Abstract
Nucleoside analogs (NAs) have been extensively examined as plausible antiviral agents in recent years, in particular since the outbreak of the global pandemic of COVID-19 in 2019. In this review, the structures and antiviral properties of over 450 NAs are collected according to the type of virus, namely SARS-CoV, SARS-CoV-2, MERS-CoV, HCoV-OC43, HCoV-229E, and HCoV-NL63. The activity of the NAs against HCoV-related enzymes is also presented. Selected studies dealing with the mode of action of the NAs are discussed in detail. The repurposing of known NAs appears to be the most extensively investigated scientific approach towards efficacious anti-HCoV agents. The recently reported de novo-designed NAs seem to open up additional approaches to new drug candidates.
1. Introduction
The family of human corona viruses (HCoVs) includes seven strains [1]. Four of them, namely HCoV-229E, HCoV-HKU1, HCoV-NL63, and HCoV-OC43, cause mild and self-limiting infections of the upper respiratory tract. However, two HCoVs are responsible for serious respiratory disease epidemics that originated in China in 2002 and 2003 (severe acute respiratory syndrome coronavirus, SARS-CoV virus) and in Saudi Arabia in 2012 (Middle East respiratory syndrome coronavirus, MERS-CoV virus) [2]. Starting in November 2002, followed by spreading to 29 countries, the SARS-CoV virus caused approximately 8000 illnesses and 700 deaths from late 2002 to late 2003 [3]. Then, it disappeared. The MERS-CoV virus is endemic to the Middle East [4]. However, due to travelers’ migration, MERS-CoV virus infections have been reported in 27 countries, resulting in 858 known deaths since 2012 [5]. The SARS-CoV-2 virus (severe acute respiratory syndrome coronavirus 2) is responsible for the pandemic of coronavirus disease (COVID-19) that started in China in 2019. As of 28 May 2023, over 767 million confirmed cases of COVID-19 and over 6.9 million deaths have been reported globally [6]. The end of COVID-19 as a public health emergency was declared in May 2023 [7]. However, continuous mutations of the SARS-CoV-2 virus and the risk of its new variants emerging elsewhere have been emphasized.
Very close similarity has been found between SARS-CoV [8], MERS-CoV [9], and SARS-CoV-2 [10] in terms of their genome sequences and the key proteins involved in the virus life cycle and entry process. These findings have provided solid arguments for postulates on the possible susceptibility of SARS-CoV, MERS-CoV, and SARS-CoV-2 viruses to the same or structurally similar antiviral agents [11,12,13].
A significant number of approved antiviral or anticancer drugs belong to the class of nucleoside analogs (NAs). Many of them show multitarget activity [14]. For this reason, considerable attention has been paid to NAs as potential anticoronavirus drug candidates since the SARS-CoV outbreak in 2002. Initial efforts to treat SARS-CoV-infected patients focused on the use of ribavirin due to its activity against a broad spectrum of RNA or DNA viruses, including RSV, influenza viruses, and parainfluenza viruses [15]. Further studies suggested the limited usefulness of ribavirin in the treatment of SARS-CoV patients [16]. A few NAs with promising activity against SARS-CoV-2 viruses were identified before or during the SARS-CoV-2 pandemic. Among them, remdesivir, molnupiravir, and VV116 have been approved for the treatment of COVID-19 in selected countries (Figure 1) [17].
Very recently, the FDA granted fast-track designation for the investigational drug bemnifosbuvir (Figure 1), which is in a global phase III clinical trial SUNRISE-3 (NCT05629962) for outpatients with high risk of disease progression [18].
The selected NAs as potential anti-HCoV agents have been discussed in several review articles within the last five years. These reviews have mainly mentioned either remdesivir or a very narrow group of NAs [19,20,21,22,23,24,25]. Particular emphasis has been on compounds with known mechanisms of action against SARS-CoV-2 (Figure 1) [26,27,28]. Although the susceptibility of HCoVs to a variety of NAs has been studied for over twenty years, only a few original publications have been included in a review discussing a relatively broad group of twenty-four NAs with micromolar activity against SARS-CoV, SARS-CoV-2 or MERS-CoV, and HCoV-NL63 [29]. The activity of NAs against HCoVs other than SARS-CoV, SARS-CoV-2, or MERS-CoV has been mentioned infrequently [20,25,29].

Figure 1.
NAs with well-recognized activity and modes of action against HCoVs. (A) RNA chain terminators, including those acting with a dual mechanism of action (remdesivir, islatravir, AT-527, and bemnifosbuvir) [26]. (B) NAs causing lethal mutagenesis. (C) NAs inhibiting viral methyltransferases.
Figure 1.
NAs with well-recognized activity and modes of action against HCoVs. (A) RNA chain terminators, including those acting with a dual mechanism of action (remdesivir, islatravir, AT-527, and bemnifosbuvir) [26]. (B) NAs causing lethal mutagenesis. (C) NAs inhibiting viral methyltransferases.
The aim of our review is to discuss a very broad panel of NAs reported to show in vitro inhibition of the replication of a specific HCoV. We collected over 450 compounds from 80 original works published from 2003 to August 2023. Our intention is to illustrate the importance of NAs (both those existing in the public domain and those newly designed) in the state-of-the-art development of anti-HCoV agents. Therefore, we also included NAs displaying moderate or low activity. All the presented compounds are summarized in Table 1, Table 2, Table 3, Table 4, Table 5 and Table 6. The NAs reported to inhibit HCoV-related enzyme(s) are collected in Table 7. In order to show research trends in the development of nucleoside anti-HCoV agents, we divided the discussed NAs into four groups. That is, depending on the reported drug research and development (DRD) approach, the NAs are arranged as those found from the following: the repurposing of known NAs, including approved drugs or their prodrugs (approach A); the development of prodrugs of known anti-HCoV NAs (approach B); structural modifications of anti-HCoV NAs, leading to their nonprodrug analogs (approach C); or the development of de novo-designed NAs (approach D). Within each defined approach A, B, C, or D, the NAs are ordered in Table 1, Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7 according to the year of publication. Within a pool of NAs depicted with a general structure, we show the antiviral activity of the most active compound, or the one demonstrating the highest SI. When applicable, International Nonproprietary Names (INNs) or abbreviations of NAs are given in order to facilitate their identification. The abovementioned NAs with well-recognized activity and their mode of action against HCoVs (Figure 1) are included in this review in order to maintain the clarity of the collected data. Owing to the fact that some of these compounds have been often employed as reference compounds in antiviral assays, only their effective concentration values reported in the original publications are cited. The goal of this review is to discuss NAs with anti-HCoV activity in the context of their chemical structure and diversity. The results of studies dealing with the mode of action of the examined compounds are presented in detail.
1.1. The Nucleoside Analogs Evaluated as In Vitro Inhibitors of SARS-CoV Replication
The NAs examined in vitro for their inhibitory activity against SARS-CoV are shown in Table 1. Two DRD approaches were employed in order to find effective agents against SARS-CoV: the repurposing of existing NAs (approach A) and the development of de novo-designed NAs (approach D). Besides NHC (entry 1.11), ribavirin (entry 1.12), remdesivir (entry 1.31), and compounds AT-511 (entry 1.32) and GS-441524 (entry 1.37), the approved nucleoside drugs (zalcitabine, entry 1.3; cidofovir, entry 1.5; mizoribine, entry 1.13; gemcitabine, entry 1.28; or acyclovir, entry 1.30) and their analogs (acyclovir analogs, entry 1.7; tenofovir analogs, entry 1.8; adefovir analogs, entry 1.10; mizoribine analog, entry 1.14; AZT analogs, entries 1.17 and 1.16; clofarabine analog, entry 1.24; and analogs of remdesivir parent nucleoside GS-441524, entry 1.36) represent a relatively large group of the examined compounds. DNA canonical nucleosides (2′-deoxycytidine, entry 1.3 and 2′-deoxyadenosine, entry 1.6) or natural noncanonical nucleosides (pyrazofurin, entry 1.2, and neplanocin A, entry 1.25) were also studied. The other repurposed NAs (approach A) were derived from typical structural modifications, such as (a) the acylation/alkylation of the functional groups in the parent nucleoside (entry 1.3), (b) the incorporation of additional substituent(s) (e.g., an alkyl group or a halogen atom) into the scaffold of the parent nucleoside (entry 1.3), (c) the replacement of the nucleobase nitrogen by a carbon atom (entries 1.4 and 1.6), (d) the replacement of the parent nucleobase or sugar with its synthetic surrogate (entries 1.9, 1.20, 1.22, and 1.27), (e) the “dehydration” of the sugar moiety (entry 1.18), and/or (f) the conjugation of an artificial glycone and an artificial nucleobase (entry 1.29). L-nucleoside analogs have also been reported (entries 1.15, 1.19, 1.21, 1.23, 1.26, and 1.29). However, comparative evaluations of the enantiomeric NAs are infrequent (entries 1.18 vs. 1.19, 1.20 vs. 1.21, and 1.25 vs. 1.26). The de novo-designed NAs (approach D) were developed through (a) the conjugation of a glycone and a nucleobase derived from different parent nucleosides (entry 1.33; ribavirin and neplanocin A, respectively) and the subsequent preparation of the nucleobase-modified analogs; (b) the conjugation of an artificial nucleobase with a furanose (entry 1.35, chloropurine) or the glycone derived from a drug (penciclovir) or its cyclobutyl analog (entry 1.34); or (c) the fluorination of the parent nucleoside (aristeromycin) followed by the preparation of the nucleobase-modified analogs or prodrugs (entry 1.38).
The majority of the NAs evaluated as anti-SARS-CoV agents featured two-digit micromolar activity (for compounds with one-digit micromolar or submicromolar activity, see entries 1.2, 1.3, 1.6, 1.11, 1.13, 1.31, 1.32, 1.37, and 1.38). Among the NAs endowed with submicromolar activity, only fluorinated analogs of aristeromycin (entry 1.38) belong to the group of de novo-designed NAs.

Table 1.
The nucleoside analogs evaluated in vitro against the replication of SARS-CoV.
Table 1.
The nucleoside analogs evaluated in vitro against the replication of SARS-CoV.
| Entry a/DRD Approach, Structure, and Number of Compounds Examined | Effective Concentration Value b; Cell Line; Virus Variant (If Given) Cytotoxic Concentration Value c; Cell Line Selectivity Index d (SI, If Given) | Ref. | ||
|---|---|---|---|---|
| 1.1/A |  | 6-azauridine | EC50 16.8 (2.9) mg/L; Vero; FFM-1 (FFM-2) CC50 104 (18) mg/L; Vero SI 6 | [30] |
| 1.2/A |  | pyrazofurin | EC50 4.2 (0.57) mg/L; Vero; FFM-1 (FFM-2) CC50 52 (9.6) mg/L; Vero SI 12 | [30] |
| EC90 27 μM; Urbani IC90 32 μM; Vero E6 SI 1 | [31] | |||
| 1.3/A |  | R1 = H or DMTr R2 = H or OH R3 = H, OH, OMe R4 = H, Me, Br, Cl R5 = H, Bz, Ac (10 compounds) | R1,R2,R4,R5 = H, R3 = OMe: EC50 4 μM; Vero 76; Urbani IC50 23 μM; Vero 76 SI 6 | [31] |
| 1.4/A |  | 3-deazaneplanocin A | EC50 > 380 μM; Vero E6; Urbani IC50 > 380 μM; Vero E6 | [31] |
| IC50 > 100 μM; Urbani CC50 > 100 μM; Vero 76 | [32] | |||
| 1.5/A |  | cidofovir | EC50 > 360 μM; Vero 76; Urbani IC50 > 360 μM; Vero 76 | [31] |
| 1.6/A |  | R1, R2, R3 = H or OH R4 = OH or NH2 R5 = H or NH2 | 2′-deoxytubercidin (R1,R2 = OH, R3,R5 = H, R4 = NH2, X = CH): EC50 0.1 μM; Vero E6; Urbani IC50 0.4 μM; Vero E6 SI 4 | [31] |
| 1.7/A |  | R1 = CN, CO2H R2 = OH, OAc, OBn, Br, P(O)(OEt)2 (5 compounds) | R1 = CN, R2 = OAc: IC50 15 μg/mL; Vero CC50 56 μg/mL; Vero | [33] |
| 1.8/A |  | X, Y = CH2 or O (2 compounds) | X = CH2, Y = O: IC50 23 μg/mL; Vero CC50 > 75 μg/mL; Vero | [33] |
| 1.9/A |  | R1 = Tol, H, Bz, Ac R2 = H, OH, OAc, OBz, R3 = CN, CO2Me, C(O)NHNH2, C(O)NH2 (7 compounds) | R1,R2 = H, R3 = CO2Me: IC50 18 μg/mL; Vero CC50 > 75 μg/mL; Vero | [33] |
| 1.10/A |  | R1 = CN, CO2H CO2Me, R2 = OH, OAc, P(O)(OH)2 (4 compounds) | R1 = CO2Me, R2 = OH: IC50 28 μg/mL; Vero CC50 > 75 μg/mL; Vero | [33] |
| 1.11/A |  | NHC | EC50 5 μM; Vero E6; Urbani IC50 50 μM; Vero E6 SI 10 | [31] |
| 1.12/A |  | ribavirin | IC50 0.1 ± 0.0 μM; Vero; Urbani CC50 12.5 ± 3.5 μM; Vero 76 SI > 125 | [32] |
| IC50 from 50 to 100 μg/mL; fRhK4; ten clinical isolates CC50 > 1000 μg/mL; fRhK4 SI from 50 to >80 | [34] | |||
| IC50 20 ± 15 μg/mL; Vero E6; Frankfurt-1 CC50 40 μg/mL; Vero E6 (2 virus variants evaluated) | [35] | |||
| IC50 210 μM; Vero 76; Urbani CC50 283 μM; Vero 76 SI 1 | [16] | |||
| IC50 0.14 μM; HAE; GFP | [36] | |||
| 1.13/A |  | mizoribine | IC50 3.5 ± 2.9 μg/mL; Vero E6; Frankfurt-1 CC50 > 200 μg/mL; Vero E6 (2 virus variants) | [35] |
| IC50 > 40 μM; Vero 76; Urbani CC50 > 40 μM; Vero 76 SI 0 e | [16] | |||
| IC50 13.5 μM; Vero E6; Frankfurt-1 CC50 > 700 μM; Vero E6 SI > 52 | [37] | |||
| 1.14/A |  | EICAR | IC50 > 40 μM; Vero 76; Urbani CC50 > 40 μM; Vero 76 SI 0 | [16] |
| 1.15/A |  | B = 5-chlorouracil-1-yl, thymin-1-yl, cytosin-1-yl, uracil-1-yl, adenin-1-yl | B = 5-chlorouracil-1-yl: EC50 < 10 μM; Vero 76 IC50 28 μM; Vero 76 SI > 2.8 | [38] |
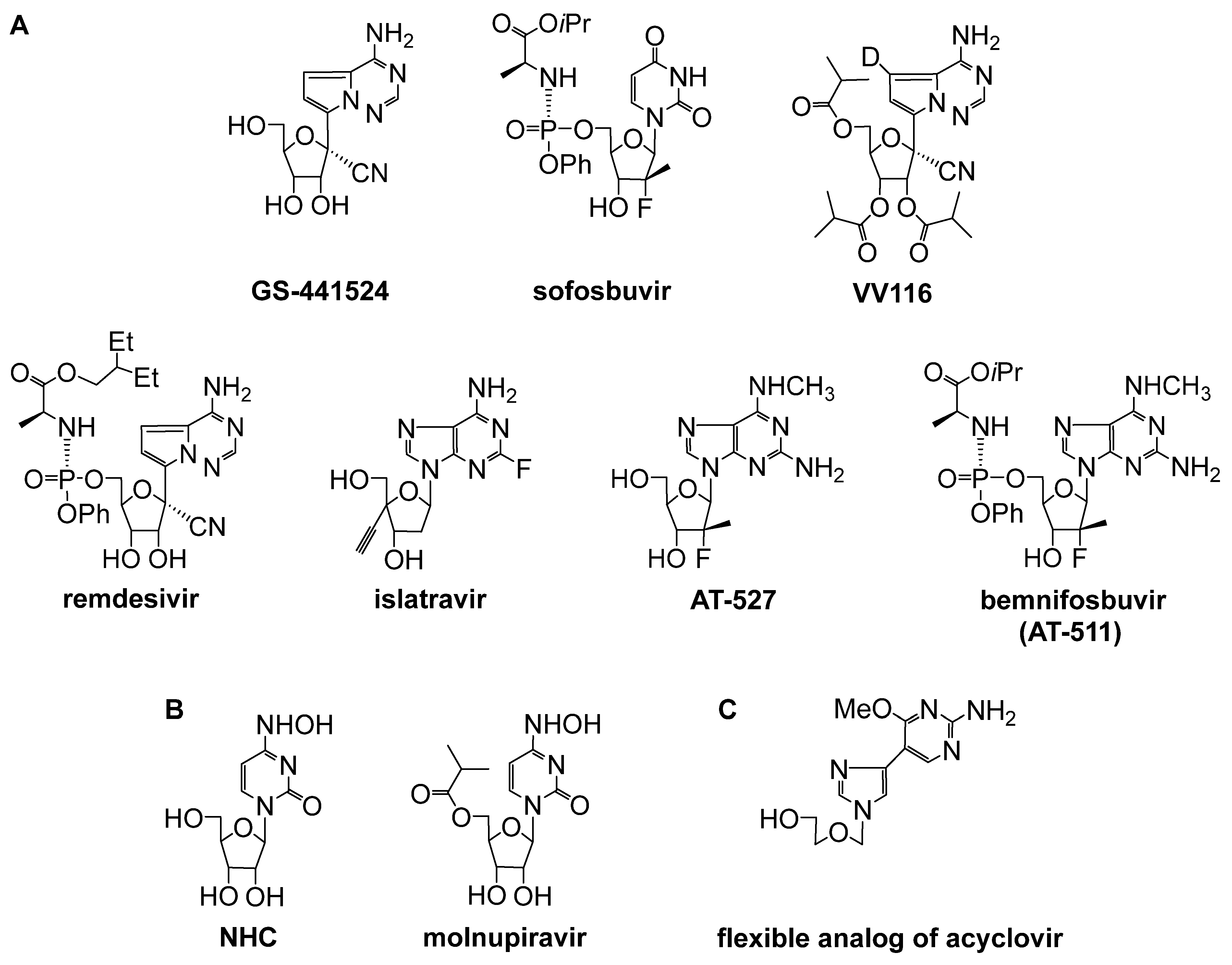
| 1.16/A | 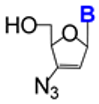 | B = thymin-1-yl, cytosin-1-yl, adenin-1-yl or guanin-1-y | B = thymin-1-yl: EC50 10.3 μM; Vero 76 IC50 16.1 μM; Vero 76 SI 1.6 | [38] |
| 1.17/A |  | B = thymin-1-yl, adenin-1-yl or guanin-1-yl | B = adenin-1-yl: EC50 11.1 μM; Vero 76 IC50 3.5 μM; Vero 76 SI 0 e | [38] |
| 1.18/A | 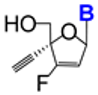 | B = cytosin-1-yl, adenin-1-yl or thymin-1-yl | B = adenin-1-yl: EC50 11.1 μM; Vero 76 IC50 14.5 μM; Vero 76 SI 1.3 | [38] |
| 1.19/A |  | B = adenin-1-yl, thymin-1-yl or cytosin-1-yl | B = adenin-1-yl: EC50 11.9 μM; Vero 76 IC50 25.6 μM; Vero 76 SI 2.2 | [38] |
| 1.20/A |  | B = thymin-1-yl, uracil-1-yl, adenin-1-yl, inosin-1-yl or guanin-1-yl | B = thymin-1-yl: EC50 20 μM; Vero 76 IC50 > 100 μM; Vero 76 SI 5 | [38] |
| 1.21/A |  | B = uracil-1-yl, cytosin-1-yl, adenin-1-yl, inosin-1-yl or guanin-1-yl | B = cytosin-1-yl: EC50 20 μM; Vero 76 IC50 20 μM; Vero 76 SI 1 | [38] |
| 1.22/A |  | dioxolane T (DOT) | EC50 24.5 μM; Vero 76 IC50 > 100 μM; Vero 76 SI 4.1 | [38] |
| 1.23/A |  | EC50 28.6 μM; Vero 76 IC50 > 18.8 μM; Vero 76 SI 0 | [38] | |
| 1.24/A |  | EC50 76.8 μM; Vero 76 IC50 > 100 μM; Vero 76 SI 1.3 | [38] | |
| 1.25/A |  | B = thymin-1-yl, N-OH-cytosin-1-yl, adenin-1-yl (neplanocin A) or inosin-1-yl | EC50 > 100 μM; Vero 76 IC50 > 10 μM; Vero 76 SI 0 | [38] |
| 1.26/A |  | B = thymin-1-yl or inosin-1-yl | EC50 > 100 μM; Vero 76 IC50 > 100 μM; Vero 76 SI 0 | [38] |
| 1.27/A |  | R = OMe or SMe | IC50 > 300 μM; Vero E6; Frankfurt-1 | [37] |
| 1.28/A |  | gemcitabine | EC50 4.957 μM; Vero E6 | [39] |
| 1.29/A e |  | galidesivir (BCX4430, immucillin-A) | EC50 57.7 μM; Urbani CC50 > 296 μM; Vero 76 SI > 5.1 | [40] |
| 1.30/A |  | acyclovir | EC50 > 1000 μM; Vero E6; Frankfurt-1 CC50 > 1000 μM; Vero E6 | [41] |
| 1.31/A |  | remdesivir | IC50 0.069 ± 0.036 μM CC50 > 10 μM; HAE SI > 100 | [42,43] |
| 1.32/A |  | AT-511 (bemnifosbuvir, free form of AT-527) | EC90 0.34 μM CC50 > 86 μM; Huh7 SI > 250 | [44] |
| 1.33/D |  | X, Y = CH or N(3 compounds) | X = CH, Y = N: EC50 21 μM IC50 > 100 μM SI > 4.8 | [45] |
| 1.34/D |  | R = H or Bz X---X = CH-CH or X = CH2 (4 compounds) | R = Bz, X---X = CH-CH: IC50 14.5 μM; Frankfurt-1 CC50 78 μM; Vero E6 SI 5.4 | [37] |
| 1.35/D |  | R1 = H or Bz R2, R3 = H or OH R4 = H or NH2 (5 compounds) | R1,R2, R3 = OH, R4 = H: IC50 48.7 μM; Vero E6; Frankfurt-1 CC50 279 μM; Vero E6 SI 5.7 | [37] |
| 1.36/D |  | R = Me, vinyl, ethynyl | R = vinyl: EC50 14.0 μM; Vero; Toronto-2 CC50 > 30 μM; Vero | [46] |
| 1.37/D |  | GS-441524 | IC50 0.18 ± 0.14 μM; HAE CC50 > 100 μM; HAE | [42,43] |
| EC50 2.4 μM; Vero; Toronto-2 CC50 > 30 μM; Vero | [46] | |||
| 1.38/D e |  | X, Y = H or F B = uracil-1-yl, cytosin-1-yl N6-Me-adenin-1-yl, adenin-1-yl, R = H or 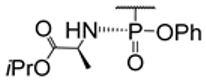 (12 compounds) (12 compounds) | X,Y = F, B = adenin-1-yl, R = H: EC50 0.5 μM; Vero E6; Frankfurt 1 CC50 5.9 μM; Vero E6 SI 11.8 | [47] |
Approach A: repurposing of the known NAs, including approved drugs or their prodrugs. Approach D: development of de novo-designed NAs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed. b Exemplified as the effective compound concentration (EC50 or EC90 or IC50). c Exemplified as the cytotoxic compound concentration (CC50 or IC50). d Defined as the ratio of the compound cytotoxic concentration and the compound effective concentration. Only values explicitly given by the authors are cited. Values SI 0 are given according to the authors. e See text for details.
Galidesivir was reported by Bavari and coworkers to be active against a wide range of ssRNA(+) and ssRNA(−) viruses, with its inhibitory effect against SARS-CoV (entry 1.29, EC50 57.7 μM, CC50 > 296 μM) and MERS-CoV (entry 3.2, EC50 68.4 μM, CC50 > 100 μM) [40]. Galidesivir’s mechanism of action was postulated to involve its rapid phosphorylation to galidesivir–triphosphate, followed by the pyrophosphate cleavage and incorporation of galidesivir–monophosphate into nascent viral RNA strands. Experiments with HCV RNA polymerase supported this hypothesis and showed that galidesivir targeted the RNA-dependent RNA polymerase (RdRp) of RNA viruses. It also inhibited the polymerase function by inducing nonobligate RNA chain termination. It was suggested that the termination occurred two bases after the incorporation of galidesivir–monophosphate. An incorporation of galidesivir–monophosphate into human RNA or DNA was not observed. A clinical trial of galidesivir for the treatment of yellow fever or COVID-19 was initiated in April 2020 in Brazil [48]. The results from the trial in COVID-19 patients showed that galidesivir was safe and well tolerated. The therapy was associated with a dose-dependent decline in viral levels of SARS-CoV-2 in the respiratory tract. However, no clinical efficacy benefit with galidesivir treatment compared to a placebo treatment was observed in the trial. Thus, a discontinuation of the pursuit of a COVID-19 indication for galidesivir was announced [49]. The potential of galidesivir against SARS-CoV-2 has been further explored (entry 2.13), including efficacy studies in an animal model [50].
Besides the 1′-branched NAs (entry 1.36) designed upon the development of the remdesivir parent nucleoside GS-441524 (entry 1.37), the NAs designed de novo in search for anti-SARS-CoV agents (approach D) include the following: the azole analogs of neplanocin A (entry 1.33), cyclic or acyclic 6-chloropurine analogs (entries 1.34 and 1.35), and the 6′-fluoro derivatives of (−)-aristeromycin (entry 1.38). Among the 6′-fluoro analogs of (−)-aristeromycin (entry 1.38), the 6′,6′-difluoro analog (X,Y = F; B = adenin-1-yl) displays submicromolar activity against the selected +RNA viruses, including SARS-CoV (entry 1.38, EC50 0.5 μM) and MERS-CoV (entry 3.10, EC50 0.2 μM) [47]. Structure–activity relationship (SAR) studies showed that an adenin-1-yl ring and two fluorine atoms were required for anti-SARS-CoV activity. However, Calu-3-based assays revealed a significant decrease in the activity of this compound, not only towards MERS-CoV but also towards SARS-CoV and SARS-CoV-2. Interestingly, the phosphoramidate prodrug of the most active compound (X,Y = F; B = adenin-1-yl) was less antivirally active (SARS-CoV, EC50 6.8 μM) and more toxic towards the host Vero E6 cells. The antiviral activity of this compound was postulated to be due to an (indirect) effect on the viral MTase activity through the inhibition of host SAH hydrolase. Preliminary assays also identified the in vitro activity of this compound against SARS-CoV-2 (entry 2.39, EC50~2 μM). Regardless of the cell line, the monophosphoramidate prodrug of the 6′,6′-difluoro analog (X,Y = F; B = adenin-1-yl) was also less active against MERS-CoV (EC50 9.3 μM) than its parent compound [51]. The 6′,6′-difluoro analog (X,Y = F; B = adenin-1-yl) demonstrated inhibition at an early stage of MERS-CoV replication. However, the development of MERS-CoV mutants strongly resistant to this compound was observed.
1.2. The Nucleoside Analogs Evaluated as In Vitro Inhibitors of SARS-CoV-2 Replication
The NAs examined in vitro for their inhibitory activity against SARS-CoV-2 are collected in Table 2. Considering that SARS-CoV-2 emerged just a short time ago, the known NAs (approach A, entries 2.1–2.30) are the broadest group of the assayed compounds. Among them, compounds endowed with submicromolar activity belong to the following groups: (a) approved drugs (gemcitabine, entry 1.19; or forodesine, entry 1.26); (b) natural noncanonical nucleosides (tubercidin and its derivatives, entries 2.10, 2.13–2.16, and 2.31; sangivamycin, entry 2.22; or riboprine, entry 2.25); and (c) drug metabolites (thioguanosine, entry 2.18) or their analogs (entry 2.20). Studies on the novel prodrugs of GS-441524 or NHC (approach B, entries 2.32–2.35) have been reported since 2021. These prodrugs were the 5′-O-phosphoric/carboxylic esters of GS-441524 (entries 2.34 and 2.33, respectively) or the 4-O-carboxylic/carbonic esters of NHC (entry 2.35). Among them, compounds with submicromolar activity were found. The structural modifications of the anti-HCoV NAs leading to their nonprodrug analogs (approach C) involved the fluorination or deuteration of the nucleobase in GS-441524 (entry 2.36 and 2.37, respectively) or the fluorination of the sugar moiety in NHC (entry 2.38). Among these modifications, the deuteration of GS-441524 was the most successful. The isobutyric triester of the 7-deutero derivative showed submicromolar activity (entry 2.37). Very few de novo-designed NAs (approach D) are represented by the 6′,6′-difluoro derivatives of (−)-aristeromycin (entry 2.39) and the 2′-branched analogs of the selected canonical nucleosides (entry 2.40). These compounds showed moderate inhibitory activity against SARS-CoV-2 in vitro, albeit accompanied by considerable cytotoxicity.

Table 2.
The nucleoside analogs evaluated in vitro against the replication of SARS-CoV-2.
Table 2.
The nucleoside analogs evaluated in vitro against the replication of SARS-CoV-2.
| Entry a/DRD Approach, Structure and Number of Compounds Examined | Effective Concentration Value b; Cell Line; Virus Variant (If Given) Cytotoxic Concentration Value c; Cell Line Selectivity Index d (SI, If Given) | Ref. | ||
|---|---|---|---|---|
| 2.1/A |  | NHC | IC50 0.08 μM; Calu-3 | [36] |
| EC50 0.3 ± 0.2 μM; Vero CC5012.6 μM; Vero SI 42 | [52] | |||
| 2.2/A |  | GS-441524 | EC50 8.2 ± 0.4 μM; Vero CC50 > 100 μM; Vero SI 12 | [52] |
| EC50 0.59 ± 0.10 μM); Vero E6 CC50 > 500 μM; Vero E6 SI > 847.46 (2 virus variants evaluated) | [53] | |||
| EC50 0.10 μM; Calu-3; WA1 CC50 > 50 μM; Calu-3 SI > 524 | [54] | |||
| 2.3/A |  | remdesivir | EC50 1.0 ± 0.1 μM; Vero CC50 > 100 μM; Vero SI > 100 | [52] |
| 2.4/A |  | R1 = Me, Cl, Br R2, R3 =  or or R2 = H and R3 =  (4 compounds) | R1 = Me, R2, R3 =  : :EC50 6.3 ± 0.1 μM; Vero CC50 16.4 μM; Vero | [52] |
| 2.5/A | 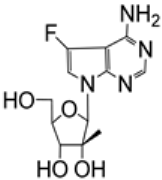 | EC50 7.6 ± 0.4 μM; Vero CC50 > 100 μM; Vero SI > 10 | [52] | |
| 2.6/A |  | 2′-MeC: R1 = H, R2 = OH, R3 = Me ALS-8112: R1 = CH2Cl, R2 = F, R3 = H | 2′-MeC: EC50 9.2 ± 0.1 μM; Vero CC50 > 100 μM; Vero SI > 10 | [52] |
| 2.7/A |  | entecavir | EC50 > 20 μM; Vero CC50 > 20 μM; Vero | [52] |
| 2.8/A |  | lamivudine: R = H emtricitabine: R = F | lamivudine and emtricitabine EC50 > 20 μM; Vero CC50 > 20 μM; Vero | [52] |
| 2.9/A |  | tenofovir alafenamide: R =  tenofovir isoproxil: R = 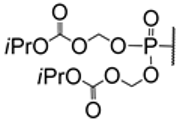 | tenofovir alafenamide and tenofovir isoproxil: EC50 > 20 μM; Vero CC50 > 20 μM; Vero | [52] |
| 2.10/A d |  | tubercidin | EC50 0.77 μM; Vero E6 CC50 > 100 μM; Vero E6 SI 129.87 | [55] |
| EC90 0.087 μM; Caco-2; WK-521 CC50 0.66 μM; MRC5 | [56] | |||
| EC50 0.05 μM; Calu-3; WA1 CC50 9.36 μM; Calu-3 SI 190 | [54] | |||
| 2.11/A |  | penciclovir | EC50 95.96 μM; Vero E6 CC50 > 400 μM; Vero E6 SI 4.17 | [55] |
| 2.12/A |  | AT-511 (bemnifosbuvir, free form of AT-527) | EC90 0.47 ± 0.12 μM; HAE CC50 > 86 μM; HAE SI > 160 | [44] |
| 2.13/A e | 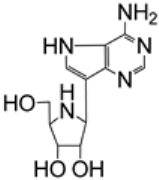 | galidesivir (BCX4430, immucillin-A) | EC50 14.15 μM; Calu-3; WA1/2020 CC50 > 50 μM; Calu-3 SI > 3.5 | [50] |
| 2.14/A e | 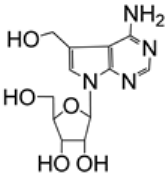 | 5-hydroxymethyltubercidin | EC90 0.57 μM; Caco-2; WK-521 CC50 > 50 μM; MRC5 | [56] |
| 2.15/A e |  | R1 = H or CN R2 = H, CH=NOH, NO2 Me, CN (5 compounds) | R1 = H, R2 = CH=NOH: EC90 0.20 μM; Caco-2; WK-521 CC50 0.74 μM; MRC5 | [56] |
| 2.16/A e |  | EC90 3.5 μM Caco-2; WK-521 CC50 24 μM; MRC5 | [56] | |
| 2.17/A |  | R = H or  | Sofosbuvir, R =  : :EC50 5.1 ± 0.8 μM, Huh-7; clinic isolate CC50 381 ± 34 μM, Huh-7 SI 74 | [57] |
| 2.18/A | 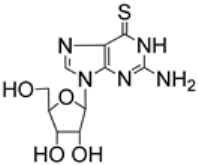 | thioguanosine | EC50 0.04 μM; Calu-3; WA1 CC50 1.14 μM; Calu-3 SI 26 | [54] |
| IC50 0.78 µM, Caco-2; clinic isolate | [58] | |||
| 2.19/A |  | gemcitabine | EC50 0.04 μM; Calu-3; WA1 CC50 > 20 μM; Calu-3 SI > 1235 | [54] |
| 2.20/A |  | R1 = H or OH R2 = H or NH2 (2 compounds) | 6-thioinosine (R1 = OH, R2 = H): EC50 0.05 μM; Calu-3; WA1 CC50 > 50 μM; Calu-3 SI > 1083 | [54] |
| 2.21/A |  | 3-deazaneplanocin A | IC50 579 nM; SARS-CoV-2; A549-nRFP-ACE2; SARS-CoV-2-MUC-IMB-1 (in combination with remdesivir) | [59] |
| 2.22/A e |  | sangivamycin | EC50 15.4 nM; Vero E6; B.1.1.7 (Alpha) SI 39.0 (8 virus variants evaluated) | [60] |
| 2.23/A e |  | cordycepin | EC50 2.01 ± 0.12 μM; Vero E6; VOC-202012/01 CC50 > 100 μM; Vero E6 | [61] |
| 2.24/A e |  | didanosine | EC50 3.10 ± μM; Vero E6; VOC-2020/01 CC50 > 100 μM; Vero E6 | [62] |
| 2.25/A e | 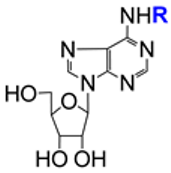 | riboprine: R = 3-methylbut-2-en-1-yl tecadenoson: R = (R)-tetrahydrofuran-3-yl | riboprine: EC50 408 ± 22 nM; Vero E6; Omicron B.1.1.529/BA.5; CC50 > 100 μM; Vero E6 (3 virus variants evaluated) | [63,64,65,66] |
| 2.26/A e |  | forodesine | EC50 657 ± 34 nM; Vero E6; Omicron B.1.1.529/BA.5 CC50 > 100 μM; Vero E6 (3 virus variants evaluated) | [63,64,65,66] |
| 2.27/A e |  | maribavir | EC50 3000 ± 131 nM; Vero E6; Omicron B.1.1.529/BA.5 CC50 > 100 μM; Vero E6 (3 virus variants evaluated) | [63,64,65] |
| 2.28/A |  | nelarabine: R1 = OMe, R2 = NH2vidarabine: R1 = NH2, R2 = H | nelarabine: EC50 1656 ± 71 nM; Vero E6; Omicron B.1.1.529/BA.5 CC50 > 100 μM; Vero E6 (3 virus variants evaluated) | [63,64,65] |
| 2.29/A | 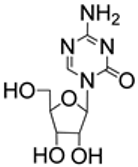 | 5-azacytidine | EC50 2.63 μM; Calu-3; Wuhan/WIV0 4/2019 CC50 25.40 μM; Calu-3 SI 9.66 | [67] |
| 2.30/A/B e |  | azvudine: R = H CL-236: R =  | CL-236: EC50 1.2 μM; Calu-3; BetaCoV/Wuhan/WIV04/2019 CC50 > 102.4 μM; Calu-3 SI 83 | [68,69,70] |
| 2.31/A/D e |  | R1 = H, R2 = furan-2-yl: EC50 0.1 ± 0.03 μM; Huh7; hCoV-19/Czech Republic/NRL_6632_2/2020 CC50 11 ± 2.5 μM; Huh7 | [71] | |
| R2 = Me, vinyl, ethynyl, cyclopropyl, furan-2-yl, thiophen-2-yl, Ph, benzofuran-2-yl, benzothiophen-2-yl, naphtalen-2-yl R2 = H,  (proTide), (proTide),  (bis(SATE), (bis(SATE),  (mono(SATE) (mono(SATE)(25 compounds) | ||||
| 2.32/B |  | molnupiravir | EC50 0.10 μM; Calu-3; WA1 CC50 > 50 μM; Calu-3 SI > 500 | [36,54] |
| 2.33/B e |  | R = 4-oxaicosan-1-yl, 3-oxahenicosan-1-yl, 2-OBn-4-oxadocosan-1-yl | R = 2-OBn-4-oxadocosan-1-yl: EC50 0.14 μM; Vero E6; USA-WA1/2020 CC50 97.9 μM; Vero E6 SI 699 | [72,73] |
| 2.34/B e |  | R1 = alkyl, cycloalkyl, CF3CH2, C6H5, α-aminoalkyl R2, R3, R4 = H, acetyl, isobutyryl (21 compounds) | obeldesivir (ATV006, R1 = isopropyl; R2,R3,R4 = H): EC50 0.106 μM; Vero E6; Omicron, B.1.1.529 CC50 128.00 μM; Vero E6 SI 1207.55 (4 virus variants evaluated) | [74] |
| 2.35/B |  | X = O or CH2 R1 = OH or F R2 = C10H21, C12H23, C14H29 (5 compounds) | X = CH2, R = C14H29: EC50 0.31 μM; Vero; B1.1 529 BA.1 (Omicron); CC50 6.32 μM; Vero (5 virus variants evaluated) | [75] |
| 2.36/C |  | EC50 3.30 μM; Vero E6 (2 virus strains evaluated) | [53] | |
| 2.37/C e |  | R1,R2,R3 = H or isobutyryl (4 compounds) | VV116 (R1,R2,R3 = isobutyryl; hydrobromide): EC50 0.35 ± 0.09 µM; Vero E6 CC50 289.19 ± 15.39 μM; Vero E6 (2 virus variants evaluated) | [53] |
| 2.38/C |  | EC50 29.91 μM; Vero; B1.1 529 BA.1 (Omicron); CC50 > 100 μM; Vero (5 virus variants evaluated) | [75] | |
| 2.39/D |  | EC50~2 μM; Calu-3 CC50 > 25 μM; Calu-3 SI > 12 μM | [47] | |
| 2.40/D | 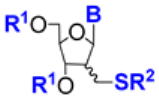 | B = thymin-1-yl, uracil-1-yl, N6-DMTr-adenin-1-yl R1 = TBDMS or (iPr)2Si-O-Si(iPr)2 R2 = C4H9, Ac, pyranos-1-yl (11 compounds) | B = thymin-1-yl, R1 = (iPr)2Si-O-Si(iPr)2, araR2 = C4H9: EC50 17 μM; Vero E6; MOI 0.05 CC50 16.6 μM; Vero E6 | [76] |
Approach A: repurposing of known NAs, including approved drugs or their prodrugs. Approach B: development of prodrugs of known anti-HCoV NAs. Approach C: structural modifications of the anti-HCoV Nas, leading to their nonprodrug analogs. Approach D: development of de novo-designed NAs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed. b Exemplified as the effective compound concentration (EC50 or EC90 or IC50). c Exemplified as the cytotoxic compound concentration (CC50). d Defined as a ratio of the compound cytotoxic concentration and the compound effective concentration. Only values explicitly given by the authors are cited. e See text for details.
Uemura et al. examined the in vitro activity of tubercidin and its derivatives against SARS-CoV-2 (entries 2.14–2.16), HCoV-OC43 (entries 4.12–4.14), and HCoV-229E (entries 5.8–5.10) [56]. Four of the tested compounds showed submicromolar activity: tubercidin, 5-hydroxymethyltubercidin, 5-((hydroxyimino)methyl)tubercidin, and 5-nitrotubercidin. However, only 5-hydroxymethyltubercidin exhibited both advantageous activity and relatively low cytotoxicity (entries 2.14, 4.13, and 5.9). 5-Hydroxymethyltubercidin inhibited the replication of the examined viruses in a dose-dependent manner. Further experiments employing ZIKV RdRp suggested that 5-hydroxymethyltubercidin was incorporated into viral RdRp and inhibited the synthesis of the viral RNA by chain termination [56].
Sangivamycin is a natural nucleoside analog endowed with anticancer, antibiotic, and antiviral activity. Recently, the anti-SARS-CoV-2 properties of sangivamycin were reported by Bennett et al. (entry 2.22) [60]. When in vitro assayed against eight variants of SARS-CoV-2, IC50 values in the low nanomolar range (14–82 nM) with the highest SI value > 39.0 for the B.1.1.7 (Alpha) variant in Vero E6/TMPRSS2 cells were obtained. The activity of sangivamycin was cell-dependent but significantly lower than that of remdesivir found in the same experiments in all the host cells employed (Vero cells, Caco-2 cells, and Calu-3 cells). These preliminary studies showed an additive effect of sangivamycin and remdesivir. The mechanism of action of sangivamycin was not established. Tonix Pharmaceuticals announced that it would develop sangivamycin for the treatment of COVID-19 [77], emphasizing its good tolerability in patients subjected to the previous clinical trials [60].
Studies on cordycepin (3′-deoxyadenosine, entry 2.23) were triggered by a recently published in silico computational docking approach [78]. The results of this simulation strongly suggested that cordycepin interacted with the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). Indeed, it was shown that cordycepin strongly inhibited virus multiplication in vitro, with potency comparable to that of remdesivir [61]. Rabie proposed that the mechanism of inhibition of virus replication originated from the similarity of cordycepin and adenosine. During viral replication, cordycepin triphosphate was incorporated instead of adenosine triphosphate into the virus RNA by the RdRp. This incorporation resulted in errors in the viral genetic code. As a consequence, a large number of mutations occurred (lethal mutagenesis). In a series of follow-up papers by Rabie and coworkers, using the nucleoside analogism approach, didanosine (2′,3′-dideoxyinosine, entry 2.24) [62], riboprine, tecadenoson (entry 2.25) [63,64,65,66], forodesine (entry 2.26) [63,64,65,66], and maribavir (entry 2.27) [63,64,65] were studied and found to strongly inhibit the replication of SARS-CoV-2. The in silico computational study suggested that these NAs are strongly bound to the main active site of the RdRp molecule or to the 3′-to-5′ exoribonuclease.
Azvudine (entry 2.30, R = H) is the first Chinese oral anti-COVID-19 drug [70,79]. This compound is almost inactive in vitro, but it is intracellularly phosphorylated to the triphosphate that displays broad-spectrum activity by inhibiting the SARS-CoV-2 RdRp. For research purposes, in order to better investigate the activity in vitro, a monophosphate prodrug of azvudine, CL-236 (entry 2.30), was also synthesized and tested. Conventional experiments proved the anticoronavirus properties of CL-236 (see also Table 4, entry 4.15) [69]. The distribution and phosphorylation in vivo of azvudine, as well as its metabolic pathway, were investigated in rats. While azvudine was detectable after oral administration in all tested organs, its triphosphate was observed only in the thymus. The clinical benefits of azvudine were demonstrated in experiments with infected rhesus macaques. The efficacy and safety of azvudine in patients with mild and common COVID-19 were evaluated in a clinical trial in China (ChiCTR2000029853) [68].
Lipid prodrugs of GS-441524 were developed as potential alternatives to remdesivir in order to overcome its metabolic instability (entry 2.33) [72]. The most active prodrug (R = 2-OBn-4-oxadocosan-1-yl) showed activity against two HCoVs (SARS-CoV-2, entry 2.33; and HCoV-229E, entry 5.12) and a variety of RNA viruses with excellent SIs ranging from 295 to 699 [72,73]. The active compound showed cell-dependent activity against SARS-CoV-2. It was significantly higher than that of remdesivir in Vero cells and equivalent to that of remdesivir in PSC-human lung cells, Calu-3 cells, Huh7.5 cells, and Caco-2 cells. In vivo studies revealed the stability of this compound in plasma for 24 h and its good tolerability during a 7-day treatment (Syrian hamsters). In contrast, the prodrug (R = 2-OBn-4-oxadocosan-1-yl) was less active against HCoV-229E than remdesivir.
Among the 21-member set of mono-, di-, tri-, or tetraacylated derivatives of GS-441524 (entry 2.34), Cao et al. identified the 5′-O-esters (R1,R2 = H, R3 = prop-2-yl, prop-1-yl, oct-1-yl, pent-3-yl, phenyl, 2-methylprop-1-yl) as endowed with higher in vitro replicon activity against SARS-CoV-2 [EC50 0.217 µM (R3 = prop-1-yl) − 0.521 (R3 = prop-2-yl) µM] then the parent GS-441524 (EC50 0.955 µM, HEK 293T cells) [74]. Moreover, the isobutyric acid ester (R3 = prop-2-yl, obeldesivir) and the nonanoic acid ester (R3 = oct-1-yl) showed higher in vitro activity than GS-441524 or remdesivir against the selected variants of SARS-CoV-2 in Vero E6 cells. However, only obeldesivir featured a satisfactory SI (1207.55, Omicron variant in Vero E6 cells). It met standards as an oral drug candidate because, compared with the parent GS-441524, it (a) featured acceptable water solubility (686.02 µg/mL), (b) reached higher oral bioavailability in animal models (macaques, about 30% vs. 8.3%, respectively), (c) achieved a broad distribution in the major target organs (C57BL/6 mice) for SARS-CoV-2 infection, and (d) showed potent anti-SARS-CoV-2 efficacy in different mouse models. Currently, obeldesivir is under clinical trials in participants that have a high risk of getting a serious COVID-19 illness (ClinicalTrials.gov Identifier: NCT05603143).
Xie et al. synthesized several derivatives of GS-441524 by introducing halogen, hydroxyl, or cyano groups at the purine 7-position (entry 2.36) [53]. Further modifications involved deuterated analogs, which, due to some synthetic problems, were obtained only at the 7-position. In this way, the compound VV116 (entry 2.37) was identified as a promising candidate. It was used as a hydrobromide salt for its stability and improved bioavailability compared to the other compounds in the series. When orally administered in mice, it showed dose-dependent efficacy. A phase 3 observer-blinded, randomized trial showed that VV116 was noninferior to paxlovid (recommended by the World Health Organization for treating mild-to-moderate COVID-19) with respect to the time to sustained clinical recovery and had fewer safety concerns [80].
Hocek and coworkers evaluated a broad panel of 7-substituted 7-deazapurine ribonucleosides, monophosphate prodrugs, and triphosphates against emerging RNA viruses, including SARS-CoV-2 (entry 2.31) [71]. The nucleosides (R1 = H) containing R2 = ethynyl, furany-2-yl, benzofurany-2-yl, benzothiophen-2-yl, or naphtalen-2-yl showed micromolar or submicromolar anti-SARS-CoV-2 activity in vitro and relatively low toxicity to the host Huh7 cells. Their prodrugs (R1 = proTide, mono(SATE) or bis(SATE)) were comparable (R1 = bis(SATE), R2 = benzofurany-2-yl) or less active in these assays. Comparative studies on the parent nucleoside (R1 = H, R2 = benzofurany-2-yl) and its prodrugs (R1 = proTide or bis(SATE)) showed that the bis(SATE) prodrug (R1 = bis(SATE)) was the best form for the cellular membrane transport and intracellular release of the nucleoside monophosphate (R1 = P(O)(OH)O−, R2 = benzofurany-2-yl) (Huh7 cells). In light of these observations, the comparable or somewhat lower in vitro activity of this prodrug than that of its parent nucleoside (R1 = H, R2 = benzofurany-2-yl) was suggested to be due to the plausible alternative mechanisms of action of the nucleoside (R1 = bis(SATE), R2 = benzofurany-2-yl), which might not involve activation through phosphorylation.
1.3. The Nucleoside Analogs Evaluated as In Vitro Inhibitors of MERS-CoV Replication
In general, the number of reported studies on NAs as potential inhibitors of MERS-CoV replication (Table 3) is much smaller than the number of those focused on SARS-CoV or SARS-CoV-2. The majority of these studies dealt with the repurposing of known NAs (approach A, entries 3.1–3.9). Among these compounds, NHC (entry 3.1) and remdesivir (entry 3.5) showed the highest activity in vitro with good SI values. The 6′-fluorinated analogs of (−)-aristeromycin (entry 3.10) are the only de novo-designed NAs. They showed promising one-digit micromolar activities, although accompanied by appreciable cytotoxicity (for details, see also entry 1.38).

Table 3.
The nucleoside analogs evaluated in vitro against replication of MERS-CoV.
Table 3.
The nucleoside analogs evaluated in vitro against replication of MERS-CoV.
| Entry a/DRD Approach, Structure, and Number of Compounds Examined | Effective Concentration Value b; Cell Line; Virus Variant (If Given) Cytotoxic Concentration Value c; Cell Line Selectivity Index d (SI, If Given) | Ref. | ||
|---|---|---|---|---|
| 3.1/A |  | ribavirin | IC50 9.99 ± 2.97 μg/mL; Vero CC50 > 1660 μg/mL; Vero SI > 152.98 | [81] |
| 3.2/A e |  | galidesivir (BCX4430, immucillin-A) | EC50 68.4 μM; Vero E6; Jordan N3 CC50 > 100 μM; Vero E6 SI > 1.5 | [40] |
| 3.3/A |  | R1 = H, Me R2 = NH2 (3 compounds) | R1 = Me, R2 = NH2: EC50 23 ± 0.6 μM; Vero; EMC/2012 CC50 71 ± 14 μM; Vero | [41] |
| 3.4/A |  | acyclovir | EC50 > 1000 μM; Vero; EMC/2012 CC50 > 1000 μM; Vero | [41] |
| 3.5/A |  | remdesivir | EC50 0.34 μM; Vero CC50 > 20 μM; Vero | [82] |
| IC50 0.025 μM; Calu-3 CC50 > 10 μM; Calu-3 SI > 400 | [42] | |||
| IC50 0.074 ± 0.023 μM; HAE CC50 > 10 μM; HAE SI > 100 | [42,43] | |||
| 3.6/A |  | GS-441524 | IC50 0.86 ± 0.78 μM; HAE CC50 > 100 μM; HAE | [42,43] |
| 3.7/A |  | NHC | IC50 0.56 μM; DBT-9 CC50 > 200 µM; DBT-9 | [83] |
| IC50 0.024 μM; HAE; RFP (2 virus variants evaluated) | [36] | |||
| 3.8/A |  | gemcitabine | EC50 1.216 μM; Vero E6 | [39] |
| 3.9/A |  | AT-511 (bemnifosbuvir, free form of AT-527) | EC90 37 ± 28μM; Huh7 CC50 > 86 μM; Huh7 SI > 3.3 | [44] |
| 3.10/D |  | X = H or F Y = H or F B = uracil-1-yl, cytosin-1-yl, adenin-1-yl, N6-Me-adenin-1-yl, R = H or  (14 compounds) | X,Y = F, B = adenin-1-yl, R = H: EC50 0.2 μM; Vero E6; EMC/2012 CC50 3.2 μM; Vero E6 SI 16 | [47,51] |
Approach A: repurposing of known NAs, including approved drugs or their prodrugs. Approach D: development of the de novo-designed NAs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed. b Exemplified as the effective compound concentration (EC50 or EC90 or IC50). c Exemplified as the cytotoxic compound concentration (CC50). d Defined as the ratio of the compound cytotoxic concentration and the compound effective concentration. Only values explicitly given by the authors are cited. e See text for details.
1.4. The Nucleoside Analogs Evaluated as In Vitro Inhibitors of HCoV-OC43 Replication
The number of studies on the potential of NAs to inhibit the replication of HCoV-OC43 is relatively limited (Table 4). All assays found in the literature featured repurposed known NAs (approach A, entries 4.1–4.14), with CL-236 (the prodrug of azvudine, approach B) as the only exception (entry 4.15, for details see also entry 2.30). Among the repurposed compounds, remdesivir was reported to show high activity in vitro (entry 4.1), although accompanied by considerable cytotoxicity in Huh7 cells. The compounds NHC (entry 4.2), AT-511 (entry 4.11), and 7-hydroxymethytubercidin (entry 4.13; for details, see also entry 2.14) displayed submicromolar activities and relatively high SI values ranging from 100 (NHC) to >170 (AT-511). Lamivudine and emtricitabine are the only examples of noncanonical L-nucleosides among the repurposed NAs (entry 4.8).

Table 4.
The nucleoside analogs evaluated in vitro against the replication of HCoV-OC43.
Table 4.
The nucleoside analogs evaluated in vitro against the replication of HCoV-OC43.
| Entry a/DRD Approach, Structure, and Number of Compounds Examined | Effective Concentration Value b; Cell Line; Virus Variant (If Given) Cytotoxic Concentration Value c; Cell Line Selectivity Index d (SI, If Given) | Ref. | ||
|---|---|---|---|---|
| 4.1/A | 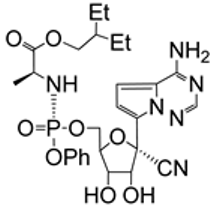 | remdesivir | EC50 0.04 ± 0.1 μM Huh7 CC50 2.1 μM; Huh7 SI 52 | [52] |
| 4.2/A | 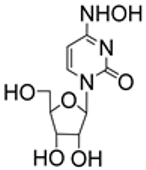 | NHC | EC50 0.8 ± 0.03 μM; Huh7 CC50 80.3 μM; Huh7 SI 100 | [52] |
| 4.3/A |  | R1 = Me, Cl, Br R2, R3 =  or R2 = H and or R2 = H andR3 =  (4 compounds) | R2 = H, R1 = Cl, R3 =  : :EC50 5.9 ± 0.6 μM; Huh7 CC50 > 100 μM; Huh7 | [52] |
| 4.4/A |  | EC50 6.7 ± 1.1 μM; Huh7 CC50 38 μM; Huh7 | [52] | |
| 4.5/A | 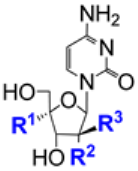 | 2′-MeC: R1 = H, R2 = OH, R3 = Me ALS-8112: R1 = CH2Cl, R2 = F, R3 = H | 2′-MeC: EC50 10 ± 0.7 μM; Huh7 CC50 > 100 μM; Huh7 | [52] |
| 4.6/A |  | entecavir | EC50 > 20 μM; Vero CC50 > 20 μM; Vero | [52] |
| 4.7/A |  | GS-441524 | EC50 > 10 μM; Vero CC50 13.2 ± 0.8 μM; Vero | [52] |
| 4.8/A |  | lamivudine: R = H emtricitabine: R = F | Lamivudine and emtricitabine: EC50 > 20 μM; Vero CC50 > 20 μM; Vero | [52] |
| 4.9/A |  | tenofovir alafenamide: R =  tenofovir isoproxil: tenofovir isoproxil: R =  | EC50 > 20 μM; Vero CC50 > 20 μM; Vero | [52] |
| 4.10/A |  | ribavirin | EC50 20.6 μM; Huh7 CC50 22.1 μM; Huh7 | [52] |
| 4.11/A |  | AT-511 (bemnifosbuvir, free form of AT-527) | EC90 0.5 μM; Huh7 CC50 > 86 μM; Huh7 SI > 170 (2 virus variants evaluated) | [44] |
| 4.12/A |  | tubercidin | EC50 0.11 μM; MRC5 CC50 0.66 μM; MRC5 | [56] |
| 4.13/A |  | 5-hydroxymethyltubercidin | EC50 0.378 ± 0.023 μM; MRC5 CC50 > 50 μM; MRC5 SI > 132 | [56] |
| 4.14/A |  | R1 = H or CN R2 = H, CH=NOH, NO2 Me, CN (5 compounds) | R1 = H, R2 = CH=NOH: EC90 0.055μM; MRC5 CC50 0.74 μM; MRC5 | [56] |
| 4.15/B |  | CL-236 | EC50 1.2 μM; H460; VR1558 SI 20 | [68] |
Approach A: repurposing of known NAs, including approved drugs or their prodrugs. Approach B: development of prodrugs of known anti-HCoV NAs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed. b Exemplified as the effective compound concentration (EC50 or EC90). c Exemplified as the cytotoxic compound concentration (CC50). d Defined as the ratio of the compound cytotoxic concentration and the compound effective concentration. Only values explicitly given by the authors are cited.
1.5. The Nucleoside Analogs Evaluated as In Vitro Inhibitors of HCoV-229E Replication
The number of reports on nucleoside inhibitors of HCoV-229E is rather limited (Table 5). The repurposing of known NAs (approach A, entries 5.1–5.11) or the examination of prodrugs of known anti-HCoV NAs (approach B, entry 5.12) were the major DRD approaches. Besides remdesivir (entry 5.1) and the compound AT-511 (entry 5.7), the derivatives of tubercidin (entries 5.8–5.10, for details, see also entries 2.10 and 2.14–2.16) and the 5′-O-lipid prodrugs of GS-441524 (entry 5.12, for details, see also 2.34) showed promising submicromolar activities. Among them, 7-hydroxymethyltubercidin featured the highest SI value (entry 5.9, SI > 94). The de novo-designed nucleoside analogs (approach D) were the 1,2,3-triazole NAs (entry 5.13) and the 2′-branched nucleosides (entry 5.14). These compounds showed moderate anti-HCoV-229E activity and suffered from appreciable cytotoxicity towards the host HEL cells.

Table 5.
The nucleoside analogs evaluated in vitro against the replication of HCoV-229E.
Table 5.
The nucleoside analogs evaluated in vitro against the replication of HCoV-229E.
| Entry a/DRD Approach, Structure, and Number of Compounds Examined | Effective Concentration Value b; Cell Line; Virus Variant (If Given) Cytotoxic Concentration Value c; Cell Line Selectivity Index d (SI, If Given) | Ref. | ||
|---|---|---|---|---|
| 5.1/A e |  | remdesivir | EC50 0.07 μM; MRC5 TC50 > 2.0 μM; MRC5 SI > 28.6 | [84] |
| 5.2/A |  | R = H (alovudine) or CH3CH2S(CH2)11C(O)– | R = CH3CH2S(CH2)11C(O)–: EC50 > 45.4 μM; MRC5 TC50 45.4 μM; MRC5 | [84] |
| 5.3/A |  | R = H (lamivudine) or CH3(CH2)12C(O)– | R = CH3(CH2)12C(O)–: EC50 > 47.5 μM; MRC5 TC50 47.5 μM; MRC5 | [84] |
| 5.4/A |  | islatravir | EC50 > 55.3 μM; MRC5 TC50 55.3 μM; MRC5 | [84] |
| 5.5/A | 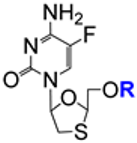 | R = H (emtricitabine) or CH3(CH2)12C(O)– | R = CH3(CH2)12C(O)–: EC50 87.5 μM; MRC5 TC50 72.8 μM; MRC5 SI 1.2 | [84] |
| 5.6/A | 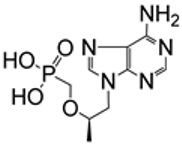 | tenofovir | EC50 > 100 μM; MRC5 TC50 > 100 μM; MRC5 | [84] |
| 5.7/A | 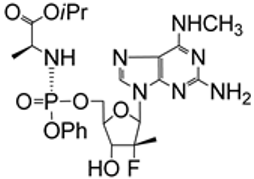 | AT-511 (bemnifosbuvir, free form of AT-527) | EC50 1.8 ± 0.3 μM; BHK-21 CC50 > 100 μM; BHK-21 SI > 55 (2 virus variants evaluated) | [44] |
| 5.8/A |  | tubercidin | EC50 < 0.078 μM; MRC5 CC50 0.66 μM; MRC5 | [56] |
| 5.9/A |  | 5-hydroxymethyltubercidin | EC50 0.528 ± 0.029 μM; MRC5 CC50 > 50 μM; MRC5 SI > 94 | [56] |
| 5.10/A |  | R1 = H or CN R2 = H, CH=NOH, NO2 Me, CN(5 compounds) | R1 = H, R2 = CH=NOH: EC90 < 0.078 μM; MRC5 CC50 0.74 μM; MRC5 | [56] |
| 5.11/A |  | EC50 4.0 μM; HEL CC50 10 μM; HEL | [76] | |
| 5.12/B |  | R = 4-oxaicosan-1-yl, 3-oxahenicosan-1-yl, 2-OBn-4-oxadocosan-1-yl | R = 2-OBn-4-oxadocosan-1-yl: EC50 0.15 μM; MRC5 CC50 > 50 μM; MRC5 | [72] |
| 5.13/D |  | R = H or F | R = H: EC50 18.5 ± 13.5 μM; HEL MCC > 20 μM; HEL | [85] |
| 5.14/D |  | B = thymin-1-yl, uracil-1-yl, N6-DMTr-adenin-1-yl R1 = TBDMS or (iPr)2Si-O-Si(iPr)2R2 = C4H9, Ac, pyranos-1-yl (11 compounds) | B = uracil-1-yl, R1 = (iPr)2Si-O-Si(iPr)2, araR2 = C4H9: EC50 4.1 μM; HEL CC50 10 μM; HEL | [76] |
Approach A: repurposing of known NAs, including approved drugs or their prodrugs. Approach B: development of prodrugs of known anti-HCoV NAs. Approach D: development of de novo-designed NAs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed table. b Exemplified as the effective compound concentration (EC50). c Exemplified as the cytotoxic compound concentration (CC50 or TC50 or MCC). d Defined as the ratio of the compound cytotoxic concentration and the compound effective concentration. Only values explicitly given by the authors are cited. e See text for details.
Parang and coworkers compared the anti-HCoV-229E activity of remdesivir (entry 5.1) with known anti-HIV agents, such as alovudine, lamividine, islatravir, emtricitabine, or tenofovir, and their derivatives (entries 5.2–5.6) [84]. Among the tested compounds, only remdesivir exhibited significant activity. These findings were tentatively explained by the unique interaction of remdesivir with the RdRp of coronaviruses compared to the mode of action of anti-HIV drugs that interfere with the reverse transcriptase.
1.6. The Nucleoside Analogs Evaluated as In Vitro Inhibitors of HCoV-NL63 Replication
We found two original studies that examined the potential of the repurposed NAs to inhibit the replication of HCoV-NL63 in vitro (Table 6, approach A). Saley-Radke and coworkers identified flexible analogs of acyclovir (entry 6.1) as moderately active in Vero 118 cells. Compared with inactive acyclovir (entry 6.2), these results confirmed the hypothesis of biological benefits resulting from introducing an additional rotatable bond into the nucleoside molecule. 6-azauridine and NHC (entries 6.3 and 6.4, respectively) showed significant nanomolar activity, with 6-azauridine having a very high SI value (2500).

Table 6.
The nucleoside analogs evaluated in vitro against the replication of HCoV-NL63.
Table 6.
The nucleoside analogs evaluated in vitro against the replication of HCoV-NL63.
| Entry a/DRD Approach, Structure, and Number of Compounds Examined | Effective Concentration Value b; Cell Line; Virus Variant (If Given) Cytotoxic Concentration Value c; Cell Line Selectivity Index d (SI, If Given) | Ref. | ||
|---|---|---|---|---|
| 6.1/A |  | R1 = H, Me R2 = NH2 (3 compounds) | R1 = Me, R2 = NH2: EC50 8.8 ± 1.5 μM; Vero 118; clinic isolate CC50 120 ± 37 μM; Vero 118 | [41] |
| 6.2/A |  | acyclovir | EC50 > 100 μM; Vero 118; clinic isolate CC50 > 100 μM; Vero 118 | [41] |
| 6.3/A |  | 6-azauridine | IC50 32 nM; LLC-MK2; Amsterdam 1 CC50 80 μM; LLC-MK2 SI 2500 | [86] |
| 6.4/A |  | NHC | IC50 400 nM; LLC-MK2; Amsterdam 1 CC50 > 100 μM; LLC-MK2 SI > 250 | [86] |
Approach A: repurposing of known NAs, including approved drugs or their prodrugs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed. b Exemplified as the effective compound concentration (EC50 or IC50). c Exemplified as the cytotoxic compound concentration (CC50). d Defined as the ratio of the compound cytotoxic concentration and the compound effective concentration. Only values explicitly given by the authors are cited.
1.7. The Nucleoside Analogs Evaluated as Inhibitors of HCoV-Related Enzymes
The NAs evaluated as potential inhibitors of HCoV-related enzymes are collected in Table 7. Two DRD approaches were employed to find active compounds: the repurposing of known NAs or their prodrugs (approach A, entries 7.1–7.10) or the development of de novo-designed NAs (approach D, entries 7.11–7.17). The NAs were evaluated as potential inhibitors of SARS-CoV (or SARS-CoV-2) nonstructural proteins, such as the RNA cap guanine-N7-methyltransferase (nsp14) or 2′-O-methyltransferase nsp16–nsp10 complex (nsp16–nsp10 complex) (entries 7.1, 7.6, and 7.11–7.16) and MERS-CoV nsp16–nsp10 complex (entry 7.2). Very recent studies by Vedadi and coworkers also involved nsp10–nsp16 complexes from HCoV-OC43, HCoV-HKU1, HCoV-NL63, HCoV-229E, and MERS-CoV as drug targets (entry 7.6). Owing to the structural relation between sinefungin and AdoHcy (entry 7.1), the NAs evaluated as potential inhibitors of the RNA cap guanine-N7-methyltransferase nsp14 or 2′-O-methyltransferase nsp16–nsp10 complex are mostly the 6′-modified-5′-adenosyl amino acids (entry 7.6), 5′-adenosyl ureides (entry 7.6), bis-adenosyl conjugates (entry 7.11), deaza-analogs of AdoHcy (entry 7.12), or non-amino acid 5′-modified adenosines (entries 7.13–7.17). The 5′-deoxy-5′-sulfonamido-adenosines (entries 7.14–7.17) or their 7-deaza counterparts (entries 7.16 and 7.17) are a very broad group of compounds. Among the de novo-designed NAs (approach D), compounds with higher inhibitory activity towards SARS-CoV nsp14 (entry 7.12) or SARS-CoV nsp14 (entries 7.13, 7.14, and 7.16) than that of sinefungin were identified. These compounds are derivatives of 7-deaza-5′-deoxy-5′-sulfonamido-adenosine (entries 7.12, 7.14, and 7.16) or 5′-deoxy-5′-thio-adenosine (entry 7.13). Molecular docking was employed to identify the mechanism of action of these inhibitors (entries 7.6, 7.11–7.14, 7.16, and 7.17). Studies focused on SARS-CoV (or SARS-CoV-2) RdRp (entries 7.3–7.5, 7.8, and 7.9) or SARS-CoV-2 3C-like protease (3CL-Pro) (entry 7.7) were also reported.

Table 7.
The nucleoside analogs assessed for their in vitro effect on HCoVs-related enzymes.
Table 7.
The nucleoside analogs assessed for their in vitro effect on HCoVs-related enzymes.
| Entry a/DRD Approach, Structure and Number of Compounds Examined | Object of Study b Antiviral Effect(s) If Given c | Ref. | ||
|---|---|---|---|---|
| 7.1/A |  | X = S or (S)-CH-NH2 n = 1 or 2 (3 compounds) | Inhibition of SARS-CoV nsp14 and SARS-CoV nsp16–nsp10 complex sinefungin (X = (S)-CH-NH2, n = 2): IC50 496 nM ± 18, nsp14 IC50 736 nM ± 71; nsp16/nsp10 AdoHcy (X = S, n = 2): IC50 16 µM ± 1.2, nsp14 IC50 12 µM ± 1.9, nsp16/nsp10 | [87,88] |
 | X = N or N+–Me Y = N or CH R1 = OH, SMe, SiBu, –O[P(O)(OH)O]3H R2 = H or Ac R3 = H or NH2 (6 compounds) | |||
 | X = N or C-C≡CH R = H or –[P(O)(OH)O]3H (3 compounds) | |||
| 7.2/A | 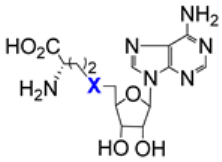 | X = S (AdoHcy) or (S)-CH-NH2 (sinefungin) | Inhibition of MERS-CoV nsp16–nsp10 complex AdoHcy: IC50 7 µM ± 0.4 sinefungin: IC50 7.4 μM ± 0.9 | [88,89] |
| 7.3/A |  R1 = H, OH, Me, F R2 = H, OH, Me B = cytosin-1-yl, uracil-1-yl, adenin-9-yl, 3-carbamoyl-5-fluoro-2-oxopyrazin-1-yl, 3-carbamoyl-1H-1,2,4-triazol-1-yl (7 compounds) | Selectivity of the SARS-CoV-2 RdRp towards incorporation of the nucleoside analog triphosphate into viral RNA over a natural nucleotide. Low discrimination of the enzyme against the cytidine triphosphate derivative (B = cytosin-1-yl, R1 = F, R2 = H) over the reference cytidine triphosphate | [89,90] | |
| 7.4/A |  | R1 = OH, N3, F R2 = H or F R3 = H or Me R4 = H or Me (3 compounds) | Proclivity of SARS-CoV RdRp or SARS-CoV-2 RdRp to incorporate the nucleoside triphosphate into the virus RNA Sofosbuvir nucleoside triphosphate (R1 = OH, R2 = F, R3 = Me, R4 = H) was incorporated by the RdRp, terminated the polymerase reaction and was resistant to SARS-CoV-2 proofreader | [90,91,92,93] |
 | triphosphate of lamivudine (R = H) or emtricitabine (R = F) | |||
 | carbovir triphosphate | |||
| 7.5/A | 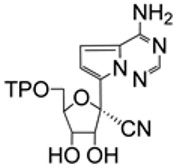 | GS-441524 triphosphate | Incorporation of nucleotide analogs to viral RNA by SARS-CoV-2 RdRp The incorporation of GS-441524 triphosphate and delay in viral RNA chain termination | [93,94] |
 R1 = F, OH, NH2, H, OMe R2 = F, H, Me, OH, N3 B = cytosin-1-yl, uracil-1-yl, purin-9-yl (6-Cl-; 6-NH2,2-Cl; 6-SMe,2-NH2; 6-SH,2- NH2), guanin-9-yl, 8-oksoguanin-9-yl, 3-carbamoyl-1H-1,2,4-triazol-1-yl, 3-carbamoyl-5-fluoro-2-oxopyrazin-1-yl (15 compounds) | ||||
 | lamivudine triphosphate | |||
 | stavudine triphosphate | |||
 | tenofovir triphosphate | |||
| 7.6/A d |  | X = N or C-CN Y = S, (S)-CHCH2NH2, N-CH2CH2C6H4-C(O)NH2-3 (4 compounds) | Inhibition of SARS-CoV-2 nsp14, SARS-CoV-2 nsp10–nsp16 complex from SARS-CoV, SARS-CoV-2, HCoV-OC43, HCoV-HKU1, HCoV-NL63, HCoV-229E, and MERS-CoV and selectivity against 20 human protein lysine methyltransferases Compound SS148 (X = C-CN, Y = S): IC50 0.07 ± 0.006 µM; SARS-CoV-2 nsp14 IC50 0.2 ± 0.04 µM; HCoV-NL63 nsp10–nsp16 complex Compound WZ16 (X = N, Y = (S)-CH-CH2NH2): IC50 0.19 ± 0.04 µM; SARS-CoV-2 nsp14 IC50 2.5 ± 0.1 µM; SARS-CoV nsp10–nsp16 complex | [94,95,96] |
 | ||||
 | R = -CH2-CH2-C6H5 (DS0464) or -CH2-c-C6H11 | |||
| 7.7/A d |  | thioguanosine | The inhibitory effect on SARS-CoV-2 3CL-Pro IC50 6.3 μM; SARS-CoV-2 3CL-Pro IC50 3.9 μM; SARS-CoV-2; Vero E6 CC50 > 20, Vero E6 | [97] |
| 7.8/A d |  | EIDD-2749 | Inhibition of SARS-CoV-2 RdRp EC50 2.47 μM; HAE; USA-WA1/2020 CC50 467.9 μM; HAE | [98] |
| 7.9/A d |  | R1 = H or –[P(O)(OH)O]3H R2,R3 = H or OH R4 = SH or NH2 R5 = NH2, F or H X = N or C-I (4 compounds) | Inhibition of SARS-CoV-2 RdRp R1,R3,R5 = H, R2 = OH, R4 = NH2, X = C-I EC50 0.75 μM; SARS-CoV-2 RdRp CC50 59.46 μM; HEK 293T EC50 1.56 μM; HCoV-OC43; HCT-8 EC50 3.62 μM; HCoV-NL63; LLCM-K2 | [99] |
| 7.10/A d |  | sinefungin | Disruption of WT SARS-CoV-2 nsp16 activity | [100] |
| 7.11/D d |  R = H, (S)-3-NH2-4-CO2H-prop-1-yl, alkyl, substituted benzyl or benzenesulfonyl (16 compounds) | Inhibition of SARS-CoV nsp14 R = –S(O)2-C6H3-3-NO2,4-Cl: IC50 0.6 ± 0.1 µM | [101] | |
| 7.12/D d | 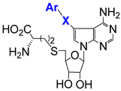 Ar = Ph, naphtyl, guinolin-3-yl, benzimidazol-2-yl, benzo[b]thiophen-5-yl X = CH2, –CH2–CH2–, –C≡C– (8 compounds) | Inhibition of SARS-CoV nsp14 R = guinolin-3-yl: IC50 3 ± 0.5 nM | [102] | |
| 7.13/D d |  | X = CH2 or C(O) Y = S, NH, N(3°), 1,2,3-triazole, piperazine R = substituted alkyl, aryl, benzyl (36 compounds) | Inhibition of SARS-CoV-2 nsp14, SARS-CoV-2 nsp16–nsp10 complex or hGNMT X = CH2, Y = S, R = CH2-C6H3-3-CO2H,5-Ph: IC50 0.58 ± 0.03 nM; nsp14 X = CH2 Y = S, R = CH2-C6H3-3-CO2H: IC50 4 ± 0.5 nM; nsp16/nsp10 | [103,104] |
| 7.14/D d |  | X = S(O)2, C(O) R1 = substituted phenyl R2 = H, Et, alkyl (39 compounds) | Inhibition of SARS-CoV-2 nsp14 X = S(O)2, R1 = C6H3-3-CN,4-OMe, R2 = Et: IC50 0.019 µM | [105] |
| 7.15/D | 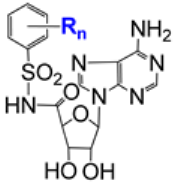 | R = NO2, NH2, CF3, Me, Cl, NHAc n = 1 or 2 (20 compounds) | Inhibition of SARS-CoV-2 nsp14 R = 4-NO2, n = 1: 40% inhibition of SARS-CoV-2 nsp14 at the compound concentration of 5 µM | [106] |
| 7.16/D d |  | R2 = H, Ac, isobutyryl R3 = H or Ac X = CH2, C(O), S(O)2 Y = N or CH Z = C or N R1 = napht-2-yl, quinolin-7-yl, 2-Cl-quinolin-6-yl (11 compounds) | Inhibition of SARS-CoV-2 nsp14 X = S(O)2, Y = CH, Z = N, R1 = napht-2-yl, R2 = H, R3 = H: IC50 0.093 µM; nsp14 EC50 0.72 µM; SARS-CoV-2; A459 AT CC50 > 100 µM; A459 AT | [107] |
| 7.17/D d |  R1 = Br, Me, OMe; R2 = Cl, NO2, CN; R3 = H, Me, Et; X = OH or NHMe; Y = Ph or quinolin-3-yl (26 compounds) | Inhibition of SARS-CoV nsp14, SARS-CoV-2 nsp14, MERS-CoV nsp14 R1 = OMe, R2 = CN, R3 = Me, Y = quinolin-3-yl: IC50 0.141 ± 0.02 nM; SARS-CoV nsp14 IC50 0.72 ± 0.4 nM; SARS-CoV-2 nsp14 R1 = OMe, R2 = CN, R3 = Me, Y = Ph: IC50 1.25 ± 0.25 nM; MERS-CoV nsp14 | [108] | |
Approach A: repurposing of known NAs, including approved drugs or their prodrugs. Approach D: development of de novo-designed NAs. a Entry numbers include the number of the table and the position of a compound (or a group of compounds) on the timeline, taking into account the year of publication of the references discussed. b Exemplified as the inhibitory compound concentration (IC50) or in a descriptive form. c Effective compound concentration (EC50), inhibitory compound concentration (IC50), or cytotoxic compound concentration (CC50). d See text for details.
Vedadi and coworkers screened a library of 161 in-house synthesized SAM-competitive MTase inhibitors and SAM analogs [94]. Among the identified screening hits (entry 7.6), compound SS148 inhibited nsp14 MTase activity (IC50 value of 70 ± 6 nM) and was selective against 20 human protein lysine MTases. Interestingly, compound DS0464, with an IC50 value of 1.1 ± 0.2 μM, showed a competitive pattern of inhibition with regard to both RNA and SAM. The docking studies suggested that DS0464 could act as a bifunctional inhibitor, occupying the cofactor site and extending into the substrate binding site of the enzyme. Moreover, compounds SS148 and WZ16 (entry 7.6)—analogs of sinefungin—are potent inhibitors of the methyltransferase nsp10–nsp16 complex [95]. The crystal structures of the SARS-CoV-2 nsp10–nsp16 2′-O-RNA methyltransferase binding compound SS148 or WZ16 revealed the binding of both compounds to the SAM/SAH binding pocket of nsp16 through interactions similar to those identified for SAM/SAH. A biochemical study indicated an SAM-competitive pattern of inhibition for both SS148 and WZ16. On the other hand, with regard to RNA, SS148 showed an apparent noncompetitive pattern of inhibition, whereas WZ16 featured an uncompetitive one. The latter suggested that RNA could bind first, and its binding increased the affinity of WZ16 for binding to the nsp10–nsp16 complex. Interestingly, SS148 and WZ16 inhibited the MTase activities of all seven HCoVs [96]. The docking studies with the inhibitors SS148 and WZ16 and the nsp16 proteins of these seven HCoVs revealed that the proteins were highly conserved and the mechanisms of their interaction with the SS148 and WZ16 inhibitors were nearly identical.
Kuzikov et al. performed a repurposing screening of compound libraries (ca. 8700 compounds) containing approved drugs, drug candidates, and small molecules considered safe in humans [97]. Apart from previously reported inhibitors of SARS-CoV-2 3CL-Pro, they found 62 compounds with IC50 values < 1 μM. Among them, thioguanosine (entry 7.7) showed a dose-dependent effect on SARS-CoV-2 3CL-Pro and demonstrated a moderate anticytopathic effect in Vero E6 cells but did not show a reduction in the virus RNA in Vero E6 cells. The docking of thioguanosine into the main catalytic cavity of 3CL-Pro revealed that the main ligand–protein interactions might involve the most probable tautomer for thioguanosine with a thiol group rather than a thio-ketonic group.
Plemper and coworkers identified 4′-fluorouridine (entry 7.8) as an oral drug candidate to treat COVID-19 [98]. SARS-CoV-2 is sensitive to 4′-fluorouridine, with EC50 values ranging from 0.2 to 0.6 µM against isolates from different donors. Studies on the mode of action of 4′-fluorouridine showed the incorporation of its triphosphate (4′-FIU-TP) into the SARS-CoV-2 RNA by the virus RdRp instead of uridine triphosphate. The multiple incorporations of 4′-FIU-TP caused delayed polymerase stalling. 4′-fluorouridine was orally efficient against SARS-CoV-2 in a ferret model [98].
Zhao et al. reported the screening of 134 compounds from the Selleckchemicals NAs library using a cell-based SARS-CoV-2 RdRp assay [99]. As a result, four potent inhibitors of SARS-CoV-2 RdRp, including 5-iodotubercidin, were identified. Notably, the four compounds also displayed a dose-dependent inhibitory effect against both the HCoV-OC43 and HCoV-NL63 viruses in HCT-8 or LLC-MK2 cells. Further studies for the most potent compound, namely 5-iodotubercidin, showed that its activity is related to the inhibition of adenosine kinase.
Menachery and coworkers explored whether SARS-CoV-2 nsp16 activity could be targeted for therapeutic treatment [100]. Testing the effect of sinefungin on wild-type (WT) SARS-CoV-2 replication (entry 7.10), they observed a dose-dependent decrease in virus replication in Vero E6 cells or Calu3 2B4 cells. Moreover, the combination of sinefungin with a constant dose of IFN-I pretreatment drove the further attenuation of WT SARS-CoV-2 in Vero E6 cells. However, a dose-dependent effect of sinefungin on the viability of Vero E6 cells was noted. The authors suggested that these results indicated dose limitations in using sinefungin as a therapeutic treatment.
Ahmed-Belkacem et al. developed a broad group of adenine dinucleosides with the O-2′→C-5′ connection via a variety of aminoethyl linkers (entry 7.11) [101]. Among them, the most active SARS-CoV nsp14 inhibitor showed submicromolar activity and was as potent as, but particularly more specific, than sinefungin. Molecular docking suggested that the inhibitor was bound to a pocket formed by the SAM and cap RNA binding sites of the examined enzyme.
Otava et al. performed detailed studies on 7-deaza-7-substituted analogs of SAH as potential inhibitors of SARS-CoV nsp14 (entry 7.12) [102]. These compounds exhibited high inhibitory activity (IC50 3 ± 0.5–37 ± 7 nM). The most active inhibitor showed an SAM-competitive but RNA-noncompetitive pattern of inhibition. The molecular modeling and docking studies with the homology model SARS-CoV nsp14 in complex with nsp10 revealed that (a) a lipophilic substituent caused a significant increase in the activity of the examined compounds; (b) derivatives with a methylene bridge were significantly less active than those with a lipophilic substituent linked via the ethynyl group or the ethylene one; and (c) derivatives bearing bicyclic lipophilic substituents have comparable activity to each other [102].
Bobileva and coworkers obtained a broad panel of SAH analogs with a variety of surrogates of the homocysteine moiety and assayed them against SARS-CoV-2 nsp14 or the SARS-CoV-2 nsp16–nsp10 complex (entry 7.13) [103,104]. The derivative with the 3-(mercaptomethyl)benzoic acid moiety as the parent compound showed one-digit nanomolar inhibitory activity towards both of the examined enzymes. Structural modifications of the parent compound, notably variations in the location of the carboxylic group and/or the additional phenyl substituent within the benzoic acid skeleton, provided an inhibitor of SARS-CoV-2 nsp14, exhibiting a very high activity of IC50 0.58 ± 0.03 nM and slightly improved cell permeability compared with the parent compound. Docking results suggested that this 3-phenylbenzoic acid derivative was a nsp14 bisubstrate inhibitor occupying both SAM and mRNA-binding sites.
Ahmed-Belkacem et al. identified several highly active inhibitors of SARS-CoV-2 nsp14 among a variety of 5′deoxy-5′-sulfonamido/amido derivatives of adenosine (entry 7.14) [105]. The docking studies with the most active inhibitor supported the hypothesis of a bisubstrate mechanism of its action. The detected orientation of this inhibitor in the enzyme binding sites suggested that the 3-cyano-4-methoxybenzenesulfonamide ring clearly occupied the capguanosine binding pocket.
Bisubstrate inhibitors of SARS-CoV-2 nsp14 were developed by Jung et al. (entry 7.16) [107]. Using a biochemical assay employing an LC-MS/MS technique, they established that highly active inhibitors could be obtained by connecting an adenosine moiety with an aromatic ring through a sulfonamide group. This study resulted in the identification of four compounds with IC50 values in the nanomolar range. Docking studies showed significant differences in the binding mode of naphthalene-containing derivatives varying in the nucleobase scaffold. Briefly, the same location of the nucleobase (adenine or its 5-aza-7-deaza counterpart) resulted in substantially different interactions between protein residues and the sugar side moiety. These in silico observations supported the in vitro results. More importantly, one of the compounds, a 2-naphthyl derivative, also displayed high antiviral activity against SARS-CoV-2 in A549 cells and low cytotoxicity in the host cells, resulting in SI > 139. Interestingly, sinefungin, used as a reference compound, did not show antiviral activity in A549 cells [107].
Debart and coworkers, using a structure-guided drug design approach based on bisubstrate inhibitors, developed a number of compounds acting as adenosine mimetics with a variously substituted arylsulfonamide substituent at the 5′-position (entry 7.17) [108]. The adenine moiety was replaced with hypoxanthine, N6-methyladenine, or C7-substituted 7-deazaadenine. In this way, a highly selective inhibitor of SARS-CoV-2 nsp14 and SARS-CoV nsp14 with subnanomolar IC50 values was discovered (R1 = OMe, R2 = CN, R3 = Me, Y = quinolin-3-yl). Docking analyses with this inhibitor suggested that C7-substituted 7-deazaadenine superimposed perfectly with the adenine of the SAM in the nsp14 structures, while the 3-quinoline or the phenyl substituent on the nucleobase protruded into the SAM entry channel. Moreover, an inhibitor of MERS-CoV nsp14 was also found (R1 = OMe, R2 = CN, R3 = Me, Y = Ph). Further in vitro studies (Vero E6 TMPRSS2 cells) on the most active compounds revealed that the C7-quinolin-ethynyl-substitued compounds showed moderate activities, reaching 50% SARS-CoV-2 (B.1) inhibition at 10 μM. Compounds with a phenyl–ethynyl substituent at the C7-position of the adenine moiety did not display any inhibitory activity. These results were interpreted in terms of delivery or stability problems [108].
2. Conclusions
Our literature survey revealed that over four hundred NAs (including prodrugs) were evaluated against HCoVs. Approach A (i.e., the repurposing of compounds existing in the public domain) appeared as the most often applied. It led to the identification of several active compounds. Some of them were approved as anti-COVID-19 drugs (e.g., remdesivir and molnupiravir). Bemnifosbuvir (entry 2.12) received an FDA fast-track status [18]. The high activities of bemnifosbuvir against SARS-CoV-1 (entry 1.32), MERS-CoV (entry 3.9), HCoV-OC43 (entry 4.11), and HCoV-229E (entry 5.7) made this compound a very attractive drug candidate for future medication for other and emerging HCoVs.
The repurposing strategy also exposed several NAs of interest, i.e., compounds with submicromolar in vitro activity: 6-azauridine (entry 1.1), pyrazofurin (entry 1.2), 2′-deoxytubercidin (entry 1.6), tubercidin (entries 2.10, 4.12, and 5.8), 5-hydroxymethyltubercidin (entries 2.14, 4.13, and 5.9), thioguanosine (entry 2.18), gemcitabine (entry 2.19), 6-thioinosine (entry 2.20), 3-deazaneplanocin A (entry 2.21), sangivamycin (entry 2.22), riboprine (entry 2.25), forodesine (entry 2.26), and 7-(furan-2-yl)tubercidin (entry 2.31). Some of these compounds suffered from low SI values (e.g., 2′-deoxytubercidin, entry 1.6) or specific limitations, e.g., serious side effects within a specific group of patients (6-azauridine [109]), or nephrotoxicity (in rodents, 3-deazaneplanocin A [110]; or in humans, tubercidin [111]). However, the reported anti-HCoV activities of these NAs might serve as starting points in the development of their further structural modifications.
The screening of large libraries of available compounds was used to search for antiviral agents—entry 1.28 (290 approved drugs with antiviral activities against other RNA viruses); entries 2.10, 2.14, 2.15, and 2.16 (753 nucleoside analogs from a compound library of Hokkaido University); and entries 2.10, 2.18, 2.19, and 2.20 (approximately 18,000 drugs, most of which have been tested in humans)—as well as for inhibitors of specific viral enzymes—entry 7.6 (161 in-house synthesized S-adenosylmethionine (SAM)-competitive MTase inhibitors and SAM analogs), entry 7.7 (8702 drugs and drug candidates from the repurposing collections), and entry 7.9 (134 compounds in the Selleckchemicals NAs library). This fast way to identify prospective drugs seemed reasonable in an emergency case, such as COVID-19, where no proven therapeutic solution existed. However, considering the overall number of NAs collected in this review, we may conclude that the library screening revealed a relatively narrow group of compounds with promising properties.
The aristeromycin fluoro-analogs, with their submicromolar in vitro activities against SARS-CoV-1 and MERS-CoV (entries 1.38 and 3.10, respectively) and activity against SARS-CoV-2 (entry 2.39), underlined the case of the development of de novo-designed NAs (approach D), even if their SI values were still unfavorable. On the other hand, the de novo-designed sulfonamide inhibitors of SARS-CoV-2 nsp14 (entry 7.16) showed both submicromolar activity against SARS-CoV-2 and low cytotoxicity in vitro.
The prodrug concept was successfully implemented in the very recently released anti-COVID-19 drug candidates (bemnifosbuvir, Figure 1; and obeldesivir, entry 2.34). Furthermore, novel lipid prodrugs of GS-441524 (entries 2.33 and 5.12) were reported. Their significantly higher or comparable in vitro activities to those of remdesivir or GS-441524, satisfactory SI values, and convenient chemical synthesis made these compounds interesting targets.
NAs offer a large number of ways for the prodrug concept to be applied: (a) introducing a progroup into the glicone (entries 2.33, 5.12, and 2.34) or the nucleobase (entry 2.35); (b) employing a proven-in-practice progroup (such as phosphoramidate or isobutyryl, Figure 1, entries 1.38, 3.10, 2.34) or an investigational one (entries 2.4, 4.3, 2.31, 2.33, 5.12, 2.35, 2.40, 5.14); and (c) the involvement of all the reactive functional groups in the parent nucleoside scaffold (Figure 1, VV116; or entries 2.4, 4.3, 2.35, 2.40, 5.14) or just the selected one (Figure 1, entries 2.4, 4.3, 2.31, 2.33, 5.12, 2.34). Owing to the high reactivity of their functional groups, in particular in combination with the prodrug concept, NAs offered the formation of multi-element sets of derivatives (entries 2.31, 7.13, 7.14, 7.15, 7.16, 7.17). This approach has been extensively applied since 2021.
NAs were valuable tools for recognizing HCoV-related enzymes as potential drug targets. NAs inhibiting HCoV-related enzymes in vitro at submicromolar or subnanomolar concentrations were reported: SARS-CoV nsp14 (entries 7.1, 7.6, 7.11, 7.12, 7.17); SARS-CoV-2 nsp14 (entries 7.6, 7.13, 7.14, 7.16, 7.17); SARS-CoV-2 RdRp (entry 7.9); the SARS-CoV-2 nsp16 complex (entry 7.10); and the HCoV-NL63 nsp10–nsp16 complex (entry 7.6). Compounds with inhibitory activity towards methyltransferases of a broad panel of HCoVs were also identified (entry 7.6). In most cases, these nucleoside inhibitors were inactive against human methyltransferases (entries 7.6, 7.11, 7.12, and 7.17).
Among inhibitors of the HCoV-related enzymes, compounds endowed with antiviral activity against HCoVs in vitro were identified: anti-SARS-CoV-2 activity (entries 7.7, 7.10, and 7.16), anti-HCoV-OC43, or anti-HCoV-NL63 activity (entry 7.9). Moreover, compounds with anti-SARS-CoV-2 activity in an animal model were found (entry 7.8).
Currently, interventional clinical trials on three NAs have been completed: remdesivir [112,113,114,115,116,117,118,119,120,121,122], molnupiravir [123,124,125,126], and AT-527 [127]. The results of these studies were posted at ClinicalTrials.gov.
Recently, a hypothesis on the involvement of more than one virus in long COVID has been postulated [128]. The reactivation of viruses that had infected the patient previously was suggested. Clinical studies conducted by the biotechnology company Virios Therapeutics showed that a combination of valacyclovir and celecoxib, designed to synergistically suppress herpes virus replication, caused clinically and statistically significant improvements in fatigue, pain, and symptoms of autonomic dysfunction and general well-being related to long COVID in patients [129]. These findings might open up new horizons for the exploration of NAs as drugs in the treatment of SARS-CoV-2-related ailments.
Author Contributions
Both authors contributed equally to this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Warsaw University of Technology. Grant Number 504/04114/1020/44.000000.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| 3CL-Pro | 3C-like protease |
| A459 cells | adenocarcinomic human alveolar basal epithelial cells |
| ATP | adenosine triphosphate |
| BHK-21 cells | baby hamster kidney fibroblast cells |
| Calu-3 cells | human lung cells |
| Caco-2 cells | human colorectal adenocarcinoma cells |
| CC50 | 50% cytotoxic concentration |
| DBT-9 cells | delayed brain tumor, murine astrocytoma clone 9 |
| DMTr | dimethoxytrityl |
| EC50 | 50% effective concentration |
| fRhK4 cells | fetal rhesus monkey kidney cells |
| H460 cells | human nonsmall cell lung cancer cells |
| HAE cells | human tracheobronchial epithelial cells |
| HCT-8 cells | human ileocecal adenocarcinoma cells |
| HEK 293T cells | human embryonic kidney cells |
| HEL cells | human embryonic lung cells |
| Huh7 cells | well differentiated hepatocyte-derived carcinoma cells |
| HSV-1 | herpes simplex virus 1 |
| HSV-2 | herpes simplex virus 2 |
| IC50 | 50% cytotoxic concentration |
| IFN-I | Interferon type I |
| KD | protein–ligand dissociation constant |
| LLC-MK2 cells | rhesus monkey kidney epithelial cells |
| MCC cells | Merkel cell carcinoma |
| MRC-5 cells | human fetal lung fibroblast cells |
| nsp14 | RNA cap guanine-N7-methyltransferase |
| nsp16–nsp10 | 2′-O-methyltransferase nsp16–nsp10 complex |
| RD cells | rhabdomyosarcoma cells |
| RdRp | RNA-dependent RNA polymerase |
| SAH | S-adenosyl-L-homocysteine |
| SAM | S-adenosyl-L-methionine |
| SCC-VII | squamous cell carcinoma |
| TBDMS | tert-butyldimethylsilyl |
| TC50 | 50% toxic concentration |
| TP | –[P(O)(OH)O]3H |
| Vero cells | Green Monkey kidney epithelial cells |
| Vero 118 cells | Vero cells clone 118 |
| Vero 76 cells | Vero cells clone 76 |
| Vero E6 cells | Vero cells clone E6 |
| Vero E6 TMPRSS2 cells | human transmembrane serine protease 2-expressing Vero E6 cells |
References
- Weiss, S.R. Forty Years with Coronaviruses. J. Exp. Med. 2020, 217, e20200537. [Google Scholar] [CrossRef]
- Guarner, J. Three Emerging Coronaviruses in Two Decades. Am. J. Clin. Pathol. 2020, 153, 420–421. [Google Scholar] [CrossRef]
- WHO Regional Office for the Western Pacific. SARS: How a Global Epidemic Was Stopped; WHO Regional Office for the Western Pacific: Manila, Philippines, 2006; ISBN 978-92-9061-213-1. [Google Scholar]
- Azhar, E.I.; Hui, D.S.C.; Memish, Z.A.; Drosten, C.; Zumla, A. The Middle East Respiratory Syndrome (MERS). Infect. Dis. Clin. N. Am. 2019, 33, 891–905. [Google Scholar] [CrossRef]
- Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Available online: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers (accessed on 5 June 2023).
- Weekly Epidemiological Update on COVID-19—1 June 2023. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---1-june-2023 (accessed on 5 June 2023).
- WHO Chief Declares End to COVID-19 as a Global Health Emergency|UN News. Available online: https://news.un.org/en/story/2023/05/1136367 (accessed on 5 June 2023).
- Marra, M.A.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.N.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef]
- Kandeil, A.; Shehata, M.M.; El Shesheny, R.; Gomaa, M.R.; Ali, M.A.; Kayali, G. Complete Genome Sequence of Middle East Respiratory Syndrome Coronavirus Isolated from a Dromedary Camel in Egypt. Genome Announc. 2016, 4, e00309-16. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.d.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Morse, J.S.; Lalonde, T.; Xu, S.; Liu, W.R. Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. ChemBioChem 2020, 21, 730–738. [Google Scholar] [CrossRef]
- Li, G.; De Clercq, E. Therapeutic Options for the 2019 Novel Coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef]
- Martinez, M.A. Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob. Agents Chemother. 2020, 64, e00399-20. [Google Scholar] [CrossRef]
- Pastuch-Gawołek, G.; Gillner, D.; Król, E.; Walczak, K.; Wandzik, I. Selected Nucleos(t)Ide-Based Prescribed Drugs and Their Multi-Target Activity. Eur. J. Pharmacol. 2019, 865, 172747. [Google Scholar] [CrossRef]
- Koren, G.; King, S.; Knowles, S.; Phillips, E. Ribavirin in the Treatment of SARS: A New Trick for an Old Drug? CMAJ Can. Med. Assoc. J. 2003, 168, 1289–1292. [Google Scholar]
- Barnard, D.L.; Day, C.W.; Bailey, K.; Heiner, M.; Montgomery, R.; Lauridsen, L.; Winslow, S.; Hoopes, J.; Li, J.K.-K.; Lee, J.; et al. Enhancement of the Infectivity of SARS-CoV in BALB/c Mice by IMP Dehydrogenase Inhibitors, Including Ribavirin. Antivir. Res. 2006, 71, 53–63. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic Strategies for COVID-19: Progress and Lessons Learned. Nat. Rev. Drug Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef]
- Horga, A.; Kuritzkes, D.R.; Kowalczyk, J.J.; Pietropaolo, K.; Belanger, B.; Lin, K.; Perkins, K.; Hammond, J. Phase II Study of Bemnifosbuvir in High-Risk Participants in a Hospital Setting with Moderate COVID-19. Future Virol. 2023, 18, 489–500. [Google Scholar] [CrossRef]
- Jordan, P.C.; Stevens, S.K.; Deval, J. Nucleosides for the Treatment of Respiratory RNA Virus Infections. Antivir. Chem. Chemother. 2018, 26, 1–19. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; Denison, M.R. Nucleoside Analogues for the Treatment of Coronavirus Infections. Curr. Opin. Virol. 2019, 35, 57–62. [Google Scholar] [CrossRef]
- Totura, A.L.; Bavari, S. Broad-Spectrum Coronavirus Antiviral Drug Discovery. Expert Opin. Drug Discov. 2019, 14, 397–412. [Google Scholar] [CrossRef]
- Yan, J.; Liu, A.; Huang, J.; Wu, J.; Fan, H. Research Progress of Drug Treatment in Novel Coronavirus Pneumonia. AAPS PharmSciTech 2020, 21, 130. [Google Scholar] [CrossRef]
- Ami, E.; Ohrui, H. Intriguing Antiviral Modified Nucleosides: A Retrospective View into the Future Treatment of COVID-19. ACS Med. Chem. Lett. 2021, 12, 510–517. [Google Scholar] [CrossRef]
- Roy, V.; Agrofoglio, L.A. Nucleosides and Emerging Viruses: A New Story. Drug Discov. Today 2022, 27, 1945–1953. [Google Scholar] [CrossRef]
- Borbone, N.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Nucleoside Analogs and Nucleoside Precursors as Drugs in the Fight against SARS-CoV-2 and Other Coronaviruses. Molecules 2021, 26, 986. [Google Scholar] [CrossRef]
- Thames, J.E.; Seley-Radtke, K.L. Comparison of the Old and New—Novel Mechanisms of Action for Anti-Coronavirus Nucleoside Analogues. CHIMIA 2022, 76, 409–417. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Lu, X.; Zhang, W.; Fang, W.; Yuan, L.; Wang, X. An Update on Inhibitors Targeting RNA-Dependent RNA Polymerase for COVID-19 Treatment: Promises and Challenges. Biochem. Pharmacol. 2022, 205, 115279. [Google Scholar] [CrossRef]
- Bekheit, M.S.; Panda, S.S.; Girgis, A.S. Potential RNA-Dependent RNA Polymerase (RdRp) Inhibitors as Prospective Drug Candidates for SARS-CoV-2. Eur. J. Med. Chem. 2023, 252, 115292. [Google Scholar] [CrossRef]
- Zenchenko, A.A.; Drenichev, M.S.; Mikhailov, S.N. Nucleoside Inhibitors of Coronaviruses. Curr. Med. Chem. 2021, 28, 5284–5310. [Google Scholar] [CrossRef]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H.W. Glycyrrhizin, an Active Component of Liquorice Roots, and Replication of SARS-Associated Coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef]
- Barnard, D.L.; Hubbard, V.D.; Burton, J.; Smee, D.F.; Morrey, J.D.; Otto, M.J.; Sidwell, R.W. Inhibition of Severe Acute Respiratory Syndrome-Associated Coronavirus (SARSCoV) by Calpain Inhibitors and Beta-D-N4-Hydroxycytidine. Antivir. Chem. Chemother. 2004, 15, 15–22. [Google Scholar] [CrossRef]
- Barnard, D.L.; Day, C.W.; Bailey, K.; Heiner, M.; Montgomery, R.; Lauridsen, L.; Chan, P.K.; Sidwell, R.W. Evaluation of Immunomodulators, Interferons and Known in Vitro SARS-CoV Inhibitors for Inhibition of SARS-Cov Replication in BALB/c Mice. Antivir. Chem. Chemother. 2006, 17, 275–284. [Google Scholar] [CrossRef]
- Günther, S.; Asper, M.; Röser, C.; Luna, L.K.S.; Drosten, C.; Becker-Ziaja, B.; Borowski, P.; Chen, H.-M.; Hosmane, R.S. Application of Real-Time PCR for Testing Antiviral Compounds against Lassa Virus, SARS Coronavirus and Ebola Virus in Vitro. Antivir. Res. 2004, 63, 209–215. [Google Scholar] [CrossRef]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.T.; Lu, H.T.; Fan, K.W.; Cheng, V.C.C.; Tsui, W.H.W.; Hung, I.F.N.; Lee, T.S.W.; et al. In Vitro Susceptibility of 10 Clinical Isolates of SARS Coronavirus to Selected Antiviral Compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef]
- Saijo, M.; Morikawa, S.; Fukushi, S.; Mizutani, T.; Hasegawa, H.; Nagata, N.; Iwata, N.; Kurane, I. Inhibitory Effect of Mizoribine and Ribavirin on the Replication of Severe Acute Respiratory Syndrome (SARS)-Associated Coronavirus. Antivir. Res. 2005, 66, 159–163. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J.; et al. An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef]
- Ikejiri, M.; Saijo, M.; Morikawa, S.; Fukushi, S.; Mizutani, T.; Kurane, I.; Maruyama, T. Synthesis and Biological Evaluation of Nucleoside Analogues Having 6-Chloropurine as Anti-SARS-CoV Agents. Bioorganic Med. Chem. Lett. 2007, 17, 2470–2473. [Google Scholar] [CrossRef]
- Chu, C.K.; Gadthula, S.; Chen, X.; Choo, H.; Olgen, S.; Barnard, D.L.; Sidwell, R.W. Antiviral Activity of Nucleoside Analogues against SARS-Coronavirus (SARS-CoV). Antivir. Chem. Chemother. 2006, 17, 285–289. [Google Scholar] [CrossRef]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of Clinically Developed Drugs for Treatment of Middle East Respiratory Syndrome Coronavirus Infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef]
- Warren, T.K.; Wells, J.; Panchal, R.G.; Stuthman, K.S.; Garza, N.L.; Van Tongeren, S.A.; Dong, L.; Retterer, C.J.; Eaton, B.P.; Pegoraro, G.; et al. Protection against Filovirus Diseases by a Novel Broad-Spectrum Nucleoside Analogue BCX4430. Nature 2014, 508, 402–405. [Google Scholar] [CrossRef]
- Peters, H.L.; Jochmans, D.; de Wilde, A.H.; Posthuma, C.C.; Snijder, E.J.; Neyts, J.; Seley-Radtke, K.L. Design, Synthesis and Evaluation of a Series of Acyclic Fleximer Nucleoside Analogues with Anti-Coronavirus Activity. Bioorganic Med. Chem. Lett. 2015, 25, 2923–2926. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Graham, R.L.; Menachery, V.D.; Gralinski, L.E.; Case, J.B.; Leist, S.R.; Pyrc, K.; Feng, J.Y.; Trantcheva, I.; et al. Broad-Spectrum Antiviral GS-5734 Inhibits Both Epidemic and Zoonotic Coronaviruses. Sci. Transl. Med. 2017, 9, eaal3653. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef]
- Good, S.S.; Westover, J.; Jung, K.H.; Zhou, X.-J.; Moussa, A.; La Colla, P.; Collu, G.; Canard, B.; Sommadossi, J.-P. AT-527, a Double Prodrug of a Guanosine Nucleotide Analog, Is a Potent Inhibitor of SARS-CoV-2 In Vitro and a Promising Oral Antiviral for Treatment of COVID-19. Antimicrob. Agents Chemother. 2021, 65, aac.02479-20. [Google Scholar] [CrossRef]
- Cho, J.H.; Bernard, D.L.; Sidwell, R.W.; Kern, E.R.; Chu, C.K. Synthesis of Cyclopentenyl Carbocyclic Nucleosides as Potential Antiviral Agents Against Orthopoxviruses and SARS. J. Med. Chem. 2006, 49, 1140–1148. [Google Scholar] [CrossRef]
- Cho, A.; Saunders, O.L.; Butler, T.; Zhang, L.; Xu, J.; Vela, J.E.; Feng, J.Y.; Ray, A.S.; Kim, C.U. Synthesis and Antiviral Activity of a Series of 1′-Substituted 4-Aza-7,9-Dideazaadenosine C-Nucleosides. Bioorganic Med. Chem. Lett. 2012, 22, 2705–2707. [Google Scholar] [CrossRef]
- Yoon, J.; Kim, G.; Jarhad, D.B.; Kim, H.-R.; Shin, Y.-S.; Qu, S.; Sahu, P.K.; Kim, H.O.; Lee, H.W.; Wang, S.B.; et al. Design, Synthesis, and Anti-RNA Virus Activity of 6′-Fluorinated-Aristeromycin Analogues. J. Med. Chem. 2019, 62, 6346–6362. [Google Scholar] [CrossRef]
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT03891420. A Study to Evaluate the Safety, Pharmacokinetics and Antiviral Effects of Galidesivir in Yellow Fever or COVID-19. 2019. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT03891420?term=NCT03891420&draw=2&rank=1 (accessed on 22 April 2024).
- BioCryst. Provides Update on Galidesivir Program|BioCryst Pharmaceuticals. Available online: https://ir.biocryst.com/news-releases/news-release-details/biocryst-provides-update-galidesivir-program (accessed on 19 June 2023).
- Taylor, R.; Bowen, R.; Demarest, J.F.; DeSpirito, M.; Hartwig, A.; Bielefeldt-Ohmann, H.; Walling, D.M.; Mathis, A.; Babu, Y.S. Activity of Galidesivir in a Hamster Model of SARS-CoV-2. Viruses 2021, 14, 8. [Google Scholar] [CrossRef]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; Jarhad, D.B.; Tripathi, S.K.; Lee, H.W.; Jeong, L.S.; Snijder, E.J.; Posthuma, C.C. 6′,6′-Difluoro-Aristeromycin Is a Potent Inhibitor of MERS-Coronavirus Replication. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zandi, K.; Amblard, F.; Musall, K.; Downs-Bowen, J.; Kleinbard, R.; Oo, A.; Cao, D.; Liang, B.; Russell, O.O.; McBrayer, T.; et al. Repurposing Nucleoside Analogs for Human Coronaviruses. Antimicrob. Agents Chemother. 2020, 65, e01652-20. [Google Scholar] [CrossRef]
- Xie, Y.; Yin, W.; Zhang, Y.; Shang, W.; Wang, Z.; Luan, X.; Tian, G.; Aisa, H.A.; Xu, Y.; Xiao, G.; et al. Design and Development of an Oral Remdesivir Derivative VV116 against SARS-CoV-2. Cell Res. 2021, 31, 1212–1214. [Google Scholar] [CrossRef]
- Schultz, D.C.; Johnson, R.M.; Ayyanathan, K.; Miller, J.; Whig, K.; Kamalia, B.; Dittmar, M.; Weston, S.; Hammond, H.L.; Dillen, C.; et al. Pyrimidine Inhibitors Synergize with Nucleoside Analogues to Block SARS-CoV-2. Nature 2022, 604, 134–140. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) in Vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Uemura, K.; Nobori, H.; Sato, A.; Sanaki, T.; Toba, S.; Sasaki, M.; Murai, A.; Saito-Tarashima, N.; Minakawa, N.; Orba, Y.; et al. 5-Hydroxymethyltubercidin Exhibits Potent Antiviral Activity against Flaviviruses and Coronaviruses, Including SARS-CoV-2. iScience 2021, 24, 103120. [Google Scholar] [CrossRef]
- Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Da Silva, A.d.P.D.; Dias, S.dS.G.; da Silva, C.d.S.; Ferreira, A.C.; Mattos, M.; Pão, C.R.R.; de Freitas, C.S.; et al. In Vitro Antiviral Activity of the Anti-HCV Drugs Daclatasvir and Sofosbuvir against SARS-CoV-2, the Aetiological Agent of COVID-19. J. Antimicrob. Chemother. 2021, 76, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, B.; Bojkova, D.; Zaliani, A.; Cinatl, J.; Claussen, C.; Westhaus, S.; Keminer, O.; Reinshagen, J.; Kuzikov, M.; Wolf, M.; et al. A SARS-CoV-2 Cytopathicity Dataset Generated by High-Content Screening of a Large Drug Repurposing Collection. Sci. Data 2021, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Bergant, V.; Yamada, S.; Grass, V.; Tsukamoto, Y.; Lavacca, T.; Krey, K.; Mühlhofer, M.-T.; Wittmann, S.; Ensser, A.; Herrmann, A.; et al. Attenuation of SARS-CoV-2 Replication and Associated Inflammation by Concomitant Targeting of Viral and Host Cap 2′-O-Ribose Methyltransferases. EMBO J. 2022, 41, e111608. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.P.; Postnikova, E.N.; Eaton, B.P.; Cai, Y.; Yu, S.; Smith, C.O.; Liang, J.; Zhou, H.; Kocher, G.A.; Murphy, M.J.; et al. Sangivamycin Is Highly Effective against SARS-CoV-2 in Vitro and Has Favorable Drug Properties. JCI Insight 2022, 7, e153165. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.M. Potent Inhibitory Activities of the Adenosine Analogue Cordycepin on SARS-CoV-2 Replication. ACS Omega 2022, 7, 2960–2969. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.M. Efficacious Preclinical Repurposing of the Nucleoside Analogue Didanosine against COVID-19 Polymerase and Exonuclease. ACS Omega 2022, 7, 21385–21396. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.M.; Abdalla, M. A Series of Adenosine Analogs as the First Efficacious Anti-SARS-CoV-2 Drugs against the B.1.1.529.4 Lineage: A Preclinical Repurposing Research Study. ChemistrySelect 2022, 7, e202201912. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.; Rabie, A.M. Dual Computational and Biological Assessment of Some Promising Nucleoside Analogs against the COVID-19-Omicron Variant. Comput. Biol. Chem. 2023, 104, 107768. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.M.; Abdalla, M. Forodesine and Riboprine Exhibit Strong Anti-SARS-CoV-2 Repurposing Potential: In Silico and In Vitro Studies. ACS Bio Med Chem Au 2022, 2, 565–585. [Google Scholar] [CrossRef] [PubMed]
- Rabie, A.M.; Abdalla, M. Evaluation of a Series of Nucleoside Analogs as Effective Anticoronaviral-2 Drugs against the Omicron-B.1.1.529/BA.2 Subvariant: A Repurposing Research Study. Med. Chem. Res. 2023, 32, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Ke, X.; Jian, X.; Xia, L.; Yang, Y.; Zhang, T.; Xiong, H.; Zhao, B.; Liu, W.; Chen, Q.; et al. Azacytidine Targeting SARS-CoV-2 Viral RNA as a Potential Treatment for COVID-19. Sci. Bull. 2022, 67, 1022–1025. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Chang, J. Azvudine (FNC): A Promising Clinical Candidate for COVID-19 Treatment. Signal Transduct. Target. Ther. 2020, 5, 236. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-L.; Li, Y.-H.; Wang, L.-L.; Liu, H.-Q.; Lu, S.-Y.; Liu, Y.; Li, K.; Liu, B.; Li, S.-Y.; Shao, F.-M.; et al. Azvudine Is a Thymus-Homing Anti-SARS-CoV-2 Drug Effective in Treating COVID-19 Patients. Signal Transduct. Target. Ther. 2021, 6, 414. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Chang, J. The First Chinese Oral Anti-COVID-19 Drug Azvudine Launched. Innov. Camb. Mass 2022, 3, 100321. [Google Scholar] [CrossRef] [PubMed]
- Milisavljevic, N.; Konkolová, E.; Kozák, J.; Hodek, J.; Veselovská, L.; Sýkorová, V.; Čížek, K.; Pohl, R.; Eyer, L.; Svoboda, P.; et al. Antiviral Activity of 7-Substituted 7-Deazapurine Ribonucleosides, Monophosphate Prodrugs, and Triphoshates against Emerging RNA Viruses. ACS Infect. Dis. 2021, 7, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Schooley, R.T.; Carlin, A.F.; Beadle, J.R.; Valiaeva, N.; Zhang, X.-Q.; Clark, A.E.; McMillan, R.E.; Leibel, S.L.; McVicar, R.N.; Xie, J.; et al. Rethinking Remdesivir: Synthesis, Antiviral Activity, and Pharmacokinetics of Oral Lipid Prodrugs. Antimicrob. Agents Chemother. 2021, 65, aac.01155-21. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Shrivastava-Ranjan, P.; Chatterjee, P.; Flint, M.; Beadle, J.R.; Valiaeva, N.; Murphy, J.; Schooley, R.T.; Hostetler, K.Y.; Montgomery, J.M.; et al. Broad-Spectrum In Vitro Antiviral Activity of ODBG-P-RVn: An Orally-Available, Lipid-Modified Monophosphate Prodrug of Remdesivir Parent Nucleoside (GS-441524). Microbiol. Spectr. 2021, 9, e01537-21. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, Y.; Yang, S.; Li, G.; Zhou, Q.; Sun, J.; Xu, T.; Yang, Y.; Liao, R.; Shi, Y.; et al. The Adenosine Analog Prodrug ATV006 Is Orally Bioavailable and Has Preclinical Efficacy against Parental SARS-CoV-2 and Variants. Sci. Transl. Med. 2022, 14, eabm7621. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.-H.; Wang, M.-M.; Li, L.-Y.; Herdewijn, P.; Snoeck, R.; Andrei, G.; Liu, Z.-P.; Liu, C. Synthesis and Anti-SARS-CoV-2 Evaluation of Lipid Prodrugs of β-D-N4-Hydroxycytidine (NHC) and a 3′-Fluoro-Substituted Analogue of NHC. Bioorganic Chem. 2023, 135, 106527. [Google Scholar] [CrossRef] [PubMed]
- Bege, M.; Kiss, A.; Bereczki, I.; Hodek, J.; Polyák, L.; Szemán-Nagy, G.; Naesens, L.; Weber, J.; Borbás, A. Synthesis and Anticancer and Antiviral Activities of C-2′-Branched Arabinonucleosides. Int. J. Mol. Sci. 2022, 23, 12566. [Google Scholar] [CrossRef] [PubMed]
- Tonix Seeks to Advance OyaGen’s COVID-19 Treatment Under New Global Licensing Deal. Available online: https://www.biospace.com/article/tonix-seeks-to-advance-oyagen-s-covid-19-treatment-under-new-global-licensing-deal/ (accessed on 24 September 2023).
- Bibi, S.; Hasan, M.M.; Wang, Y.-B.; Papadakos, S.P.; Yu, H. Cordycepin as a Promising Inhibitor of SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp). Curr. Med. Chem. 2022, 29, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y. China Approves First Homegrown COVID Antiviral. In Nature; Springer Nature Limited: London, UK, 26 July 2022. [Google Scholar] [CrossRef]
- McCarthy, M.W. VV116 as a Potential Treatment for COVID-19. Expert Opin. Pharmacother. 2023, 24, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Chan, K.-H.; Kao, R.Y.T.; To, K.K.W.; Zheng, B.-J.; Li, C.P.Y.; Li, P.T.W.; Dai, J.; Mok, F.K.Y.; Chen, H.; et al. Broad-Spectrum Antivirals for the Emerging Middle East Respiratory Syndrome Coronavirus. J. Infect. 2013, 67, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic Efficacy of the Small Molecule GS-5734 against Ebola Virus in Rhesus Monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Pruijssers, A.J.; Chappell, J.D.; Gribble, J.; Lu, X.; Andres, E.L.; Bluemling, G.R.; Lockwood, M.A.; Sheahan, T.P.; Sims, A.C.; et al. Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019, 93, e01348-19. [Google Scholar] [CrossRef] [PubMed]
- Parang, K.; El-Sayed, N.S.; Kazeminy, A.J.; Tiwari, R.K. Comparative Antiviral Activity of Remdesivir and Anti-HIV Nucleoside Analogs against Human Coronavirus 229E (HCoV-229E). Molecules 2020, 25, 2343. [Google Scholar] [CrossRef] [PubMed]
- Głowacka, I.E.; Andrei, G.; Schols, D.; Snoeck, R.; Gawron, K. Design, Synthesis, and the Biological Evaluation of a New Series of Acyclic 1,2,3-Triazole Nucleosides. Arch. Pharm. 2017, 350, e1700166. [Google Scholar] [CrossRef] [PubMed]
- Pyrc, K.; Bosch, B.J.; Berkhout, B.; Jebbink, M.F.; Dijkman, R.; Rottier, P.; Hoek, L. van der Inhibition of Human Coronavirus NL63 Infection at Early Stages of the Replication Cycle. Antimicrob. Agents Chemother. 2006, 50, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, M.; Debarnot, C.; Imbert, I.; Selisko, B.; Snijder, E.J.; Canard, B.; Decroly, E. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLoS Pathog. 2010, 6, e1000863, Correction in PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef]
- Aouadi, W.; Blanjoie, A.; Vasseur, J.-J.; Debart, F.; Canard, B.; Decroly, E. Binding of the Methyl Donor S-Adenosyl-l-Methionine to Middle East Respiratory Syndrome Coronavirus 2′-O-Methyltransferase Nsp16 Promotes Recruitment of the Allosteric Activator Nsp10. J. Virol. 2016, 91, e02217-16. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir Is a Direct-Acting Antiviral That Inhibits RNA-Dependent RNA Polymerase from Severe Acute Respiratory Syndrome Coronavirus 2 with High Potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.; Li, X.; Kumar, S.; Jockusch, S.; Chien, M.; Tao, C.; Morozova, I.; Kalachikov, S.; Kirchdoerfer, R.N.; Russo, J.J. Nucleotide Analogues as Inhibitors of SARS-CoV Polymerase. Pharmacol. Res. Perspect. 2020, 8, e00674. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.; Anderson, T.K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. [Google Scholar] [CrossRef] [PubMed]
- Jockusch, S.; Tao, C.; Li, X.; Chien, M.; Kumar, S.; Morozova, I.; Kalachikov, S.; Russo, J.J.; Ju, J. Sofosbuvir Terminated RNA Is More Resistant to SARS-CoV-2 Proofreader than RNA Terminated by Remdesivir. Sci. Rep. 2020, 10, 16577. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Zhang, X.; Zheng, W.; Sun, J.; Hua, L.; Xu, L.; Chu, X.; Ding, S.; Xiong, W. Development of a Simple In Vitro Assay To Identify and Evaluate Nucleotide Analogs against SARS-CoV-2 RNA-Dependent RNA Polymerase. Antimicrob. Agents Chemother. 2020, 65, e01508-20. [Google Scholar] [CrossRef] [PubMed]
- Devkota, K.; Schapira, M.; Perveen, S.; Khalili Yazdi, A.; Li, F.; Chau, I.; Ghiabi, P.; Hajian, T.; Loppnau, P.; Bolotokova, A.; et al. Probing the SAM Binding Site of SARS-CoV-2 Nsp14 In Vitro Using SAM Competitive Inhibitors Guides Developing Selective Bisubstrate Inhibitors. SLAS Discov. Adv. Life Sci. R D 2021, 26, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Klima, M.; Khalili Yazdi, A.; Li, F.; Chau, I.; Hajian, T.; Bolotokova, A.; Kaniskan, H.Ü.; Han, Y.; Wang, K.; Li, D.; et al. Crystal Structure of SARS-CoV-2 Nsp10–Nsp16 in Complex with Small Molecule Inhibitors, SS148 and WZ16. Protein Sci. 2022, 31, e4395. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ghiabi, P.; Hajian, T.; Klima, M.; Li, A.S.M.; Khalili Yazdi, A.; Chau, I.; Loppnau, P.; Kutera, M.; Seitova, A.; et al. SS148 and WZ16 Inhibit the Activities of Nsp10-Nsp16 Complexes from All Seven Human Pathogenic Coronaviruses. Biochim. Biophys. Acta Gen. Subj. 2023, 1867, 130319. [Google Scholar] [CrossRef] [PubMed]
- Kuzikov, M.; Costanzi, E.; Reinshagen, J.; Esposito, F.; Vangeel, L.; Wolf, M.; Ellinger, B.; Claussen, C.; Geisslinger, G.; Corona, A.; et al. Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen. ACS Pharmacol. Transl. Sci. 2021, 4, 1096–1110. [Google Scholar] [CrossRef] [PubMed]
- Sourimant, J.; Lieber, C.M.; Aggarwal, M.; Cox, R.M.; Wolf, J.D.; Yoon, J.-J.; Toots, M.; Ye, C.; Sticher, Z.; Kolykhalov, A.A.; et al. 4′-Fluorouridine Is an Oral Antiviral That Blocks Respiratory Syncytial Virus and SARS-CoV-2 Replication. Science 2022, 375, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, Q.; Yi, D.; Li, Q.; Guo, S.; Ma, L.; Zhang, Y.; Dong, D.; Guo, F.; Liu, Z.; et al. 5-Iodotubercidin Inhibits SARS-CoV-2 RNA Synthesis. Antivir. Res. 2022, 198, 105254. [Google Scholar] [CrossRef] [PubMed]
- Schindewolf, C.; Lokugamage, K.; Vu, M.N.; Johnson, B.A.; Scharton, D.; Plante, J.A.; Kalveram, B.; Crocquet-Valdes, P.A.; Sotcheff, S.; Jaworski, E.; et al. SARS-CoV-2 Uses Nonstructural Protein 16 To Evade Restriction by IFIT1 and IFIT3. J. Virol. 2023, 97, e01532-22. [Google Scholar] [CrossRef] [PubMed]
- Ahmed-Belkacem, R.; Sutto-Ortiz, P.; Guiraud, M.; Canard, B.; Vasseur, J.-J.; Decroly, E.; Debart, F. Synthesis of Adenine Dinucleosides SAM Analogs as Specific Inhibitors of SARS-CoV Nsp14 RNA Cap Guanine-N7-Methyltransferase. Eur. J. Med. Chem. 2020, 201, 112557. [Google Scholar] [CrossRef] [PubMed]
- Otava, T.; Šála, M.; Li, F.; Fanfrlík, J.; Devkota, K.; Perveen, S.; Chau, I.; Pakarian, P.; Hobza, P.; Vedadi, M.; et al. The Structure-Based Design of SARS-CoV-2 Nsp14 Methyltransferase Ligands Yields Nanomolar Inhibitors. ACS Infect. Dis. 2021, 7, 2214–2220. [Google Scholar] [CrossRef] [PubMed]
- Bobiļeva, O.; Bobrovs, R.; Kaņepe, I.; Patetko, L.; Kalniņš, G.; Šišovs, M.; Bula, A.L.; Gri Nberga, S.; Borodušķis, M.R.; Ramata-Stunda, A.; et al. Potent SARS-CoV-2 mRNA Cap Methyltransferase Inhibitors by Bioisosteric Replacement of Methionine in SAM Cosubstrate. ACS Med. Chem. Lett. 2021, 12, 1102–1107. [Google Scholar] [CrossRef]
- Bobileva, O.; Bobrovs, R.; Sirma, E.E.; Kanepe, I.; Bula, A.L.; Patetko, L.; Ramata-Stunda, A.; Grinberga, S.; Jirgensons, A.; Jaudzems, K. 3-(Adenosylthio)Benzoic Acid Derivatives as SARS-CoV-2 Nsp14 Methyltransferase Inhibitors. Molecules 2023, 28, 768. [Google Scholar] [CrossRef] [PubMed]
- Ahmed-Belkacem, R.; Hausdorff, M.; Delpal, A.; Sutto-Ortiz, P.; Colmant, A.M.G.; Touret, F.; Ogando, N.S.; Snijder, E.J.; Canard, B.; Coutard, B.; et al. Potent Inhibition of SARS-CoV-2 Nsp14 N7-Methyltransferase by Sulfonamide-Based Bisubstrate Analogues. J. Med. Chem. 2022, 65, 6231–6249. [Google Scholar] [CrossRef] [PubMed]
- Amador, R.; Delpal, A.; Canard, B.; Vasseur, J.-J.; Decroly, E.; Debart, F.; Clavé, G.; Smietana, M. Facile Access to 4′-(N-Acylsulfonamide) Modified Nucleosides and Evaluation of Their Inhibitory Activity against SARS-CoV-2 RNA Cap N7-Guanine-Methyltransferase Nsp14. Org. Biomol. Chem. 2022, 20, 7582–7586. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.; Soto-Acosta, R.; Xie, J.; Wilson, D.J.; Dreis, C.D.; Majima, R.; Edwards, T.C.; Geraghty, R.J.; Chen, L. Bisubstrate Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 Nsp14 Methyltransferase. ACS Med. Chem. Lett. 2022, 13, 1477–1484. [Google Scholar] [CrossRef] [PubMed]
- Hausdorff, M.; Delpal, A.; Barelier, S.; Nicollet, L.; Canard, B.; Touret, F.; Colmant, A.; Coutard, B.; Vasseur, J.-J.; Decroly, E.; et al. Structure-Guided Optimization of Adenosine Mimetics as Selective and Potent Inhibitors of Coronavirus Nsp14 N7-Methyltransferases. Eur. J. Med. Chem. 2023, 256, 115474. [Google Scholar] [CrossRef] [PubMed]
- Elis, J.; Rašková, H. New Indications for 6-Azauridine Treatment in Man. A Review. Eur. J. Clin. Pharmacol. 1972, 4, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Lee, L.; Zhang, Z.; Wang, X.; Yu, Q.; Duan, X.; Chan, E. Preclinical Pharmacokinetic Studies of 3-Deazaneplanocin A, a Potent Epigenetic Anticancer Agent, and Its Human Pharmacokinetic Prediction Using GastroPlusTM. Eur. J. Pharm. Sci. 2015, 77, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Bisel, H.F.; Ansfield, F.; Mason, J.H.; Wilson, W. Clinical Studies with Tubercidin Administered by Direct Intravenous Injection. Cancer Res. 1970, 30, 76–78. [Google Scholar] [PubMed]
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04280705. Adaptive COVID-19 Treatment Trial (ACTT). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04280705?term=NCT04280705&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04292730. Study to Evaluate the Safety and Antiviral Activity of Remdesivir (GS-5734™) in Participants with Moderate Coronavirus Disease (COVID-19) Compared to Standard of Care Treatment. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04292730?term=NCT04292730&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04292899. Study to Evaluate the Safety and Antiviral Activity of Remdesivir (GS-5734™) in Participants with Severe Coronavirus Disease (COVID-19). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04292899?term=NCT04292899&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04401579. Adaptive COVID-19 Treatment Trial 2 (ACTT-2). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04401579?term=NCT04401579&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04409262. A Study to Evaluate the Efficacy and Safety of Remdesivir Plus Tocilizumab Compared with Remdesivir Plus Placebo in Hospitalized Participants with Severe COVID-19 Pneumonia (REMDACTA). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04409262?term=NCT04409262&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04492475. Adaptive COVID-19 Treatment Trial 3 (ACTT-3). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04492475?term=NCT04492475&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04539262. Study in Participants with Early Stage Coronavirus Disease 2019 (COVID-19) to Evaluate the Safety, Efficacy, and Pharmacokinetics of Remdesivir Administered by Inhalation. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04539262?term=NCT04539262&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04546581. Inpatient Treatment of COVID-19 with Anti-Coronavirus Immunoglobulin (ITAC). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04546581?term=NCT04546581&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04583956. ACTIV-5/Big Effect Trial (BET-A) for the Treatment of COVID-19. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04583956?term=NCT04583956&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Med-icine. Identifier NCT04593940. Immune Modulators for Treating COVID-19 (ACTIV-1 IM). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04593940?term=NCT04593940&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04988035. ACTIV-5/Big Effect Trial (BET-C) for the Treatment of COVID-19. 2021. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04988035?term=NCT04988035&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04405570. Safety, Tolerability and Efficacy of Molnupiravir (EIDD-2801) to Eliminate Infectious Virus Detection in Persons with COVID-19. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04405570?term=NCT04405570&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04392219. COVID-19 First in Human Study to Evaluate Safety, Tolerability, and Pharmacokinetics of EIDD-2801 in Healthy Volunteers. 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04392219?term=NCT04392219&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04405739. The Safety of Molnupiravir (EIDD-2801) and Its Effect on Viral Shedding of SARS-CoV-2 (END-COVID). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04405739?term=NCT04405739&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04575597. Efficacy and Safety of Molnupiravir (MK-4482) in Non-Hospitalized Adult Participants with COVID-19 (MK-4482-002). 2020. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04575597?term=NCT04575597&draw=2&rank=1 (accessed on 22 April 2024).
- ClinicalTrials.gov [Internet]. U.S. National Library of Medicine. Identifier NCT04709835. Study to Evaluate the Effects of AT-527 in Non-Hospitalized Adult Patients with Mild or Moderate COVID-19. 2021. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04709835?term=NCT04709835&draw=2&rank=1 (accessed on 22 April 2024).
- Kim, S.E. Long COVID: The Hunt for Causes and Cures. Available online: https://cen.acs.org/biological-chemistry/infectious-disease/Long-COVID-hunt-causes-cures/101/i30 (accessed on 19 September 2023).
- Virios Therapeutics Announces Positive Data Demonstrating Improvement in Multiple Long-COVID Symptoms Following Treatment with a Combination of Valacyclovir and Celecoxib in an Exploratory, Open-Label, Proof of Concept Study. Available online: https://ir.virios.com/news/press-releases/detail/100/virios-therapeutics-announcespositive-data-demonstrating (accessed on 18 September 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
