The Relevance of Circadian Clocks to Stem Cell Differentiation and Cancer Progression
Abstract
1. Introduction
1.1. The Master Circadian Pacemaker in the SCN
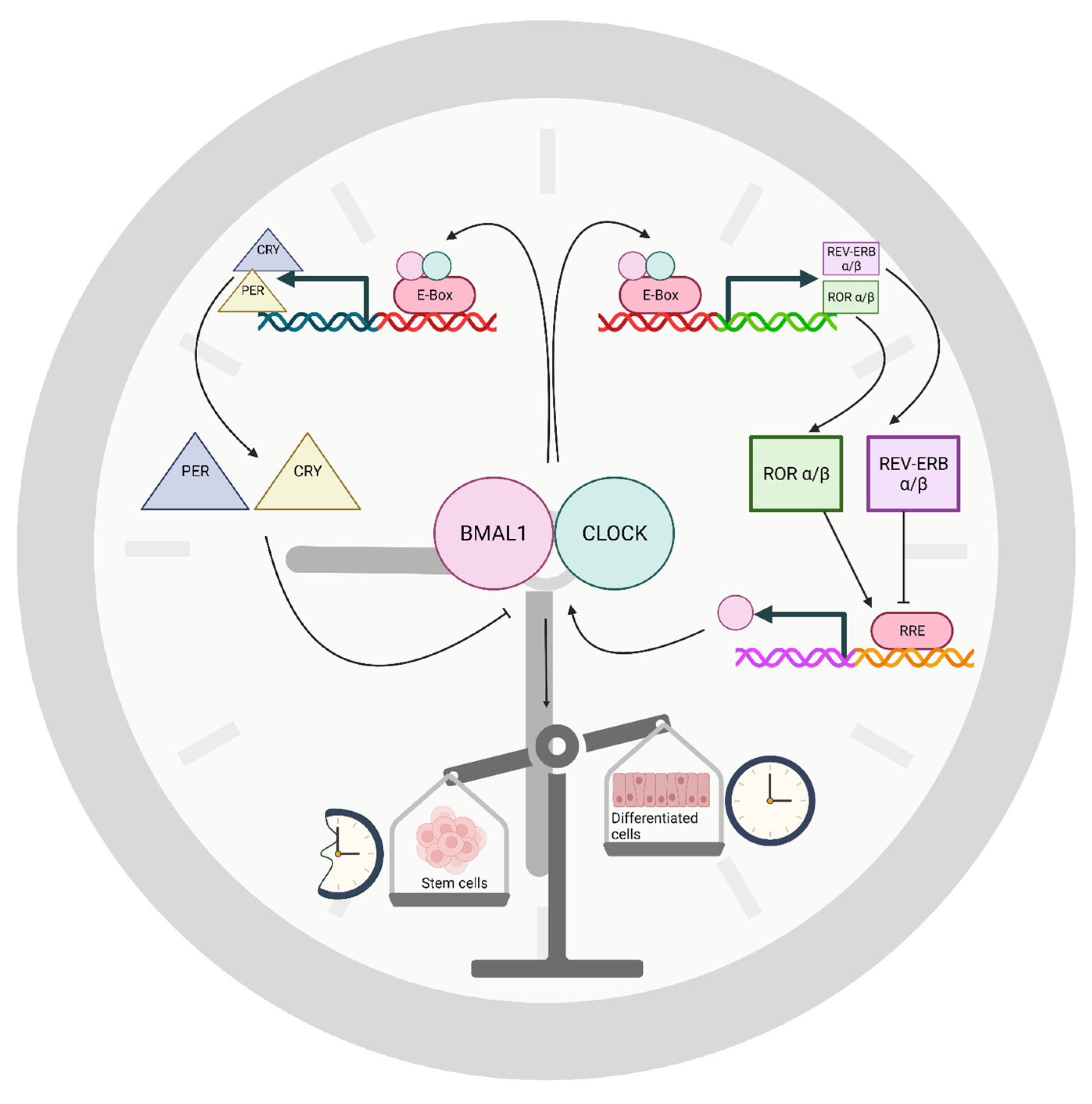
1.2. The Role of TTFLs in the Molecular Clock Mechanism
1.3. Stem Cells
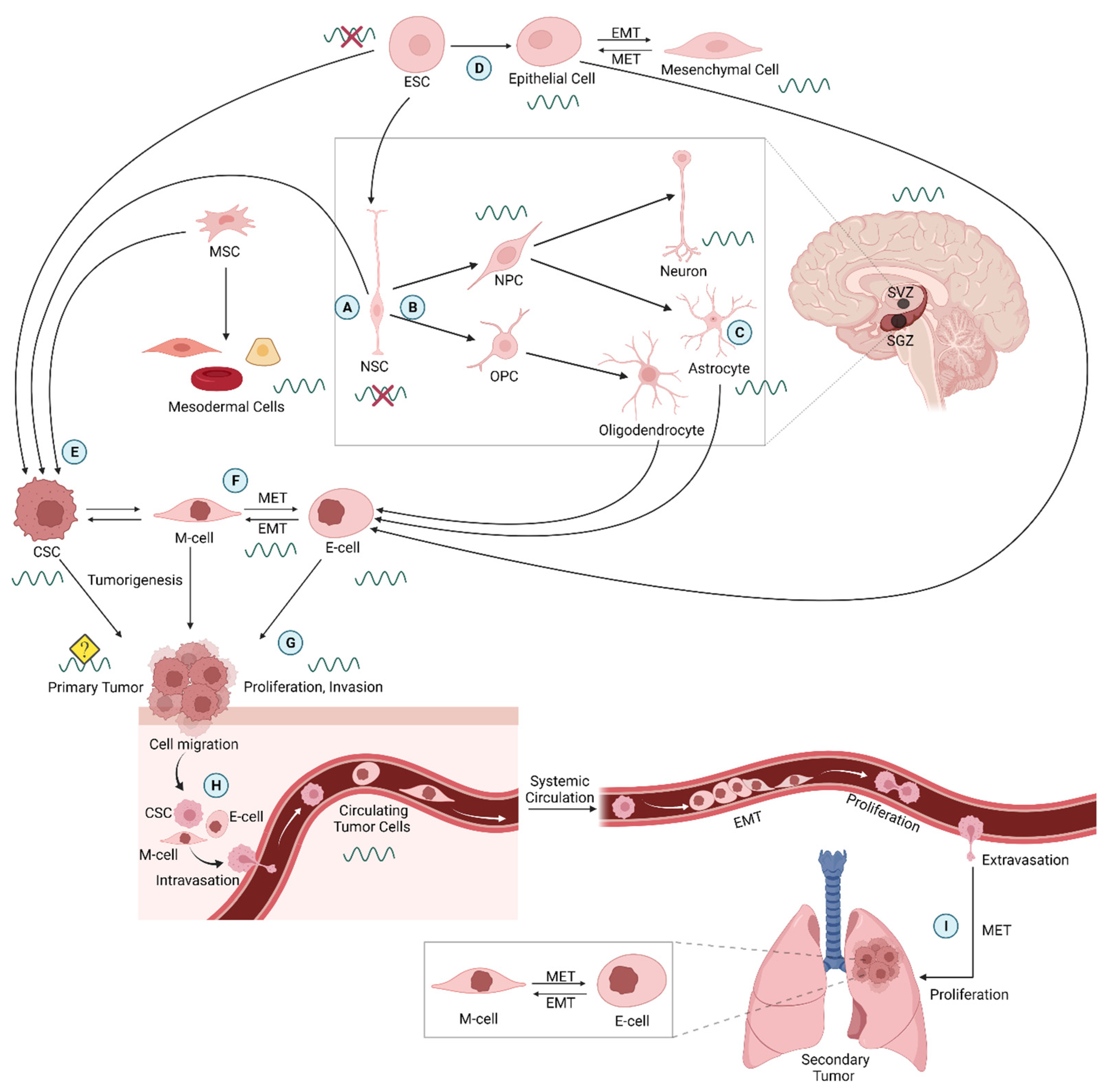
2. Embryonic Stem Cells and Circadian Rhythms
3. Neural Stem Cells and Circadian Rhythms
3.1. Characteristics and Functions
3.2. Circadian Regulation of Adult Neural Stem Cells
3.3. Additional Influences on Neural Stem Cell Circadian Clocks
3.4. Circadian Clock Plasticity in Neurons and Glial Cells
4. Circadian Clocks Acting in Tumorigenesis and Metastasis
4.1. Cancer Cells within the Circadian Timing System
4.2. Metastasis, EMT, and Circulating Tumor Cells
4.3. Cancer Stem Cells and Circadian Rhythms
4.4. Evidence of Circadian Clocks Regulating Stem Cell Properties of Cancer Cells
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| SCN | Suprachiasmatic nucleus |
| TTFL | Transcription translation feedback loop |
| CLOCK | Circadian locomotor output cycles kaput |
| BMAL1 | Brain and muscle arnt-like protein |
| PER | Period |
| CRY | Cryptochrome |
| CSC | Cancer stem cells |
| MSC | Mesenchymal stem cells |
| ESC | Embryonic stem cells |
| NSC | Neural stem cells |
| SVZ | Sub ventricular zone |
| SGZ | Sub granular zone |
| OPC | Oligodendrocyte progenitor cells |
| EMT | Epithelial to mesenchymal transition |
| MET | Mesenchymal to epithelial transition |
| CTC | Circulating tumor cells |
| M cells | Mesenchymal cancer cell |
| E cells | Epithelial cancer stem cell |
References
- Ma, M.A.; Morrison, E.H. Neuroanatomy, Nucleus Suprachiasmatic; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Honma, S. The mammalian circadian system: A hierarchical multi-oscillator structure for generating circadian rhythm. J. Physiol. Sci. 2018, 68, 207–219. [Google Scholar] [CrossRef]
- Weger, M.; Diotel, N.; Dorsemans, A.C.; Dickmeis, T.; Weger, B.D. Stem cells and the circadian clock. Dev. Biol. 2017, 431, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990, 247, 975–978. [Google Scholar] [CrossRef]
- Page, T.L. Transplantation of the cockroach circadian pacemaker. Science 1982, 216, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Granados-Fuentes, D.; Saxena, M.T.; Prolo, L.M.; Aton, S.J.; Herzog, E.D. Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur. J. NeuroSci. 2004, 19, 898–906. [Google Scholar] [CrossRef]
- Pezuk, P.; Mohawk, J.A.; Yoshikawa, T.; Sellix, M.T.; Menaker, M. Circadian organization is governed by extra-SCN pacemakers. J. Biol. Rhythm. 2010, 25, 432–441. [Google Scholar] [CrossRef]
- Plikus, M.V.; Van Spyk, E.N.; Pham, K.; Geyfman, M.; Kumar, V.; Takahashi, J.S.; Andersen, B. The circadian clock in skin: Implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J. Biol. Rhythm. 2015, 30, 163–182. [Google Scholar] [CrossRef] [PubMed]
- Scheving, L.A. Biological clocks and the digestive system. Gastroenterology 2000, 119, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Atger, F.; Mauvoisin, D.; Weger, B.; Gobet, C.; Gachon, F. Regulation of Mammalian Physiology by Interconnected Circadian and Feeding Rhythms. Front. Endocrinol. 2017, 8, 42. [Google Scholar] [CrossRef]
- Kinouchi, K.; Sassone-Corsi, P. Metabolic rivalry: Circadian homeostasis and tumorigenesis. Nat. Rev. Cancer 2020, 20, 645–661. [Google Scholar] [CrossRef]
- Guilding, C.; Hughes, A.T.; Brown, T.M.; Namvar, S.; Piggins, H.D. A riot of rhythms: Neuronal and glial circadian oscillators in the mediobasal hypothalamus. Mol. Brain 2009, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Moore, H.A.; Whitmore, D. Circadian rhythmicity and light sensitivity of the zebrafish brain. PLoS ONE 2014, 9, e86176. [Google Scholar] [CrossRef] [PubMed]
- Weger, M.; Weger, B.D.; Diotel, N.; Rastegar, S.; Hirota, T.; Kay, S.A.; Strahle, U.; Dickmeis, T. Real-time in vivo monitoring of circadian E-box enhancer activity: A robust and sensitive zebrafish reporter line for developmental, chemical and neural biology of the circadian clock. Dev. Biol. 2013, 380, 259–273. [Google Scholar] [CrossRef]
- Rosenwasser, A.M.; Turek, F.W. Neurobiology of Circadian Rhythm Regulation. Sleep Med. Clin. 2015, 10, 403–412. [Google Scholar] [CrossRef]
- Yang, X.; Lamia, K.A.; Evans, R.M. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 387–394. [Google Scholar] [CrossRef]
- Finger, A.M.; Jaschke, S.; Del Olmo, M.; Hurwitz, R.; Granada, A.E.; Herzel, H.; Kramer, A. Intercellular coupling between peripheral circadian oscillators by TGF-beta signaling. Sci. Adv. 2021, 7, eabg5174. [Google Scholar] [CrossRef]
- Liu, A.C.; Welsh, D.K.; Ko, C.H.; Tran, H.G.; Zhang, E.E.; Priest, A.A.; Buhr, E.D.; Singer, O.; Meeker, K.; Verma, I.M.; et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 2007, 129, 605–616. [Google Scholar] [CrossRef]
- Brown, L.S.; Doyle, F.J., 3rd. A dual-feedback loop model of the mammalian circadian clock for multi-input control of circadian phase. PLoS Comput. Biol. 2020, 16, e1008459. [Google Scholar] [CrossRef] [PubMed]
- Hergenhan, S.; Holtkamp, S.; Scheiermann, C. Molecular Interactions between Components of the Circadian Clock and the Immune System. J. Mol. Biol. 2020, 432, 3700–3713. [Google Scholar] [CrossRef]
- Roenneberg, T.; Merrow, M. The Circadian Clock and Human Health. Curr. Biol. 2016, 26, R432–R443. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Perez, E.; Gonzalez-Pujana, A.; Igartua, M.; Santos-Vizcaino, E.; Hernandez, R.M. Mesenchymal Stromal Cell Secretome for the Treatment of Immune-Mediated Inflammatory Diseases: Latest Trends in Isolation, Content Optimization and Delivery Avenues. Pharmaceutics 2021, 13, 1802. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzynski, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Yagita, K.; Horie, K.; Koinuma, S.; Nakamura, W.; Yamanaka, I.; Urasaki, A.; Shigeyoshi, Y.; Kawakami, K.; Shimada, S.; Takeda, J.; et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 3846–3851. [Google Scholar] [CrossRef]
- Myung, J.; Nakamura, T.J.; Jones, J.R.; Silver, R.; Ono, D. Editorial: Development of Circadian Clock Functions. Front. NeuroSci. 2021, 15, 735007. [Google Scholar] [CrossRef]
- Ali, A.A.H.; Schwarz-Herzke, B.; Mir, S.; Sahlender, B.; Victor, M.; Gorg, B.; Schmuck, M.; Dach, K.; Fritsche, E.; Kremer, A.; et al. Deficiency of the clock gene Bmal1 affects neural progenitor cell migration. Brain Struct. Funct. 2019, 224, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Umemura, Y.; Koike, N.; Tsuchiya, Y.; Watanabe, H.; Kondoh, G.; Kageyama, R.; Yagita, K. Circadian key component CLOCK/BMAL1 interferes with segmentation clock in mouse embryonic organoids. Proc. Natl. Acad. Sci. USA 2022, 119, e2114083119. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Jamasbi, R.J.; Kondratov, R.V.; Geusz, M.E. Development of circadian oscillators in neurosphere cultures during adult neurogenesis. PLoS ONE 2015, 10, e0122937. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, M.; Wolfes, A.C.; Ness, N. Astrocyte Circadian Timekeeping in Brain Health and Neurodegeneration. Adv. Exp. Med. Biol. 2021, 1344, 87–110. [Google Scholar] [CrossRef] [PubMed]
- Colwell, C.S.; Ghiani, C.A. Potential Circadian Rhythms in Oligodendrocytes? Working Together through Time. Neurochem. Res. 2020, 45, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Chi-Castaneda, D.; Ortega, A. Glial Cells in the Genesis and Regulation of Circadian Rhythms. Front. Physiol. 2018, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, J.H.; Kim, K.; Leem, J.; Park, K.K. Melatonin Inhibits Transforming Growth Factor-beta1-Induced Epithelial-Mesenchymal Transition in AML12 Hepatocytes. Biology 2019, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wu, X.; Wang, Y.; Ji, S.; Zhang, W.; Kang, J.; Li, J.; Fei, G. Melatonin prevents LPS-induced epithelial-mesenchymal transition in human alveolar epithelial cells via the GSK-3beta/Nrf2 pathway. BioMed. Pharmacother. 2020, 132, 110827. [Google Scholar] [CrossRef]
- Qi, S.; Yan, L.; Liu, Z.; Mu, Y.L.; Li, M.; Zhao, X.; Chen, Z.J.; Zhang, H. Melatonin inhibits 17beta-estradiol-induced migration, invasion and epithelial-mesenchymal transition in normal and endometriotic endometrial epithelial cells. Reprod. Biol. Endocrinol. 2018, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Biserova, K.; Jakovlevs, A.; Uljanovs, R.; Strumfa, I. Cancer Stem Cells: Significance in Origin, Pathogenesis and Treatment of Glioblastoma. Cells 2021, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, C.; Wang, X.S.; Manoranjan, B.; McFarlane, N.; Nolte, S.; Li, M.; Murty, N.; Siu, K.W.; Singh, S.K. GBM secretome induces transient transformation of human neural precursor cells. J. Neurooncol. 2012, 109, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Matarredona, E.R.; Pastor, A.M. Neural Stem Cells of the Subventricular Zone as the Origin of Human Glioblastoma Stem Cells. Therapeutic Implications. Front. Oncol. 2019, 9, 779. [Google Scholar] [CrossRef] [PubMed]
- Colangelo, T.; Carbone, A.; Mazzarelli, F.; Cuttano, R.; Dama, E.; Nittoli, T.; Albanesi, J.; Barisciano, G.; Forte, N.; Palumbo, O.; et al. Loss of circadian gene Timeless induces EMT and tumor progression in colorectal cancer via Zeb1-dependent mechanism. Cell Death Differ. 2022. [Google Scholar] [CrossRef] [PubMed]
- Hwang-Verslues, W.W.; Chang, P.H.; Jeng, Y.M.; Kuo, W.H.; Chiang, P.H.; Chang, Y.C.; Hsieh, T.H.; Su, F.Y.; Lin, L.C.; Abbondante, S.; et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc. Natl. Acad. Sci. USA 2013, 110, 12331–12336. [Google Scholar] [CrossRef]
- Zhang, Y.; Devocelle, A.; Desterke, C.; de Souza, L.E.B.; Hadadi, E.; Acloque, H.; Foudi, A.; Xiang, Y.; Ballesta, A.; Chang, Y.; et al. BMAL1 Knockdown Leans Epithelial-Mesenchymal Balance toward Epithelial Properties and Decreases the Chemoresistance of Colon Carcinoma Cells. Int. J. Mol. Sci. 2021, 22, 5247. [Google Scholar] [CrossRef]
- De, A.; Beligala, D.H.; Sharma, V.P.; Burgos, C.A.; Lee, A.M.; Geusz, M.E. Cancer stem cell generation during epithelial-mesenchymal transition is temporally gated by intrinsic circadian clocks. Clin. Exp. Metastasis 2020, 37, 617–635. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, G.; Qu, M.; Gimple, R.C.; Wu, Q.; Qiu, Z.; Prager, B.C.; Wang, X.; Kim, L.J.Y.; Morton, A.R.; et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov. 2019, 9, 1556–1573. [Google Scholar] [CrossRef]
- Sharma, V.P.; Anderson, N.T.; Geusz, M.E. Circadian properties of cancer stem cells in glioma cell cultures and tumorspheres. Cancer Lett. 2014, 345, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, A.; Takashima, N.; Shigeyoshi, Y. Circadian rhythm generation in a glioma cell line. Biochem. Biophys. Res. Commun. 2006, 346, 169–174. [Google Scholar] [CrossRef]
- Pendergast, J.S.; Yeom, M.; Reyes, B.A.; Ohmiya, Y.; Yamazaki, S. Disconnected circadian and cell cycles in a tumor-driven cell line. Commun. Integr. Biol. 2010, 3, 536–539. [Google Scholar] [CrossRef]
- Sarkar, S.; Porter, K.I.; Dakup, P.P.; Gajula, R.P.; Koritala, B.S.C.; Hylton, R.; Kemp, M.G.; Wakamatsu, K.; Gaddameedhi, S. Circadian clock protein BMAL1 regulates melanogenesis through MITF in melanoma cells. Pigment. Cell Melanoma Res. 2021, 34, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Sotak, M.; Sumova, A.; Pacha, J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann. Med. 2014, 46, 221–232. [Google Scholar] [CrossRef]
- Xuan, W.; Khan, F.; James, C.D.; Heimberger, A.B.; Lesniak, M.S.; Chen, P. Circadian regulation of cancer cell and tumor microenvironment crosstalk. Trends Cell Biol. 2021, 31, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Suo, Y.; Fu, Y.; Zhang, F.; Ding, N.; Pang, K.; Xie, C.; Weng, X.; Tian, M.; He, H.; et al. In vivo flow cytometry reveals a circadian rhythm of circulating tumor cells. Light Sci. Appl. 2021, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Jayarama-Naidu, R.; Gallus, E. Abnormal Schwannoma-like Growth of multiple, multifocal BRAF V600E-positive Glioblastoma in the Interior Acoustic Canal with Leptomeningeal Infiltration: A case report. J. Med. Case Rep. 2022, 16, 50. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Yang, Y.; Zhao, R.; Hua, B.; Xu, C.; Yan, Z.; Sun, N.; Qian, R. Role of circadian gene Clock during differentiation of mouse pluripotent stem cells. Protein Cell 2016, 7, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Dolatshad, H.; Cary, A.J.; Davis, F.C. Differential expression of the circadian clock in maternal and embryonic tissues of mice. PLoS ONE 2010, 5, e9855. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, E.; Moriggi, E.; Bauer, C.; Dibner, C.; Brown, S.A. The circadian clock starts ticking at a developmentally early stage. J. Biol. Rhythm. 2010, 25, 442–449. [Google Scholar] [CrossRef]
- Paulose, J.K.; Rucker, E.B., 3rd; Cassone, V.M. Toward the beginning of time: Circadian rhythms in metabolism precede rhythms in clock gene expression in mouse embryonic stem cells. PLoS ONE 2012, 7, e49555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kriegstein, A.; Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. NeuroSci. 2009, 32, 149–184. [Google Scholar] [CrossRef] [PubMed]
- Fontan-Lozano, A.; Morcuende, S.; Davis-Lopez de Carrizosa, M.A.; Benitez-Temino, B.; Mejias, R.; Matarredona, E.R. To Become or Not to Become Tumorigenic: Subventricular Zone versus Hippocampal Neural Stem Cells. Front. Oncol. 2020, 10, 602217. [Google Scholar] [CrossRef] [PubMed]
- Goergen, E.M.; Bagay, L.A.; Rehm, K.; Benton, J.L.; Beltz, B.S. Circadian control of neurogenesis. J. NeuroBiol. 2002, 53, 90–95. [Google Scholar] [CrossRef]
- Tamai, S.; Sanada, K.; Fukada, Y. Time-of-day-dependent enhancement of adult neurogenesis in the hippocampus. PLoS ONE 2008, 3, e3835. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bouchard-Cannon, P.; Mendoza-Viveros, L.; Yuen, A.; Kaern, M.; Cheng, H.Y. The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep. 2013, 5, 961–973. [Google Scholar] [CrossRef]
- Kimiwada, T.; Sakurai, M.; Ohashi, H.; Aoki, S.; Tominaga, T.; Wada, K. Clock genes regulate neurogenic transcription factors, including NeuroD1, and the neuronal differentiation of adult neural stem/progenitor cells. Neurochem. Int. 2009, 54, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Borgs, L.; Beukelaers, P.; Vandenbosch, R.; Nguyen, L.; Moonen, G.; Maquet, P.; Albrecht, U.; Belachew, S.; Malgrange, B. Period 2 regulates neural stem/progenitor cell proliferation in the adult hippocampus. BMC NeuroSci. 2009, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Rakai, B.D.; Chrusch, M.J.; Spanswick, S.C.; Dyck, R.H.; Antle, M.C. Survival of adult generated hippocampal neurons is altered in circadian arrhythmic mice. PLoS ONE 2014, 9, e99527. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Meijer, J.H.; Michel, S.; Vanderleest, H.T.; Rohling, J.H. Daily and seasonal adaptation of the circadian clock requires plasticity of the SCN neuronal network. Eur. J. NeuroSci. 2010, 32, 2143–2151. [Google Scholar] [CrossRef]
- Herrero, A.; Yoshii, T.; Ispizua, J.I.; Colque, C.; Veenstra, J.A.; Muraro, N.I.; Ceriani, M.F. Coupling Neuropeptide Levels to Structural Plasticity in Drosophila Clock Neurons. Curr. Biol. 2020, 30, 3154–3166.e4. [Google Scholar] [CrossRef]
- Rohr, K.E.; Pancholi, H.; Haider, S.; Karow, C.; Modert, D.; Raddatz, N.J.; Evans, J. Seasonal plasticity in GABAA signaling is necessary for restoring phase synchrony in the master circadian clock network. Elife 2019, 8, e49578. [Google Scholar] [CrossRef]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. NeuroSci. 2018, 19, 453–469. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Tsunekawa, Y.; Nomura, T.; Suto, F.; Matsumata, M.; Tsuchiya, S.; Osumi, N. Differential proliferation rhythm of neural progenitor and oligodendrocyte precursor cells in the young adult hippocampus. PLoS ONE 2011, 6, e27628. [Google Scholar] [CrossRef]
- Artiushin, G.; Sehgal, A. The Glial Perspective on Sleep and Circadian Rhythms. Annu. Rev. NeuroSci. 2020, 43, 119–140. [Google Scholar] [CrossRef]
- McCauley, J.P.; Petroccione, M.A.; D’Brant, L.Y.; Todd, G.C.; Affinnih, N.; Wisnoski, J.J.; Zahid, S.; Shree, S.; Sousa, A.A.; De Guzman, R.M.; et al. Circadian Modulation of Neurons and Astrocytes Controls Synaptic Plasticity in Hippocampal Area CA1. Cell Rep. 2020, 33, 108255. [Google Scholar] [CrossRef] [PubMed]
- Gorska-Andrzejak, J. Glia-related circadian plasticity in the visual system of Diptera. Front. Physiol. 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Perez-Catalan, N.A.; Doe, C.Q.; Ackerman, S.D. The role of astrocyte-mediated plasticity in neural circuit development and function. Neural Dev. 2021, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.S.; Wang, S.; Harrison-Restelli, C.; Benraiss, A.; Fraser, R.A.; Gravel, M.; Braun, P.E.; Goldman, S.A. Identification, isolation, and promoter-defined separation of mitotic oligodendrocyte progenitor cells from the adult human subcortical white matter. J. NeuroSci. 1999, 19, 9986–9995. [Google Scholar] [CrossRef] [PubMed]
- Boulanger, J.J.; Messier, C. Unbiased stereological analysis of the fate of oligodendrocyte progenitor cells in the adult mouse brain and effect of reference memory training. Behav. Brain Res. 2017, 329, 127–139. [Google Scholar] [CrossRef]
- Mi, S.; Miller, R.H.; Tang, W.; Lee, X.; Hu, B.; Wu, W.; Zhang, Y.; Shields, C.B.; Zhang, Y.; Miklasz, S.; et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann. Neurol. 2009, 65, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Liang, X.; Dou, X.; Qi, Y.; Yang, C.; Luo, Y.; Chao, F.; Zhang, L.; Xiao, Q.; Jiang, L.; et al. Exercise rather than fluoxetine promotes oligodendrocyte differentiation and myelination in the hippocampus in a male mouse model of depression. Transl. Psychiatry 2021, 11, 622. [Google Scholar] [CrossRef]
- Gaughwin, P.M.; Caldwell, M.A.; Anderson, J.M.; Schwiening, C.J.; Fawcett, J.W.; Compston, D.A.; Chandran, S. Astrocytes promote neurogenesis from oligodendrocyte precursor cells. Eur. J. NeuroSci. 2006, 23, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Maeda, Y.; Ma, J.; Xu, J.; Horiuchi, M.; Miers, L.; Vaccarino, F.; Pleasure, D. Pyramidal neurons are generated from oligodendroglial progenitor cells in adult piriform cortex. J. NeuroSci. 2010, 30, 12036–12049. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Molecular Architecture of the Circadian Clock in Mammals. In A Time for Metabolism and Hormones; Sassone-Corsi, P., Christen, Y., Eds.; Springer: Cham, Switzerland, 2016; pp. 13–24. [Google Scholar] [CrossRef]
- Morin, L.P. SCN organization reconsidered. J. Biol. Rhythm. 2007, 22, 3–13. [Google Scholar] [CrossRef]
- James, P.; Bertrand, K.A.; Hart, J.E.; Schernhammer, E.S.; Tamimi, R.M.; Laden, F. Outdoor Light at Night and Breast Cancer Incidence in the Nurses’ Health Study II. Environ. Health Perspect. 2017, 125, 087010. [Google Scholar] [CrossRef] [PubMed]
- Madden, M.H.; Anic, G.M.; Thompson, R.C.; Nabors, L.B.; Olson, J.J.; Browning, J.E.; Monteiro, A.N.; Egan, K.M. Circadian pathway genes in relation to glioma risk and outcome. Cancer Causes Control 2014, 25, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.A.; Weaver, D.R. Disrupting the circadian clock: Gene-specific effects on aging, cancer, and other phenotypes. Aging 2011, 3, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Puram, R.V.; Kowalczyk, M.S.; de Boer, C.G.; Schneider, R.K.; Miller, P.G.; McConkey, M.; Tothova, Z.; Tejero, H.; Heckl, D.; Jaras, M.; et al. Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell 2016, 165, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, S.; Marks, T.; Hudson, D.J.; Menaker, M. Genetic control of melatonin synthesis in the pineal gland of the mouse. Science 1986, 231, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Roseboom, P.H.; Namboodiri, M.A.; Zimonjic, D.B.; Popescu, N.C.; Rodriguez, I.R.; Gastel, J.A.; Klein, D.C. Natural melatonin ‘knockdown’ in C57BL/6J mice: Rare mechanism truncates serotonin N-acetyltransferase. Brain Res. Mol. Brain Res. 1998, 63, 189–197. [Google Scholar] [CrossRef]
- DiGiovanni, J.; Bhatt, T.S.; Walker, S.E. C57BL/6 mice are resistant to tumor promotion by full thickness skin wounding. Carcinogenesis 1993, 14, 319–321. [Google Scholar] [CrossRef]
- Granda, T.G.; Liu, X.H.; Smaaland, R.; Cermakian, N.; Filipski, E.; Sassone-Corsi, P.; Levi, F. Circadian regulation of cell cycle and apoptosis proteins in mouse bone marrow and tumor. FASEB J. 2005, 19, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Pretzmann, C.P.; Fahrenkrug, J.; Georg, B. Differentiation of PC12 cells results in enhanced VIP expression and prolonged rhythmic expression of clock genes. J. Mol. NeuroSci. 2008, 36, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, S.; Beaulieu-Laroche, L.; Blum, I.D.; Landgraf, D.; Welsh, D.K.; Storch, K.F.; Labrecque, N.; Cermakian, N. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Vollmers, C.; Panda, S.; DiTacchio, L. A high-throughput assay for siRNA-based circadian screens in human U2OS cells. PLoS ONE 2008, 3, e3457. [Google Scholar] [CrossRef] [PubMed]
- Slat, E.A.; Sponagel, J.; Marpegan, L.; Simon, T.; Kfoury, N.; Kim, A.; Binz, A.; Herzog, E.D.; Rubin, J.B. Cell-intrinsic, Bmal1-dependent Circadian Regulation of Temozolomide Sensitivity in Glioblastoma. J. Biol. Rhythm. 2017, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, F.; Matsunaga, N.; Okazaki, H.; Azuma, H.; Hamamura, K.; Tsuruta, A.; Tsurudome, Y.; Ogino, T.; Hara, Y.; Suzuki, T.; et al. Circadian Clock in a Mouse Colon Tumor Regulates Intracellular Iron Levels to Promote Tumor Progression. J. Biol. Chem. 2016, 291, 7017–7028. [Google Scholar] [CrossRef]
- Hadadi, E.; Acloque, H. Role of circadian rhythm disorders on EMT and tumour-immune interactions in endocrine-related cancers. Endocr. Relat. Cancer 2021, 28, R67–R80. [Google Scholar] [CrossRef] [PubMed]
- Geusz, M.E.; Malik, A.; De, A. Insights into Oncogenesis from Circadian Timing in Cancer Stem Cells. Crit. Rev. Oncog. 2021, 26, 1–17. [Google Scholar] [CrossRef]
- Wang, Z.; Li, F.; He, S.; Zhao, L.; Wang, F. Period circadian regulator 2 suppresses drug resistance to cisplatin by PI3K/AKT pathway and improves chronochemotherapeutic efficacy in cervical cancer. Gene 2022, 809, 146003. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Marcucci, F.; Ghezzi, P.; Rumio, C. The role of autophagy in the cross-talk between epithelial-mesenchymal transitioned tumor cells and cancer stem-like cells. Mol. Cancer 2017, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Matsu-Ura, T.; Dovzhenok, A.; Aihara, E.; Rood, J.; Le, H.; Ren, Y.; Rosselot, A.E.; Zhang, T.; Lee, C.; Obrietan, K.; et al. Intercellular Coupling of the Cell Cycle and Circadian Clock in Adult Stem Cell Culture. Mol. Cell 2016, 64, 900–912. [Google Scholar] [CrossRef]
- Moradkhani, F.; Moloudizargari, M.; Fallah, M.; Asghari, N.; Heidari Khoei, H.; Asghari, M.H. Immunoregulatory role of melatonin in cancer. J. Cell Physiol. 2020, 235, 745–757. [Google Scholar] [CrossRef]
- Reiter, R.J.; Sharma, R.; Rodriguez, C.; Martin, V.; Rosales-Corral, S.; Zuccari, D.; Chuffa, L.G.A. Part-time cancers and role of melatonin in determining their metabolic phenotype. Life Sci. 2021, 278, 119597. [Google Scholar] [CrossRef] [PubMed]
- Tarin, D.; Thompson, E.W.; Newgreen, D.F. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005, 65, 5996–6000, discussion 5991–6000. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Weinberg, R.A. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015, 25, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Brabletz, T.; Jung, A.; Reu, S.; Porzner, M.; Hlubek, F.; Kunz-Schughart, L.A.; Knuechel, R.; Kirchner, T. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA 2001, 98, 10356–10361. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Langley, R.R.; Fidler, I.J. The seed and soil hypothesis revisited—The role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 2011, 128, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Mittal, V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, T.; Jinno, H.; Kitagawa, Y.; Kitajima, M. Cooperation of cancer stem cell properties and epithelial-mesenchymal transition in the establishment of breast cancer metastasis. J. Oncol. 2011, 2011, 591427. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, Y.T.; Cheng, J.; Feng, X.; He, S.J.; Wang, Y.W.; Huang, Q. The viable circulating tumor cells with cancer stem cells feature, where is the way out? J. Exp. Clin. Cancer Res. 2018, 37, 38. [Google Scholar] [CrossRef] [PubMed]
- Tinhofer, I.; Saki, M.; Niehr, F.; Keilholz, U.; Budach, V. Cancer stem cell characteristics of circulating tumor cells. Int. J. Radiat. Biol. 2014, 90, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Masuda, T.; Hayashi, N.; Iguchi, T.; Ito, S.; Eguchi, H.; Mimori, K. Clinical and biological significance of circulating tumor cells in cancer. Mol. Oncol. 2016, 10, 408–417. [Google Scholar] [CrossRef]
- Stobiecka, M.; Ratajczak, K.; Jakiela, S. Toward early cancer detection: Focus on biosensing systems and biosensors for an anti-apoptotic protein survivin and survivin mRNA. Biosens. Bioelectron. 2019, 137, 58–71. [Google Scholar] [CrossRef]
- Gramantieri, L.; Giovannini, C.; Suzzi, F.; Leoni, I.; Fornari, F. Hepatic Cancer Stem Cells: Molecular Mechanisms, Therapeutic Implications, and Circulating Biomarkers. Cancers 2021, 13, 4550. [Google Scholar] [CrossRef]
- Brown, T.C.; Sankpal, N.V.; Gillanders, W.E. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules 2021, 11, 956. [Google Scholar] [CrossRef]
- Golan, T.; Messer, A.R.; Amitai-Lange, A.; Melamed, Z.; Ohana, R.; Bell, R.E.; Kapitansky, O.; Lerman, G.; Greenberger, S.; Khaled, M.; et al. Interactions of Melanoma Cells with Distal Keratinocytes Trigger Metastasis via Notch Signaling Inhibition of MITF. Mol. Cell 2015, 59, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.; Levine, H. Cluster size distribution of cells disseminating from a primary tumor. PLoS Comput. Biol. 2021, 17, e1009011. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Hernandez, L.E.; Eslami, S.Z.; Dujon, A.M.; Giraudeau, M.; Ujvari, B.; Thomas, F.; Alix-Panabieres, C. Do malignant cells sleep at night? Genome Biol. 2020, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.L.; Fitzgerald, J.E.; Ivich, F.; Sontag, E.D.; Niedre, M. Short-Term Circulating Tumor Cell Dynamics in Mouse Xenograft Models and Implications for Liquid Biopsy. Front. Oncol. 2020, 10, 601085. [Google Scholar] [CrossRef]
- Zhu, X.; Suo, Y.; Fu, Y.; Zhang, F.; Ding, N.; Pang, K.; Xie, C.; Weng, X.; Tian, M.; He, H.; et al. Reply to Comment on “In vivo flow cytometry reveals a circadian rhythm of circulating tumor cells”. Light Sci. Appl. 2021, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Hrushesky, W.J.; Lester, B.; Lannin, D. Circadian coordination of cancer growth and metastatic spread. Int. J. Cancer 1999, 83, 365–373. [Google Scholar] [CrossRef]
- Lathia, J.; Liu, H.; Matei, D. The Clinical Impact of Cancer Stem Cells. Oncologist 2020, 25, 123–131. [Google Scholar] [CrossRef]
- Morse, D.; Cermakian, N.; Brancorsini, S.; Parvinen, M.; Sassone-Corsi, P. No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol. Endocrinol. 2003, 17, 141–151. [Google Scholar] [CrossRef]
- Malik, A.; Kondratov, R.V.; Jamasbi, R.J.; Geusz, M.E. Circadian Clock Genes Are Essential for Normal Adult Neurogenesis, Differentiation, and Fate Determination. PLoS ONE 2015, 10, e0139655. [Google Scholar] [CrossRef]
- Noguchi, T.; Wang, L.L.; Welsh, D.K. Fibroblast PER2 circadian rhythmicity depends on cell density. J. Biol. Rhythm. 2013, 28, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Brena, D.; Huang, M.B.; Bond, V. Extracellular vesicle-mediated transport: Reprogramming a tumor microenvironment conducive with breast cancer progression and metastasis. Transl. Oncol. 2022, 15, 101286. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.C.; Guo, S.C. Extracellular Vesicles: Potential Participants in Circadian Rhythm Synchronization. Int. J. Biol. Sci. 2018, 14, 1610–1620. [Google Scholar] [CrossRef]
- Sancar, A.; Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science 2021, 371, 101068. [Google Scholar] [CrossRef]
- Yang, Y.; Lindsey-Boltz, L.A.; Vaughn, C.M.; Selby, C.P.; Cao, X.; Liu, Z.; Hsu, D.S.; Sancar, A. Circadian clock, carcinogenesis, chronochemotherapy connections. J. Biol. Chem. 2021, 297, 101068. [Google Scholar] [CrossRef]
- Huang, Z.; He, A.; Wang, J.; Lu, H.; Zhang, R.; Wu, L.; Feng, Q. The circadian clock is associated with prognosis and immune infiltration in stomach adenocarcinoma. Aging 2021, 13, 16637–16655. [Google Scholar] [CrossRef]
- Lee, Y. Roles of circadian clocks in cancer pathogenesis and treatment. Exp. Mol. Med. 2021, 53, 1529–1538. [Google Scholar] [CrossRef]
- Sarma, A.; Sharma, V.P.; Sarkar, A.B.; Sekar, M.C.; Samuel, K.; Geusz, M.E. The circadian clock modulates anti-cancer properties of curcumin. BMC Cancer 2016, 16, 759. [Google Scholar] [CrossRef]
- Hassan, S.A.; Ali, A.A.H.; Sohn, D.; Flogel, U.; Janicke, R.U.; Korf, H.W.; von Gall, C. Does timing matter in radiotherapy of hepatocellular carcinoma? An experimental study in mice. Cancer Med. 2021, 10, 7712–7725. [Google Scholar] [CrossRef]
- Sinha, D.; Saha, P.; Samanta, A.; Bishayee, A. Emerging Concepts of Hybrid Epithelial-to-Mesenchymal Transition in Cancer Progression. Biomolecules 2020, 10, 1561. [Google Scholar] [CrossRef]
- Leise, T.L.; Harrington, M.E. Wavelet-based time series analysis of circadian rhythms. J. Biol. Rhythm. 2011, 26, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Rosselot, A.E.; Park, M.; Kim, M.; Matsu-Ura, T.; Wu, G.; Flores, D.E.; Subramanian, K.R.; Lee, S.; Sundaram, N.; Broda, T.R.; et al. Ontogeny and function of the circadian clock in intestinal organoids. EMBO J. 2022, 41, e106973. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malik, A.; Nalluri, S.; De, A.; Beligala, D.; Geusz, M.E. The Relevance of Circadian Clocks to Stem Cell Differentiation and Cancer Progression. NeuroSci 2022, 3, 146-165. https://doi.org/10.3390/neurosci3020012
Malik A, Nalluri S, De A, Beligala D, Geusz ME. The Relevance of Circadian Clocks to Stem Cell Differentiation and Cancer Progression. NeuroSci. 2022; 3(2):146-165. https://doi.org/10.3390/neurosci3020012
Chicago/Turabian StyleMalik, Astha, Shreya Nalluri, Arpan De, Dilshan Beligala, and Michael E. Geusz. 2022. "The Relevance of Circadian Clocks to Stem Cell Differentiation and Cancer Progression" NeuroSci 3, no. 2: 146-165. https://doi.org/10.3390/neurosci3020012
APA StyleMalik, A., Nalluri, S., De, A., Beligala, D., & Geusz, M. E. (2022). The Relevance of Circadian Clocks to Stem Cell Differentiation and Cancer Progression. NeuroSci, 3(2), 146-165. https://doi.org/10.3390/neurosci3020012






