1. Introduction
In December 2019, a new respiratory illness began to spread throughout Wuhan, China. The virus responsible for this illness is the SARS-CoV-2 and the disease is called COVID-19 [
1]. It quickly spread through Wuhan, a city of 11 million people in Hubei province. It infected tens of thousands of people over the ensuing weeks. China imposed major restrictions on travel and work, and by the end of February, cases of COVID-19 had slowed inside the country while spiking all over the world. COVID-19 data from different countries reflects various mitigation measures [
2,
3], such as lockdown, social distancing, early detection of infectives, contact tracing, and vaccination [
4,
5,
6]. Many data-driven approaches in infectious disease modeling are linear models. When using linear regression, statistical methods such as Auto Regressive Moving Average (ARIMA) and Moving Average (MA) rely on assumptions which make it impossible to forecast transmission rate at any given time during a pandemic [
7]. Time-varying transmission rates have been suggested to efficiently model the spread of COVID-19. For example, fast methods for estimating time-varying transmission rate were introduced in [
8]; however, they reported that their method suffers from extreme sensitivity to noise. In [
9], a first-principle machine learning approach was presented to predict time-dependent parameters, but these parameters require good initial guesses. In March and April 2020, many countries instituted widespread lockdown [
10]. A model-fitting approach for lockdown and lockdown relaxation is presented in [
11], which requires good estimation of the model parameters as well as quantification of the impact of relaxation. In [
12], the time-varying reproduction number
is estimated for counties in Georgia, USA, with a
confidence credible interval.
The first epidemiology model, the SIR model, was presented by Kermack and McKendrick in 1927 [
13]. The SIR model has inspired several epidemiological studies of diseases like, Malaria and Dengue fever [
14] and recently COVID-19. A widely used threshold parameter for the spread or extinction of an infectious disease in an epidemiology model is the basic reproduction number [
15]. It is defined as the average number of persons an infected person can infect. When the basic reproduction number is less than one, the infectious disease vanishes. In the SIR model [
13], the basic reproduction number is computed as the ratio of the transmission rate to the recovery rate. In this paper, we adopt a variant of the asymptomatic-SIR model presented in [
16]. When the transmission and recovery rates are constants, the basic reproduction number is given by the ratio of the transmission rate to a weighted sum of the symptomatic and asymptomatic recovery rates. However, When the transmission rate is time-varying, we use a modified reproduction, which we call the time-varying reproduction
. This time-varying reproduction number,
, demonstrates the spread pattern of COVID-19 throughout the duration of the pandemic.
There is an asymptomatic period for every infective individual in the range of 7 to 14 days [
17]. There are also asymptomatic infectives that never show symptoms but are infectious [
16]. Early studies of the spread of COVID-19 shows that some of the infectives are asymptomatic infectives [
18,
19] and they are mostly unreported in the publicly available data [
16]. In [
20], it was reported that the asymptomatic infectives can spread the virus efficiently, and they are the silent spreaders of COVID-19, which has caused difficulties in the control of the pandemic. Early in the pandemic, the Centers for Disease Control and Prevention (CDC) estimates the proportion of the asymptomatic infectives to be
of the total infectives in the USA [
19]. A high population proportion of asymptomatic infectives was estimated in [
18] for China and Singapore. In [
20], the proportion of Asymptomatic infectious patients in Wanzhou district before 10 April 2020 was
. [
16] reported
of the total infectives were asymptomatic in northern Italy. In a study conducted in England from June through September 2020 and in Spain from 27 April to 11 May 2020, the proportions of asymptomatic infectives in England and Spain were reported to be
and
respectively [
21].
Deep learning [
22] and Neural networks have found applications in function approximation tasks, since neural networks are known to be universal approximators of continuous functions [
23,
24]. Feedforward neural networks (FNN) have been used to learn approximate solutions of differential equations. In [
25], FNN was combined with the traditional Cox model for survival analysis to predict the clinical outcome of COVID-19 patients. In [
26], FNN was used to develop differential equation solvers and parameter estimators by constraining the residual. This FNN is called the Physics Informed Neural Network (PINN). PINN has been used to simulate pandemic spread, see [
27], where the model parameters were taken to be constants [
26,
28], PINN was used to solve nonlinear partial differential equations from data. PINN has been used to solve system of ordinary differential equations [
29] and system of fractional differential equations [
30]. In [
31], an algorithm that combines PINN together with LSTM is presented to solve an epidemiological model and identify weekly and daily time-varying parameters.
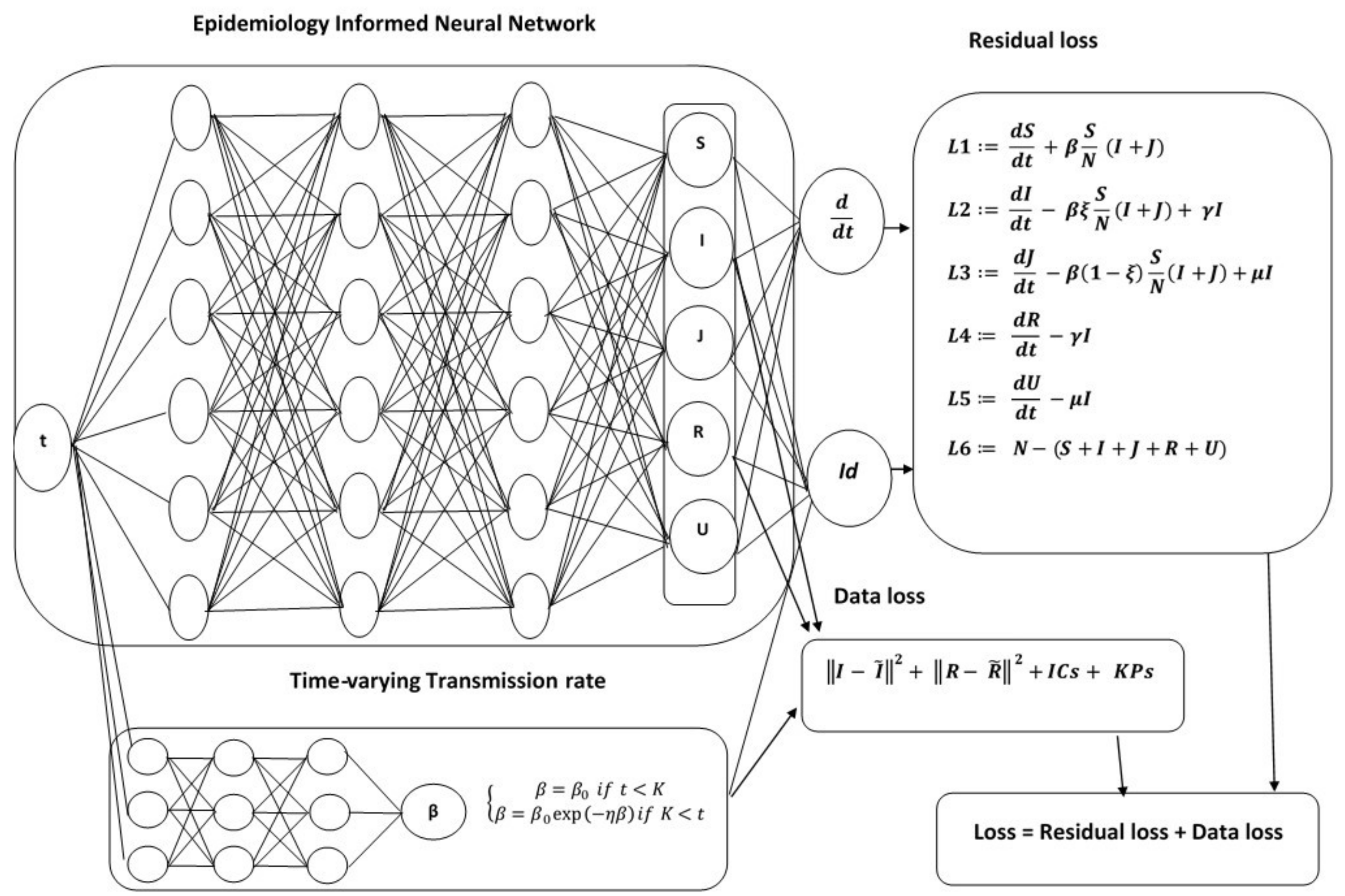
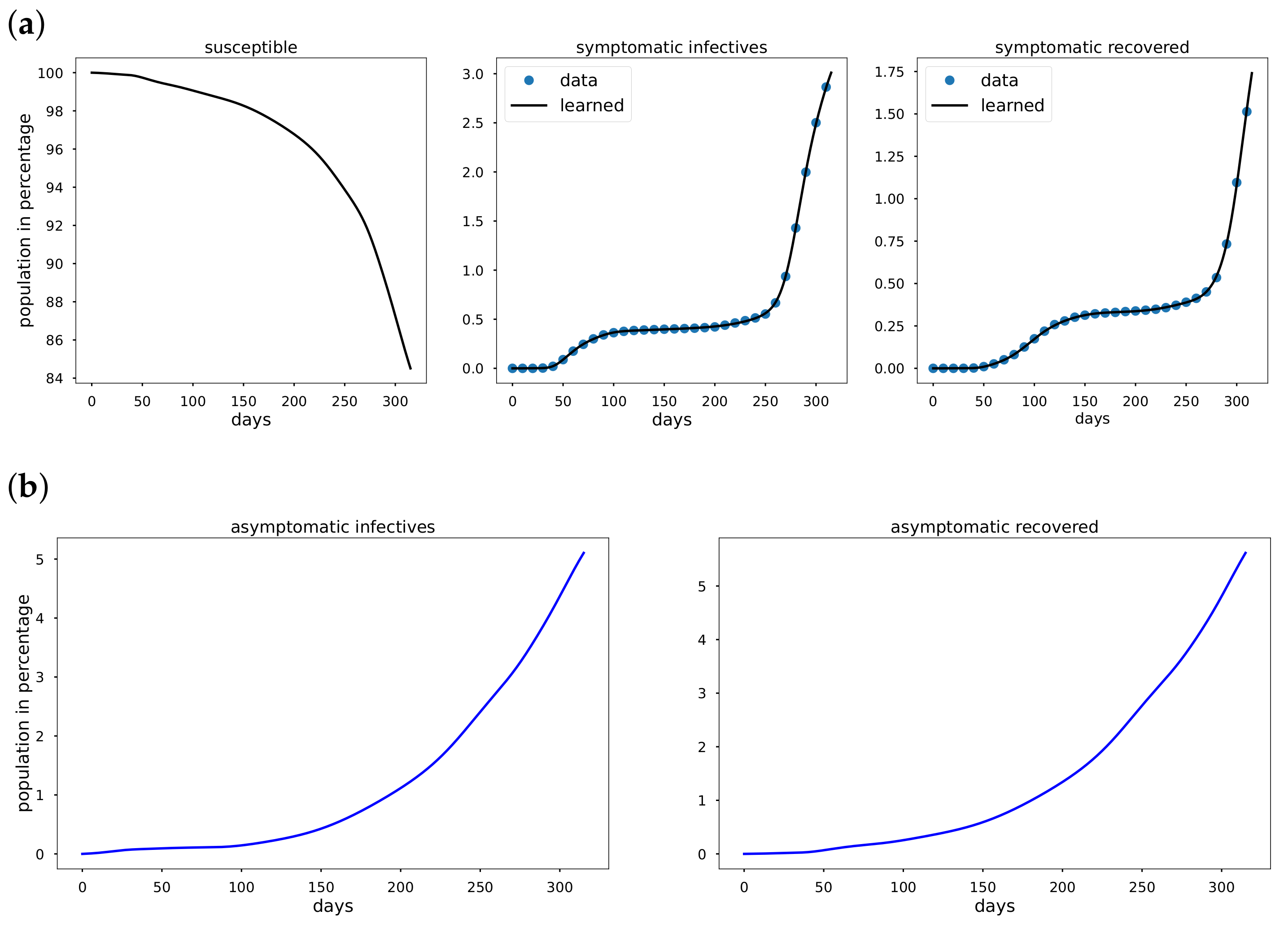
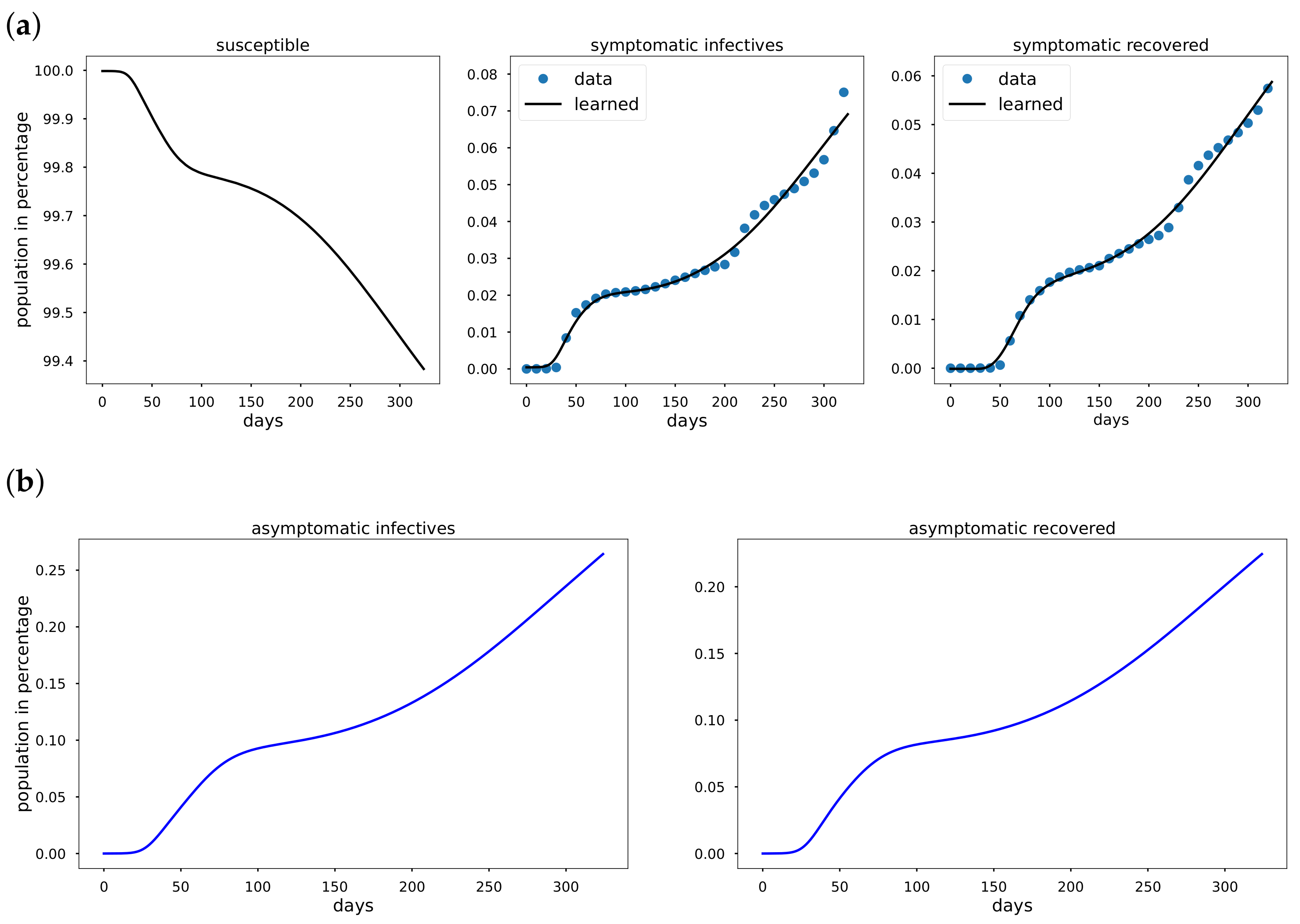
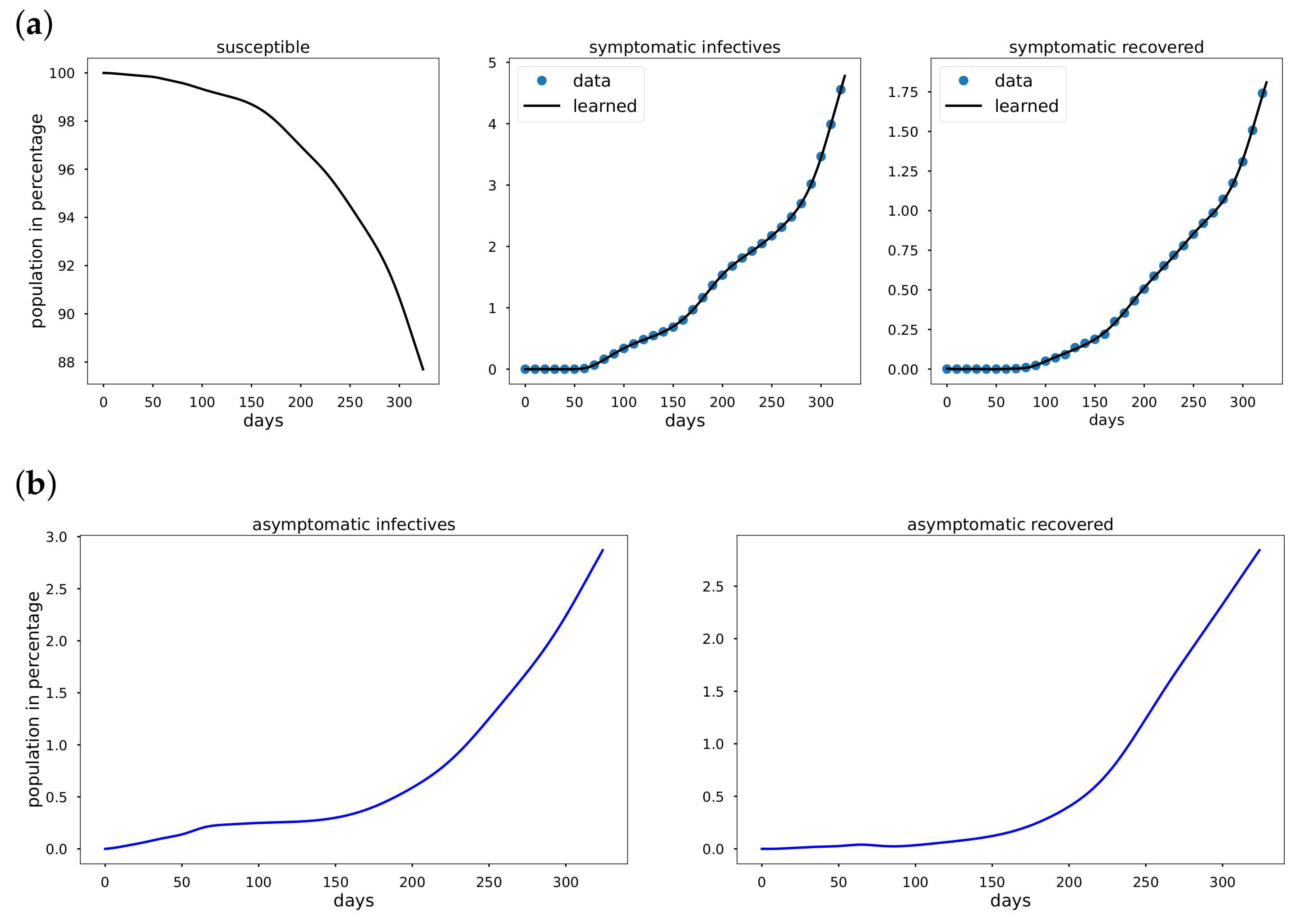
To overcome the limitations of statistical approaches, we present an Epidemiology-Informed Neural Network (EINN) inspired by applying a PINN to epidemiology models. Given that it may not be possible to know the most accurate form of a time-varying transmission rate, EINN algorithms is a viable option to learn time-varying transmission rate and to detect the impact of mitigation measures from data. The EINN loss function is extended to include some known epidemiology facts about infectious diseases. To detect hidden details in the training data, a cubic spline interpolation is used to generate sufficient training data. The proposed EINN algorithm can capture the dynamics of the spread of the disease and the influence of various mitigation measure. Since asymptomatic infectives population is unreported in the publicly available data [
32]. EINN algorithm learns asymptomatic infectives population by training on symptomatic infectives data that are available in the reported public data.
The paper is organized as follows. In
Section 2, we introduce and discuss the asymptomatic-SIR model, the neural network structure of EINN and the EINN algorithm for time-varying transmission rate. In
Section 3, data-driven simulation results for constant transmission rates, data-driven simulation results for pharmaceutical and non-pharmaceutical mitigation measures, and data-driven simulation results for time-varying transmission rates are presented. In
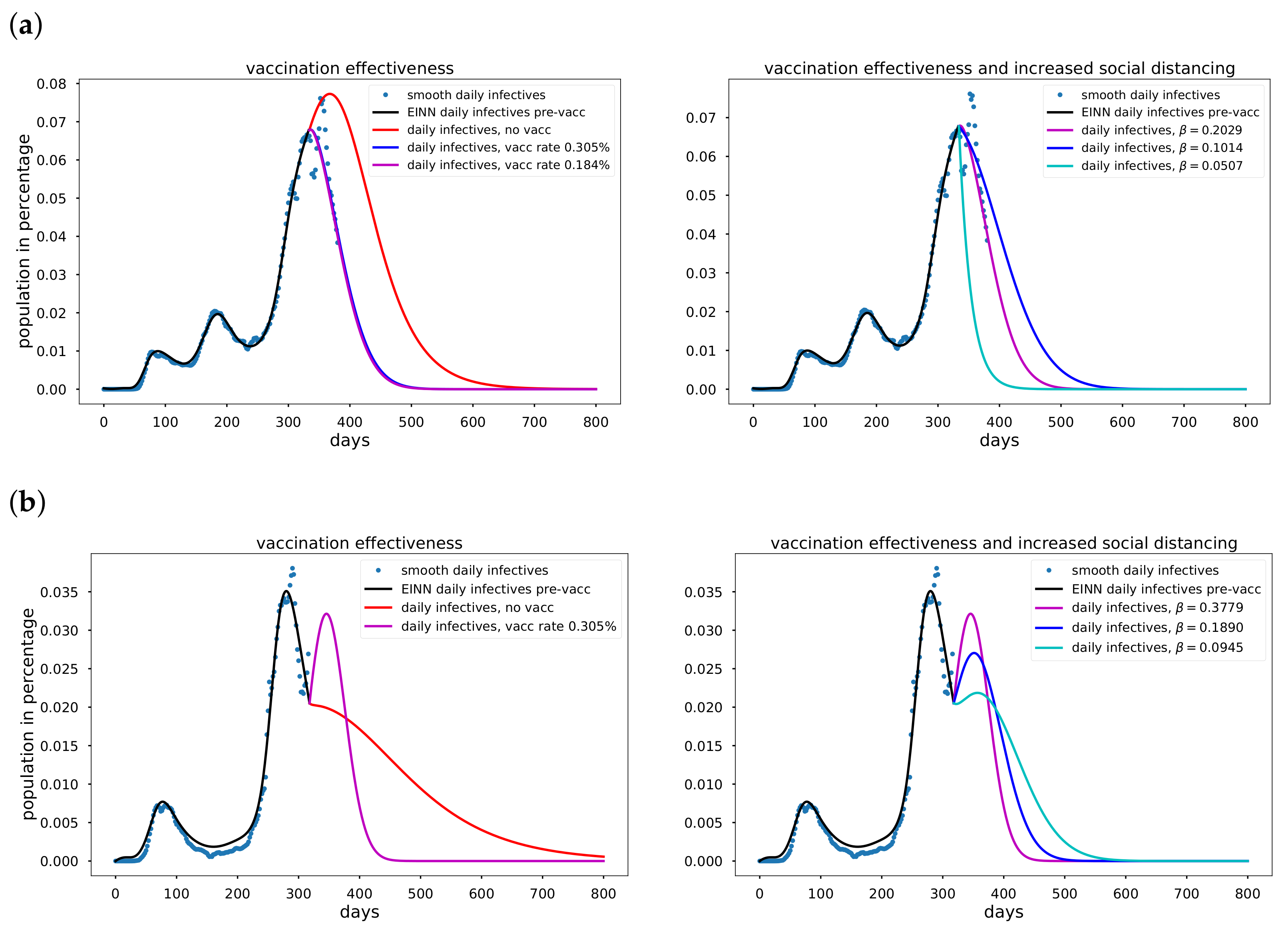
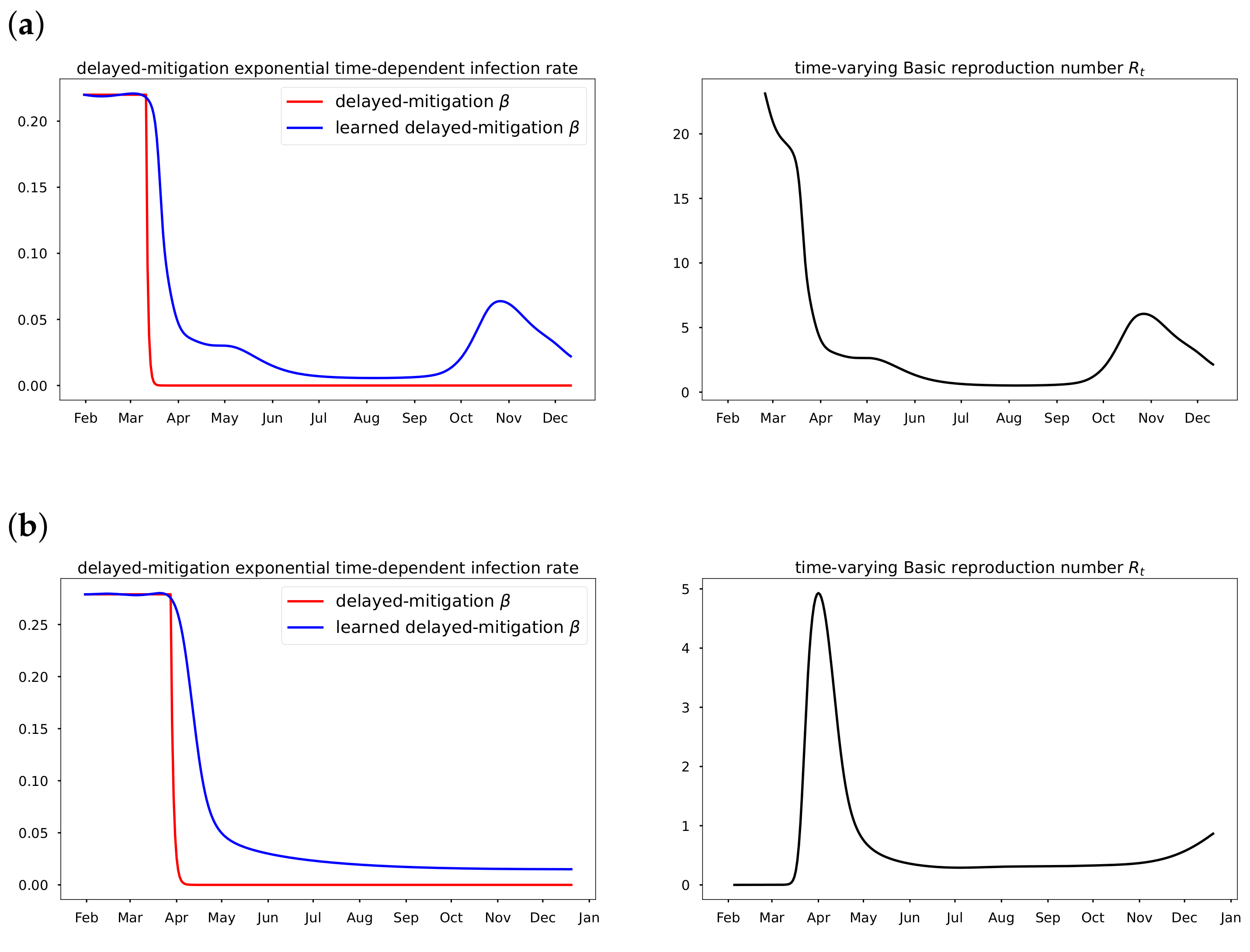
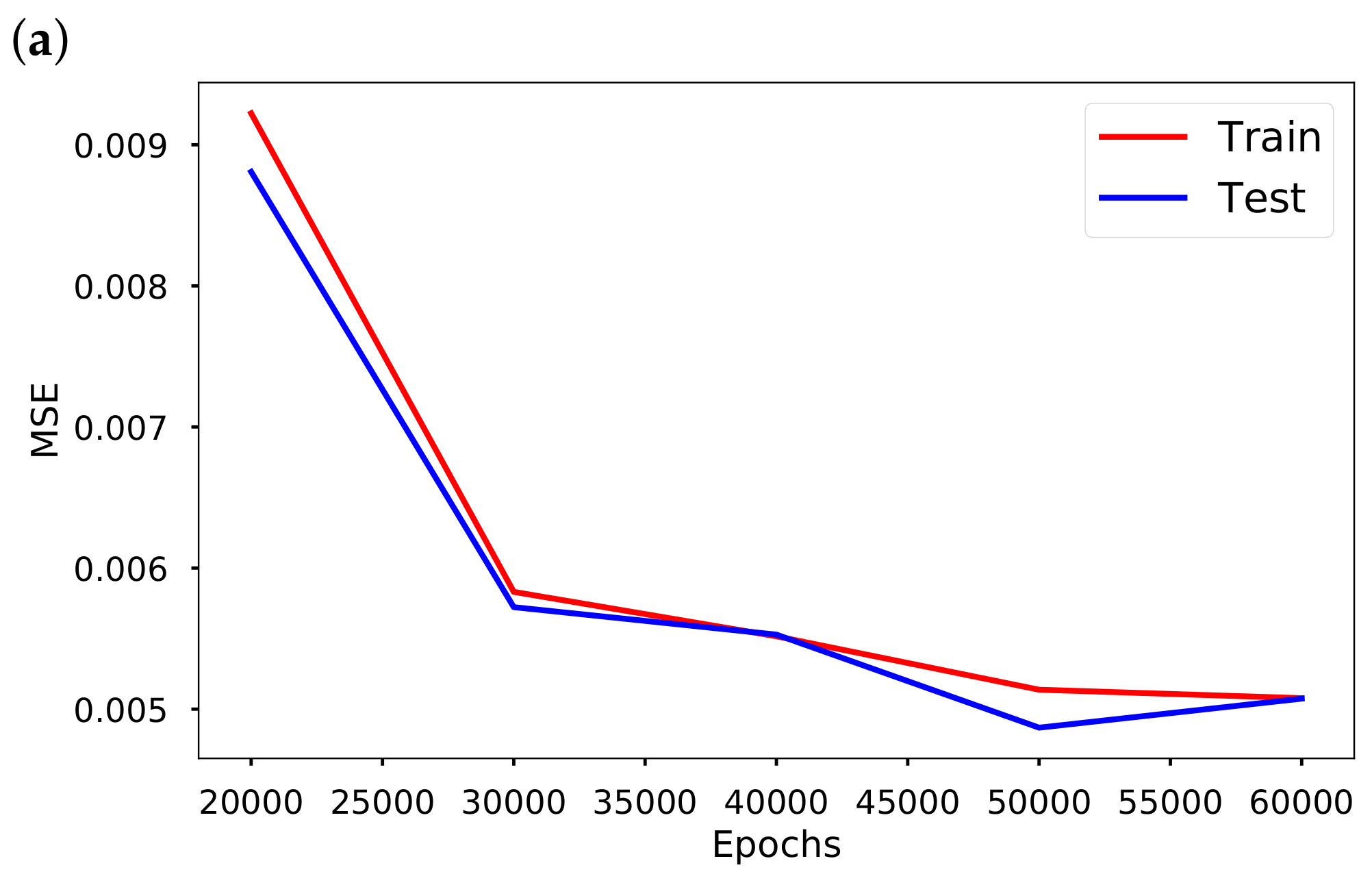
Section 4, we discuss the mitigation measures, vaccination efficacy, the time-varying transmission results and error metrics for data-driven simulation. Finally, a summary of the results in this paper is presented in
Section 5.