiatry Neurol. 2010, 2010, 728052. [Google Scholar] [CrossRef]
- Hagmeyer, S.; Cristóvão, J.S.; Mulvihill, J.J.E.; Boeckers, T.M.; Gomes, C.M.; Grabrucker, A.M. Zinc Binding to S100B Affords Regulation of Trace Metal Homeostasis and Excitotoxicity in the Brain. Front. Mol. Neurosci. 2018, 10, 456. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting mod-ern challenges in visualization and analysis. Protein Sci. 2017, 27, 14–25. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2022, 51, D933–D941. [Google Scholar] [CrossRef] [PubMed]
- Sharova, L.V.; Sharov, A.A.; Nedorezov, T.; Piao, Y.; Shaik, N.; Ko, M.S. Database for mRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells. DNA Res. 2009, 16, 45–58. [Google Scholar] [CrossRef]
- Leclerc, E.; Sturchler, E.; Vetter, S.W. The S100B/RAGE Axis in Alzheimer’s Disease. Cardiovasc. Psychiatry Neurol. 2010, 2010, 539581. [Google Scholar] [CrossRef] [PubMed]
- Gasparotto, J.; Girardi, C.S.; Somensi, N.; Ribeiro, C.T.; Moreira, J.C.F.; Michels, M.; Sonai, B.; Rocha, M.; Steckert, A.V.; Barichello, T.; et al. Receptor for advanced glycation end products mediates sepsis-triggered amyloid-β accumulation, Tau phosphorylation, and cognitive impairment. J. Biol. Chem. 2018, 293, 226–244. [Google Scholar] [CrossRef]
- Bianchi, R.; Giambanco, I.; Donato, R. S100B/RAGE-dependent activation of microglia via NF-κB and AP-1. Neurobiol. Aging 2010, 31, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Meffert, M.K. Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006, 13, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Sun, L.; Chen, X.; Zhang, D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 2013, 8, 2003–2014. [Google Scholar] [CrossRef]
- Barger, S.; Van Eldik, L. S100 beta stimulates calcium fluxes in glial and neuronal cells. J. Biol. Chem. 1992, 267, 9689–9694. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.L.; Camozzato, A.L.; Ferreira, E.D.; Piazenski, I.; Kochhann, R.; Dall’Igna, O.; Mazzini, G.S.; Souza, D.O.; Portela, L.V. Serum levels of S100B and NSE proteins in Alzheimer’s disease patients. J. Neuroinflamm. 2010, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Cristóvão, J.S.; Morris, V.K.; Cardoso, I.; Leal, S.S.; Martínez, J.; Botelho, H.M.; Göbl, C.; David, R.; Kierdorf, K.; Alemi, M.; et al. The neuronal S100B protein is a calcium-tuned suppressor of amyloid-β aggregation. Sci. Adv. 2018, 4, eaaq1702. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Q.; Xing, D. Focal Adhesion Kinase Activates NF-κB via the ERK1/2 and p38MAPK Pathways in Amyloid-β25-35-Induced Apoptosis in PC12 Cells. J. Alzheimer’s Dis. 2012, 32, 77–94. [Google Scholar] [CrossRef]
- Selinfreund, R.H.; Barger, S.W.; Pledger, W.J.; Van Eldik, L.J. Neurotrophic protein S100 beta stimulates glial cell proliferation. Proc. Natl. Acad. Sci. USA 1991, 88, 3554–3558. [Google Scholar] [CrossRef]
- Morozzi, G.; Beccafico, S.; Bianchi, R.; Riuzzi, F.; Bellezza, I.; Giambanco, I.; Arcuri, C.; Minelli, A.; Donato, R. Oxidative stress-induced S100B accumulation converts myoblasts into brown adipocytes via an NF-κB/YY1/miR-133 axis and NF-κB/YY1/BMP-7 axis. Cell Death Differ. 2017, 24, 2077–2088. [Google Scholar] [CrossRef]
- Reali, C.; Scintu, F.; Pillai, R.; Donato, R.; Michetti, F.; Sogos, V. S100b counteracts effects of the neurotoxicant trimethyltin on astrocytes and microglia. J. Neurosci. Res. 2005, 81, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, D.S.; Lenz, G.; Karl, J.; Gonçalves, C.A.; Rodnight, R. Extracellular S100B protein modulates ERK in astrocyte cultures. NeuroReport 2000, 11, 807–809. [Google Scholar] [CrossRef]
- Guo, S.; Wang, H.; Yin, Y. Microglia Polarization From M1 to M2 in Neurodegenerative Diseases. Front. Aging Neurosci. 2022, 14, 815347. [Google Scholar] [CrossRef]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, W.; Zhang, Y.; Pan, S.; Bao, J. S100B promotes microglia M1 polarization and migration to aggravate cerebral ischemia. Inflamm. Res. 2018, 67, 937–949. [Google Scholar] [CrossRef]
- Sathe, K.; Maetzler, W.; Lang, J.D.; Mounsey, R.B.; Fleckenstein, C.; Martin, H.L.; Schulte, C.; Mustafa, S.; Synofzik, M.; Vukovic, Z.; et al. S100B is increased in Parkinson’s disease and ablation protects against MPTP-induced toxicity through the RAGE and TNF-α pathway. Brain 2012, 135, 3336–3347. [Google Scholar] [CrossRef]
- Brandt, R.; Bakota, L. Microtubule dynamics and the neurodegenerative triad of Alzheimer’s disease: The hidden connection. J. Neurochem. 2017, 143, 409–417. [Google Scholar] [CrossRef]
- Esposito, G.; Scuderi, C.; Lu, J.; Savani, C.; De Filippis, D.; Iuvone, T.; Jr, L.S.; Sheen, V.; Steardo, L. S100B induces tau protein hyperphosphorylation via Dickopff-1 up-regulation and disrupts the Wnt pathway in human neural stem cells. J. Cell. Mol. Med. 2008, 12, 914–927. [Google Scholar] [CrossRef]
- Alonso, A.D.C.; Grundke-Iqbal, I.; Barra, H.S.; Iqbal, K. Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: Sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc. Natl. Acad. Sci. USA 1997, 94, 298–303. [Google Scholar] [CrossRef]
- Rahmati, M.; Azarpazhooh, M.R.; Ehteram, H.; Ferns, G.A.; Ghayour-Mobarhan, M.; Ghannadan, H.; Mobarra, N. The elevation of S100B and downregulation of circulating miR-602 in the sera of ischemic stroke (IS) patients: The emergence of novel diagnostic and prognostic markers. Neurol. Sci. 2020, 41, 2185–2192. [Google Scholar] [CrossRef] [PubMed]
- Riuzzi, F.; Sorci, G.; Donato, R. S100B protein regulates myoblast proliferation and differentiation by activating FGFR1 in a bFGF-dependent manner. J. Cell Sci. 2011, 124, 2389–2400. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ye, M.; Zhang, G. Aberrant expression of miR-199a in newborns with hypoxic-ischemic encephalopathy and its diagnostic and prognostic significance when combined with S100B and NSE. Acta Neurol. Belg. 2021, 121, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yang, Q.; Wilder, P.T.; Carrier, F.; Weber, D.J. The Calcium-binding Protein S100B Down-regulates p53 and Apoptosis in Malignant Melanoma. J. Biol. Chem. 2010, 285, 27487–27498. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Chao, Y.; Wang, F.; Wang, Y.; Han, B. Correlation analysis of miRNA-124, miRNA-210 with brain injury and inflammatory response in patients with craniocerebral injury. Am. J. Transl. Res. 2022, 14, 285–294. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Seok, H.; Ham, J.; Jang, E.-S.; Chi, S.W. MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions. Mol. Cells 2016, 39, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Wightman, F.F.; Giono, L.E.; Fededa, J.P.; de la Mata, M. Target RNAs Strike Back on MicroRNAs. Front. Genet. 2018, 9, 435. [Google Scholar] [CrossRef] [PubMed]
- Chopra, N.; Wang, R.; Maloney, B.; Nho, K.; Beck, J.S.; Pourshafie, N.; Niculescu, A.; Saykin, A.J.; Rinaldi, C.; Counts, S.E.; et al. MicroRNA-298 reduces levels of human amyloid-β precursor protein (APP), β-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol. Psychiatry 2020, 26, 5636–5657. [Google Scholar] [CrossRef]
- Wang, R.; Chopra, N.; Nho, K.; Maloney, B.; Obukhov, A.G.; Nelson, P.T.; Counts, S.E.; Lahiri, D.K. Human microRNA (miR-20b-5p) modulates Alzheimer’s disease pathways and neuronal function, and a specific polymorphism close to the MIR20B gene influences Alzheimer’s biomarkers. Mol. Psychiatry 2022, 27, 1256–1273. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Maloney, B.; Rogers, J.T.; Lahiri, D.K. Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: Implications in Alzheimer’s disease. Mol. Psychiatry 2018, 24, 345–363. [Google Scholar] [CrossRef] [PubMed]
- Abd-Aziz, N.; Kamaruzman, N.I.; Poh, C.L. Development of MicroRNAs as Potential Therapeutics against Cancer. J. Oncol. 2020, 2020, 8029721. [Google Scholar] [CrossRef]
- Ma, C.; Yang, L.; Gao, Q.; Wang, L.; Wan, X. miR-602 Activates NRF2 Antioxidant Pathways to Protect HBMECs from OGD/R-Induced Oxidative Stress via Targeting KEAP1 and HRD1. Dis. Markers 2022, 2022, 6967573. [Google Scholar] [CrossRef]
- Buendia, I.; Michalska, P.; Navarro, E.; Gameiro, I.; Egea, J.; León, R. Nrf2–ARE pathway: An emerging target against oxidative stress and neuroinflammation in neurodegenerative diseases. Pharmacol. Ther. 2016, 157, 84–104. [Google Scholar] [CrossRef] [PubMed]
- Haines, D.D.; Lekli, I.; Teissier, P.; Bak, I.; Tosaki, A. Role of haeme oxygenase-1 in resolution of oxidative stress-related pathologies: Focus on cardiovascular, lung, neurological and kidney disorders. Acta Physiol. 2012, 204, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, L.; Qiao, Y.; Zhou, X.; Wu, G.; Wang, L.; Peng, Y.; Dong, X.; Huang, H.; Si, L.; et al. Heme Oxygenase-1 Prevents Cardiac Dysfunction in Streptozotocin-Diabetic Mice by Reducing Inflammation, Oxidative Stress, Apoptosis and Enhancing Autophagy. PLoS ONE 2013, 8, e75927. [Google Scholar] [CrossRef]
- Sun, X.; Wang, L.; Huang, X.; Zhou, S.; Jiang, T. Regulatory mechanism of the SNHG3/miR-302a-3p/E2F1 feedback loop in nerve repair post cerebral ischemic stroke. Curr. Neurovasc. Res. 2021, 18, 515–524. [Google Scholar] [CrossRef]
- Lucas, R.M.; Luo, L.; Stow, J.L. ERK1/2 in immune signalling. Biochem. Soc. Trans. 2022, 50, 1341–1352. [Google Scholar] [CrossRef]
- Perner, F.; Perner, C.; Ernst, T.; Heidel, F.H. Roles of JAK2 in Aging, Inflammation, Hematopoiesis and Malignant Transformation. Cells 2019, 8, 854. [Google Scholar] [CrossRef]
- Arora, S.; Ranade, A.R.; Tran, N.L.; Nasser, S.; Sridhar, S.; Korn, R.L.; Ross, J.T.; Dhruv, H.; Foss, K.M.; Sibenaller, Z.; et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int. J. Cancer 2011, 129, 2621–2631. [Google Scholar] [CrossRef]
- Chen, L.-T.; Xu, S.-D.; Xu, H.; Zhang, J.-F.; Ning, J.-F.; Wang, S.-F. MicroRNA-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med. Oncol. 2011, 29, 1673–1680. [Google Scholar] [CrossRef]
- Singh, R.K.; Kumar, S.; Gautam, P.K.; Tomar, M.S.; Verma, P.K.; Singh, S.P.; Acharya, A. Protein kinase C-α and the regulation of diverse cell responses. Biomol. Concepts 2017, 8, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, P.P.; Deng, X.; Carr, B.K.; May, W.S. A Functional Role for Mitochondrial Protein Kinase Cα in Bcl2 Phosphorylation and Suppression of Apoptosis. J. Biol. Chem. 1998, 273, 25436–25442. [Google Scholar] [CrossRef]
- Wang, S.; Rosengren, L.E.; Franlund, M.; Hamberger, A.; Haglid, K.G. Bcl-2 expression regulates cell sensitivity to S100β-mediated apoptosis. Mol. Brain Res. 1999, 70, 167–176. [Google Scholar] [CrossRef]
- Wu, W.; Liang, D. Expression and related mechanisms of miR-330-3p and S100B in an animal model of cartilage injury. J. Int. Med. Res. 2021, 49, 03000605211039471. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, X.; Chen, L.; Chen, W.; Zhang, Y.; Chen, J.; Wu, X.; Zhao, Y.; Wu, X.; Sun, G. The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem. Biophys. Res. Commun. 2018, 505, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Guo, Y.; Zhang, Y.; Wang, D.; Zhang, G.; Hou, J.; Yang, J. Circ_MUC16 attenuates the effects of Propofol to promote the aggressive behaviors of ovarian cancer by mediating the miR-1182/S100B signaling pathway. BMC Anesthesiol. 2021, 21, 297. [Google Scholar] [CrossRef]
- Cunha, C.; Santos, C.; Gomes, C.; Fernandes, A.; Correia, A.M.; Sebastião, A.M.; Vaz, A.R.; Brites, D. Downregulated Glia Interplay and Increased miRNA-155 as Promising Markers to Track ALS at an Early Stage. Mol. Neurobiol. 2017, 55, 4207–4224. [Google Scholar] [CrossRef]
- Gomes, C.; Cunha, C.; Nascimento, F.; Ribeiro, J.A.; Vaz, A.R.; Brites, D. Cortical Neurotoxic Astrocytes with Early ALS Pathology and miR-146a Deficit Replicate Gliosis Markers of Symptomatic SOD1G93A Mouse Model. Mol. Neurobiol. 2018, 56, 2137–2158. [Google Scholar] [CrossRef]
- Li, M.; Luan, L.; Liu, Q.; Liu, Y.; Lan, X.; Li, Z.; Liu, W. MiRNA-199a-5p Protects Against Cerebral Ischemic Injury by Down-Regulating DDR1 in Rats. World Neurosurg. 2019, 131, e486–e494. [Google Scholar] [CrossRef] [PubMed]
- TargetScanHuman 8.0. Available online: https://www.targetscan.org/vert_80/ (accessed on 21 June 2025).
- Wen, L.; Sun, J.; Chen, X.; Du, R. miR-135b-dependent downregulation of S100B promotes neural stem cell differentiation in a hypoxia/ischemia-induced cerebral palsy rat model. Am. J. Physiol. Physiol. 2020, 319, C955–C966. [Google Scholar] [CrossRef]
- Lee, J.C.; Seong, J.; Kim, S.H.; Lee, S.J.; Cho, Y.J.; An, J.; Nam, D.-H.; Joo, K.M.; Cha, C.I. Replacement of microglial cells using Clodronate liposome and bone marrow transplantation in the central nervous system of SOD1G93A transgenic mice as an in vivo model of amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2012, 418, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Chun, S.J.; Boillee, S.; Fujimori-Tonou, N.; Yamashita, H.; Gutmann, D.H.; Takahashi, R.; Misawa, H.; Cleveland, D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008, 11, 251–253. [Google Scholar] [CrossRef]
- Clement, A.M.; Nguyen, M.D.; Roberts, E.A.; Garcia, M.L.; Boillee, S.; Rule, M.; McMahon, A.P.; Doucette, W.; Siwek, D.; Ferrante, R.J.; et al. Wild-Type Nonneuronal Cells Extend Survival of SOD1 Mutant Motor Neurons in ALS Mice. Science 2003, 302, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Brites, D.; Vaz, A.R. Microglia centered pathogenesis in ALS: Insights in cell interconnectivity. Front. Cell. Neurosci. 2014, 8, 117. [Google Scholar] [CrossRef]
- Barbosa, M.; Gomes, C.; Sequeira, C.; Gonçalves-Ribeiro, J.; Pina, C.C.; Carvalho, L.A.; Moreira, R.; Vaz, S.H.; Vaz, A.R.; Brites, D. Recovery of Depleted miR-146a in ALS Cortical Astrocytes Reverts Cell Aberrancies and Prevents Paracrine Pathogenicity on Microglia and Motor Neurons. Front. Cell Dev. Biol. 2021, 9, 634355. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.; Santos, M.; de Sousa, N.; Duarte-Silva, S.; Vaz, A.R.; Salgado, A.J.; Brites, D. Intrathecal Injection of the Secretome from ALS Motor Neurons Regulated for miR-124 Expression Prevents Disease Outcomes in SOD1-G93A Mice. Biomedicines 2022, 10, 2120. [Google Scholar] [CrossRef]
- Han, B.; Chao, J.; Yao, H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol. Ther. 2018, 187, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Li, J.; Yu, S.; Chen, X.; She, Y.; Lu, Y.; Shu, H. RAGE mediates hippocampal pericyte responses and neurovascular unit lesions after TBI. Exp. Neurol. 2024, 380, 114912. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, T.; Yang, Y.; Lu, D.; Xu, A.; Li, K. FPS-ZM1 Alleviates Neuroinflammation in Focal Cerebral Ischemia Rats via Blocking Ligand/RAGE/DIAPH1 Pathway. ACS Chem. Neurosci. 2020, 12, 63–78. [Google Scholar] [CrossRef]
- Saglam, E.; Zırh, S.; Aktas, C.C.; Muftuoglu, S.F.; Bilginer, B. Papaverine provides neuroprotection by suppressing neuroinflammation and apoptosis in the traumatic brain injury via RAGE- NF-B pathway. J. Neuroimmunol. 2021, 352, 577476. [Google Scholar] [CrossRef] [PubMed]
- Balança, B.; Desmurs, L.; Grelier, J.; Perret-Liaudet, A.; Lukaszewicz, A.-C. DAMPs and RAGE Pathophysiology at the Acute Phase of Brain Injury: An Overview. Int. J. Mol. Sci. 2021, 22, 2439. [Google Scholar] [CrossRef]
- Reddy, V.P.; Aryal, P.; Soni, P. RAGE Inhibitors in Neurodegenerative Diseases. Biomedicines 2023, 11, 1131. [Google Scholar] [CrossRef]
- Gao, T.-L.; Yuan, X.-T.; Yang, D.; Dai, H.-L.; Wang, W.-J.; Peng, X.; Shao, H.-J.; Jin, Z.-F.; Fu, Z.-J. Expression of HMGB1 and RAGE in rat and human brains after traumatic brain injury. J. Trauma Acute Care Surg. 2012, 72, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, S.; Okabe, J.; Ziemann, M.; Drew, B.; Murakoshi, M.; Sourris, K.C.; McClelland, A.D.; Bose, M.; Ekinci, E.I.; Coughlan, M.T.; et al. miR-214 and Its Primary Transcript Dnm3os Regulate Fibrosis and Inflammation Through RAGE Signaling in Diabetic Kidney Disease. Diabetes 2025, 74, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Zhang, G.; Sun, R.; Pan, X.; Wang, X.; Li, H.; Sun, Y. miR-185-5p inhibits F-actin polymerization and reverses epithelial mesenchymal transition of human breast cancer cells by modulating RAGE. Mol. Med. Rep. 2018, 18, 2621–2630. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.-C.; Lin, S.-S.; Yuan, L.-J.; Lu, M.-L.; Ueng, S.W.N.; Yang, C.-Y.; Tsai, T.-T.; Lai, P.-L. Upregulation of miR-107 expression following hyperbaric oxygen treatment suppresses HMGB1/RAGE signaling in degenerated human nucleus pulposus cells. Arthritis Res. Ther. 2019, 21, 42. [Google Scholar] [CrossRef]
- Onyeagucha, B.C.; Mercado-Pimentel, M.E.; Hutchison, J.; Flemington, E.K.; Nelson, M.A. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Exp. Cell Res. 2013, 319, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-S.; Yuan, L.-J.; Niu, C.-C.; Tu, Y.-K.; Yang, C.-Y.; Ueng, S. Hyperbaric oxygen inhibits the HMGB1/RAGE signaling pathway by upregulating Mir-107 expression in human osteoarthritic chondrocytes. Osteoarthr. Cartil. 2019, 27, 1372–1381. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, S.; Marei, O.; Elalfy, O.; Zaben, M. Neurogenesis after traumatic brain injury—The complex role of HMGB1 and neuroinflammation. Neuropharmacology 2021, 183, 108400. [Google Scholar] [CrossRef] [PubMed]
- Mitusova, K.; Peltek, O.O.; Karpov, T.E.; Muslimov, A.R.; Zyuzin, M.V.; Timin, A.S. Overcoming the blood–brain barrier for the therapy of malignant brain tumor: Current status and prospects of drug delivery approaches. J. Nanobiotechnol. 2022, 20, 412. [Google Scholar] [CrossRef] [PubMed]
- NIH. Clinical Trials.gov. Available online: https://clinicaltrials.gov/ (accessed on 1 March 2025).
- Chen, J.S.; Revilla, A.C.; Guerrero, M.; Gumbayan, A.M.; Zeller, R.W. Properties and kinetics of microRNA regulation through canonical seed sites. J. RNAi Gene Silenc. 2015, 11, 507–514. [Google Scholar]
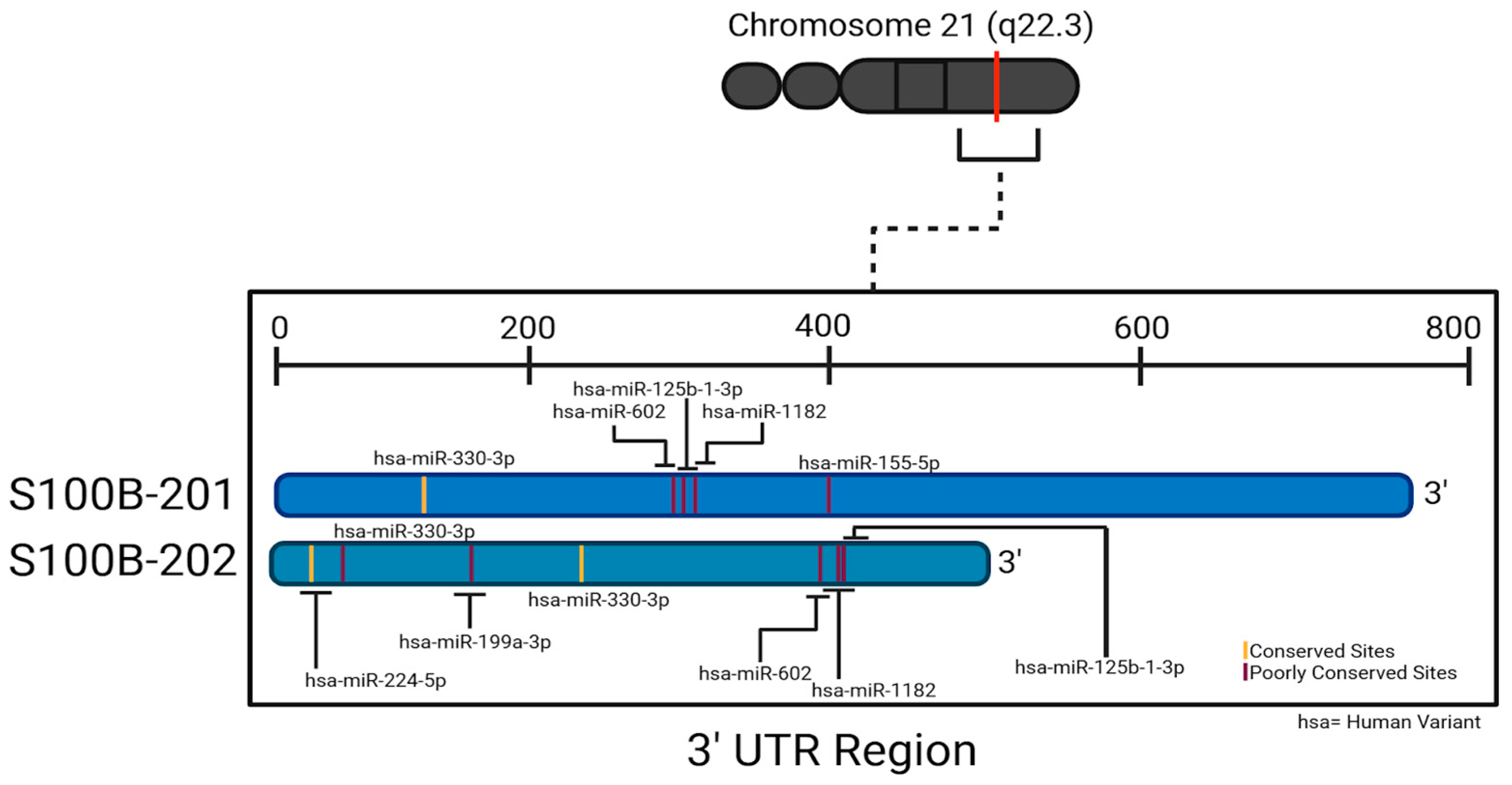
| mRNA Species | miRNA Name | Binding Pattern * | 3′ UTR Position | Seed Match | Site Type | Reference |
|---|---|---|---|---|---|---|
| S100B-201 | hsa-miR-330-3p |  | 127-133 | 7mer-m8 | Conserved | [57,76,77] |
| hsa-miR-1182 |  | 308-315 | 8mer | Poorly Conserved | [57,78] | |
| hsa-miR-602 |  | 302-308 | 7mer-A1 | Poorly Conserved | [46,57,64] | |
| hsa-miR-125b-1-3p |  | 314-320 | 7mer-m8 | Poorly Conserved | [57,79,80] | |
| hsa-miR-155-5p |  | 398-404 | 7mer-m8 | Poorly Conserved | [57] | |
| S100B-202 | hsa-miR-330-3p |  | 215-221 | 7mer-m8 | Conserved | [56,75,76] |
| hsa-miR-1182 |  | 396-403 | 8mer | Poorly Conserved | [57,78] | |
| hsa-miR-602 |  | 390-396 | 7mer-A1 | Poorly Conserved | [46,57,64] | |
| hsa-miR-125b-1-3p |  | 402-408 | 7mer-m8 | Poorly Conserved | [57,79,80] | |
| hsa-miR-224-5p |  | 15-21 | 7mer-m8 | Conserved | [57] | |
| hsa-miR-199a-3p |  | 53-59 | 7mer-m8 | Poorly Conserved | [48,57,81] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dali, A.; Basnyat, S.; Delancey, R.; Chopra, N. MiRNA-Mediated Regulation of S100B: A Review. NeuroSci 2025, 6, 75. https://doi.org/10.3390/neurosci6030075
Dali A, Basnyat S, Delancey R, Chopra N. MiRNA-Mediated Regulation of S100B: A Review. NeuroSci. 2025; 6(3):75. https://doi.org/10.3390/neurosci6030075
Chicago/Turabian StyleDali, Animesh, Suhana Basnyat, Rachel Delancey, and Nipun Chopra. 2025. "MiRNA-Mediated Regulation of S100B: A Review" NeuroSci 6, no. 3: 75. https://doi.org/10.3390/neurosci6030075
APA StyleDali, A., Basnyat, S., Delancey, R., & Chopra, N. (2025). MiRNA-Mediated Regulation of S100B: A Review. NeuroSci, 6(3), 75. https://doi.org/10.3390/neurosci6030075






