Finite-Element Based Image Registration for Endovascular Aortic Aneurysm Repair
Abstract
:1. Introduction
2. Materials and Methods
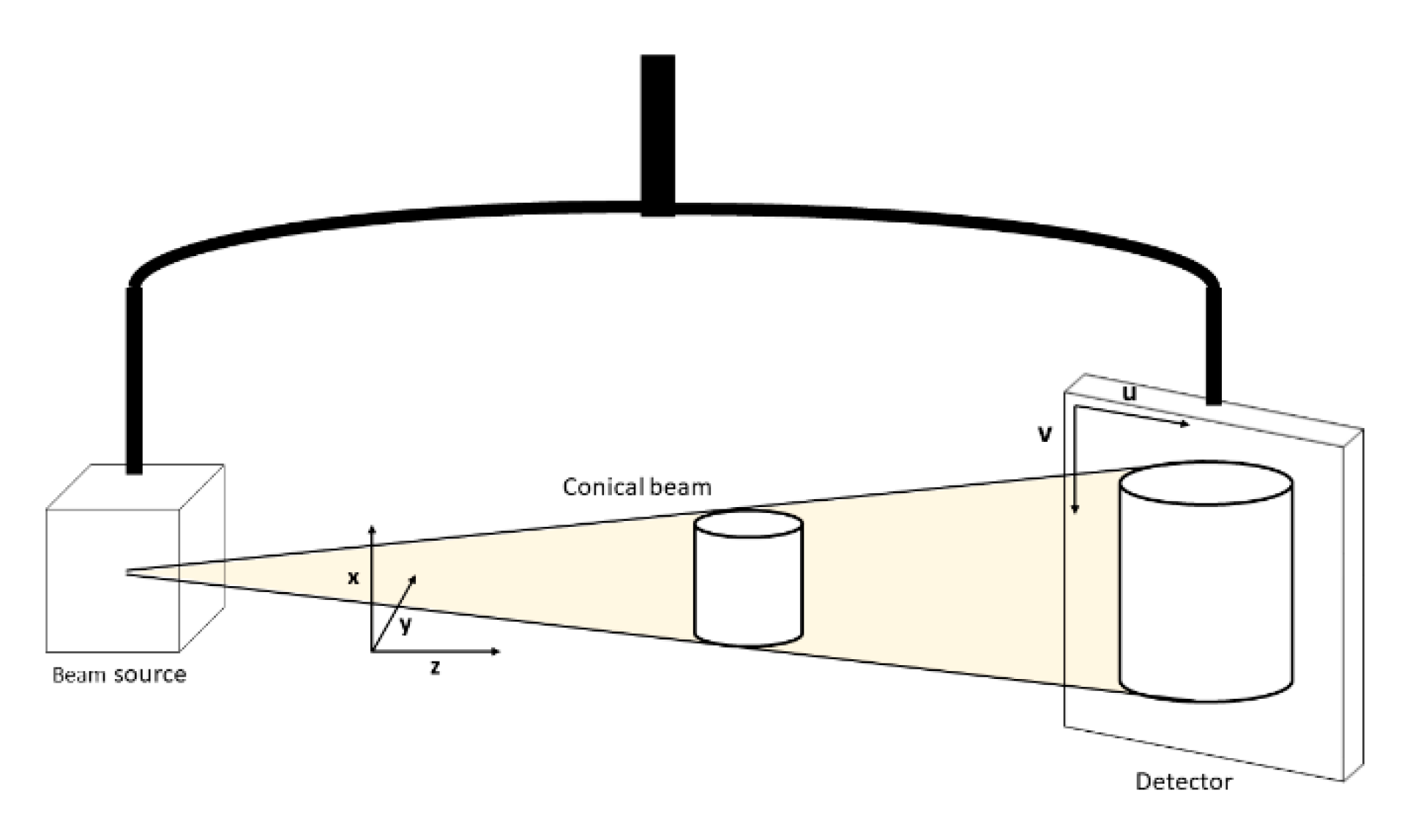
2.1. Data Acquisition
2.2. Non-Rigid Registration (NR) and Measurements
2.2.1. Non-Rigid Registration (NR) Method
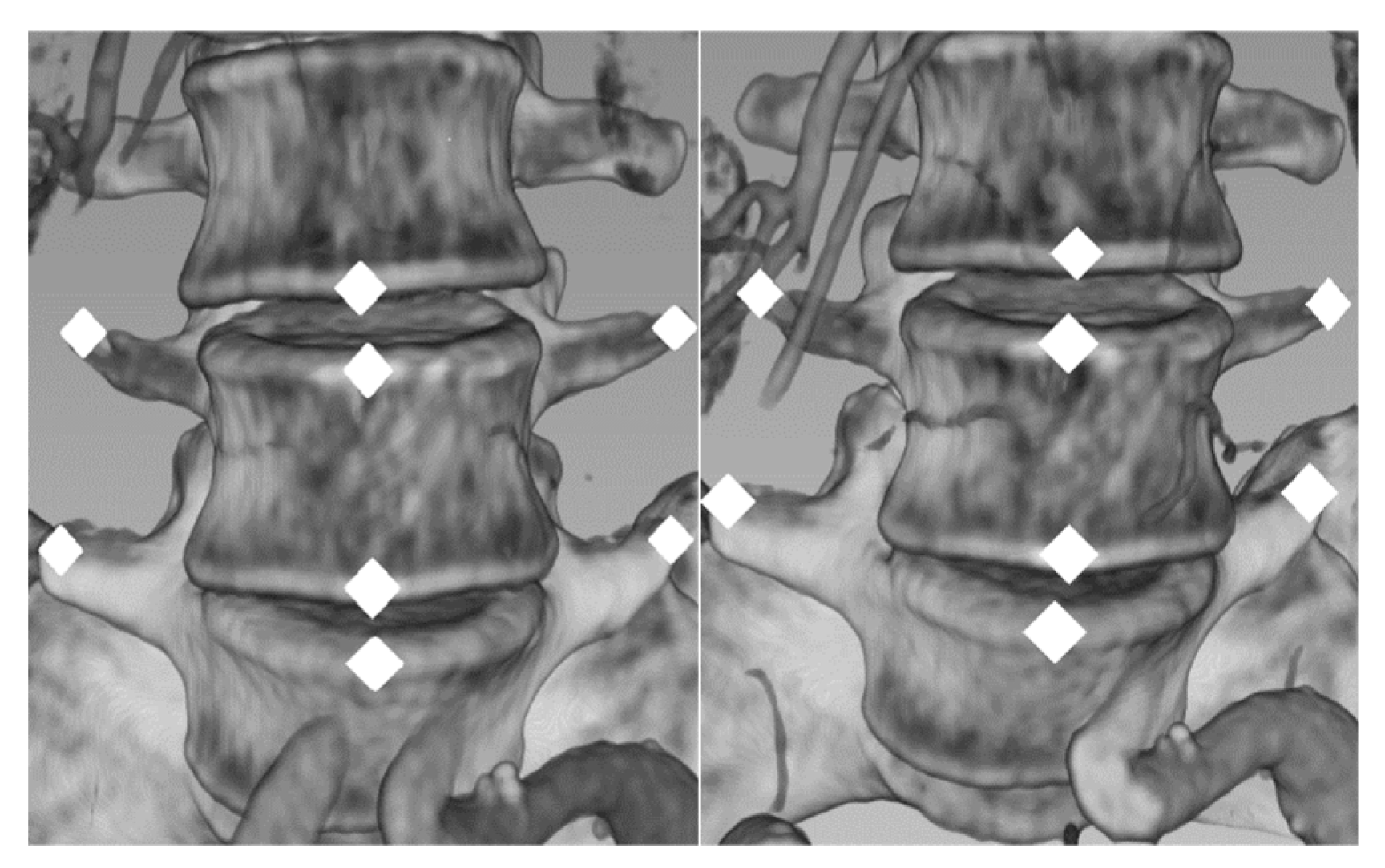
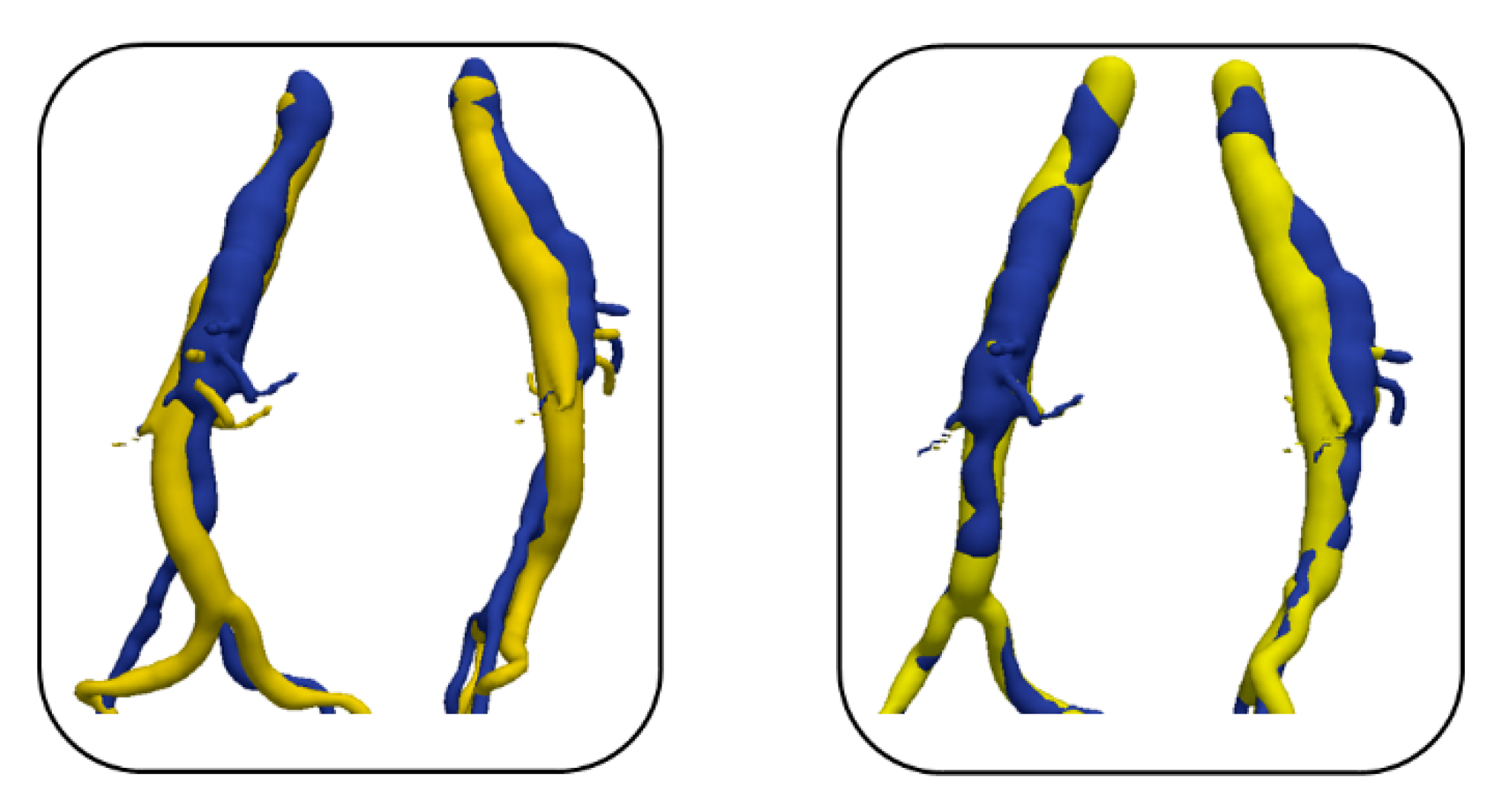
2.2.2. Evaluation
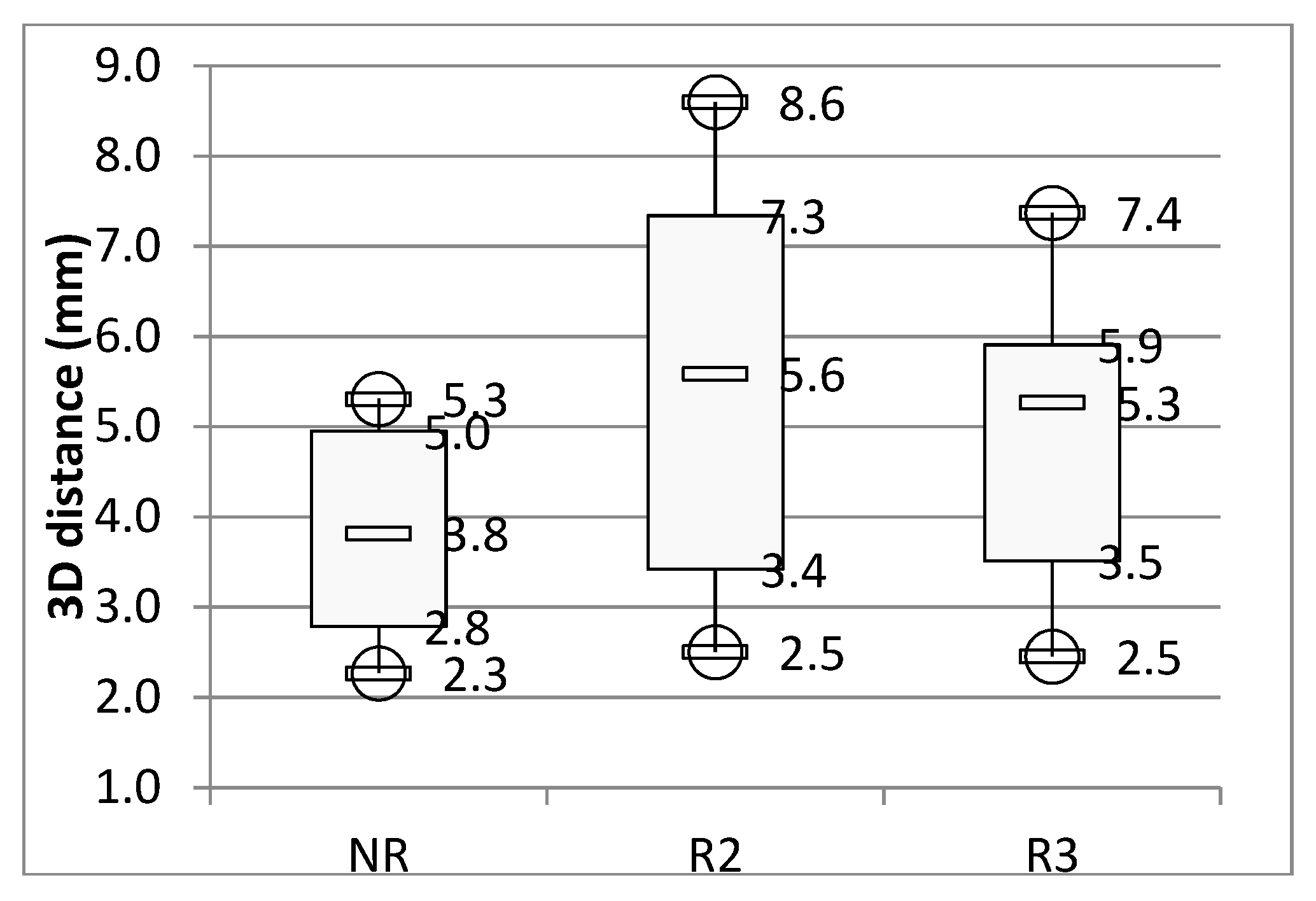
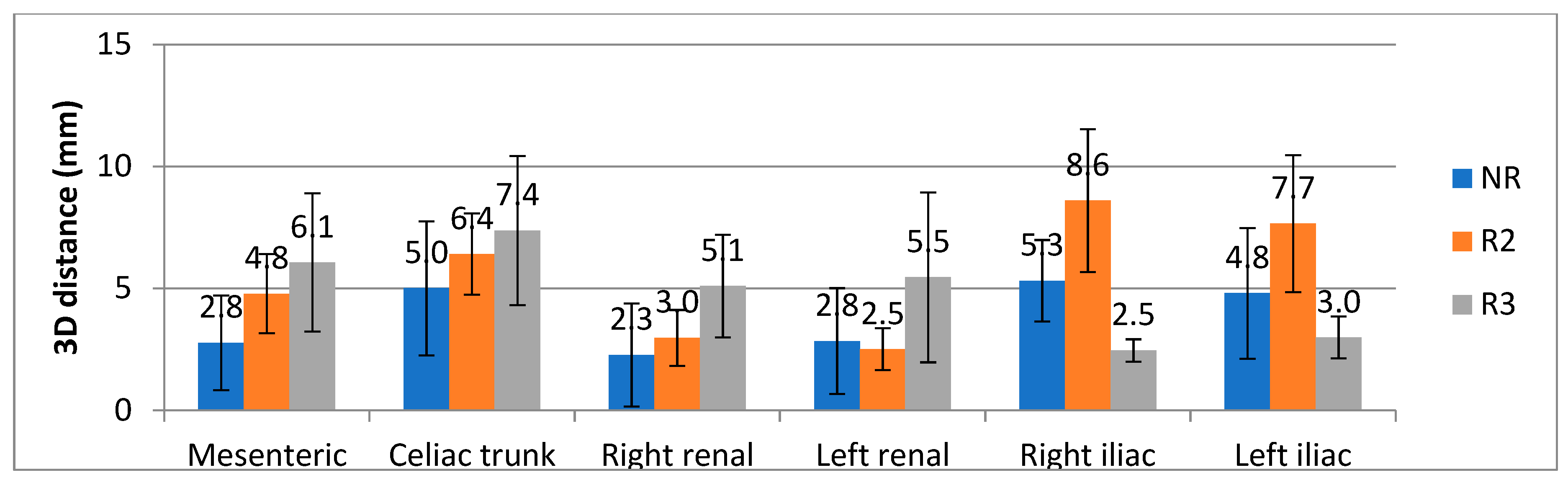
3. Results
4. Discussion
5. Patents
Author Contributions
Funding
Conflicts of Interest
Appendix A
Appendix A.1. Non-Rigid Registration (NR)
Appendix A.1.1. Initial Configuration
Appendix A.1.2. Model Constrains
Appendix A.1.3. 2D/2D Non-Rigid Registration
Appendix A.1.4. Back Projection Lines
Appendix A.2. Finite Element Model Resolution
Appendix A.2.1. Model Properties
Appendix A.2.2. FEM Resolution with Lagrange Multipliers
Appendix A.2.3. 3D Reconstruction
Appendix B
Appendix B.1. Evaluation
Appendix B.2. 3D Rigid Registration (R3D)
Appendix B.3. 2D Rigid Registration (R2D)
Appendix B.4. Measurement
References
- Anahita, D.; SreyRam, K.; Cheong, J.L.; Gilbert, R.U.; Sapan, S.D. Epidemiology of aortic aneurysm repair in the United States from 2000 to 2010. J. Vasc. Surg. 2014, 59, 1512–1517. [Google Scholar]
- Sajid, M.S.; Desai, M.; Haider, Z.; Baker, D.M.; Hamilton, G. Endovascular aortic aneurysm repair (EVAR) has significantly lower perioperative mortality in comparison to open repair: A systematic review. Asian J. Surg. 2008, 31, 119–123. [Google Scholar] [CrossRef]
- Halliday, A.W.; The United Kingdom EVAR Trial Investigators. Endovascular Repair of Aortic Aneurysm in Patients Physically Ineligible for Open Repair. N. Engl. J. Med. 2010, 362, 1863–1871. [Google Scholar]
- Macdonald, S.; Lee, R.; Williams, R.; Stansby, G. Towards safer carotid artery stenting: A scoring system for anatomic suitability. Stroke 2009, 40, 1698–1703. [Google Scholar] [CrossRef]
- Kauffmann, C.; Douane, F.; Therasse, E.; Lessard, S.; Elkouri, S.; Gilbert, P.; Beaudoin, N.; Pfister, M.; Blair, J.-F.; Soulez, G. Source of Errors and Accuracy of a Two-Dimensional/Three-Dimensional Fusion Road Map for Endovascular Aneurysm Repair of Abdominal Aortic Aneurysms. J. Vasc. Interv. Radiol. 2015. [Google Scholar] [CrossRef]
- Markelj, P.; Tomaževič, D.; Likar, B.; Pernuš, F. A review of 3D/2D registration methods for image-guided interventions. Med Image Anal. 2012, 16, 642–661. [Google Scholar] [CrossRef]
- Yu, W.; Tannast, M.; Zheng, G. Non-rigid free-form 2D–3D registration using a B-spline-based statistical deformation model. Pattern Recognit. 2017, 63, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Miao, S.; Wang, Z.J.; Liao, R. A CNN Regression Approach for Real-Time 2D/3D Registration. IEEE Trans. Med. Imaging 2016, 35, 1352–1363. [Google Scholar] [CrossRef]
- Chen, F.; Liu, J.; Liao, H. 3D Catheter Shape Determination for Endovascular Navigation Using a Two-Ste. IEEE Trans. Med. Imaging 2017, 36, 685–695. [Google Scholar] [CrossRef]
- Gorges, S.; Kerrien, E.; Berger, M.-O.; Trousset, Y.; Pescatore, J.; Anxionnat, R.; Picard, L. Model of a Vascular C-Arm for 3D Augmented Fluoroscopy in Interventional Radiology. Med. Image Comput. Comput. -Assist. Interv. Miccai 2005, 8, 214–222. [Google Scholar]
- Tacher, V.; Lin, M.; Desgranges, P.; Deux, J.-F.; Grünhagen, T.; Becquemin, J.; Luciani, A.; Rahmouni, A.; Kobeiter, H. Image Guidance for Endovascular Repair of Complex Aortic Aneurysms: Comparison of Two-dimensional and Three-dimensional Angiography and Image Fusion. J. Vasc. Interv. Radiol. 2013, 24, 1698–1706. [Google Scholar] [CrossRef] [Green Version]
- Hertault, A.; Maurel, B.; Sobocinski, J.; Gonzalez, T.M.; le Roux, M.; Azzaoui, R.; Midulla, M.; Haulon, S. Impact of Hybrid Rooms with Image Fusion on Radiation Exposure during Endovascular Aortic Repair. Eur. J. Vasc. Endovasc. Surg. 2014, 48, 382–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Ruiter, Q.M.B.; Reitsma, J.B.; Mol, F.L.; van Herwaarden, J.A. Meta-analysis of Cumulative Radiation Duration and Dose During EVAR Using Mobile, Fixed, or Fixed/3D Fusion C-Arms. J. Endovasc. Ther. 2016, 23, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Varnavas, A.; Carrell, T.; Penney, G. Fully automated 2D–3D registration and verification. Med. Image Anal. 2015, 26, 108–119. [Google Scholar] [CrossRef]
- Maurel, B.; Hertault, A.; Gonzalez, T.M.; Sobocinski, J.; le Roux, M.; Delaplace, J.; Azzaoui, R.; Midulla, M.; Haulon, S. Evaluation of Visceral Artery Displacement by Endograft Delivery System Insertion. J. Endovasc. Ther. 2014, 21, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Kaladji, A.; Dumenil, A.; Castro, M.; Cardon, A.; Becquemin, J.; Bou-Saïd, B.; Lucas, A.; Haigron, P. Prediction of deformations during endovascular aortic aneurysm repair using finite element simulation. Comput. Med. Imaging Graph. 2013, 37, 142–149. [Google Scholar] [CrossRef] [Green Version]
- Kaladji, A.; Villena, A.; Pascot, R.; Lalys, F.; Daoudal, A.; Clochard, E.; Lucas, A.; Cardon, A. Fusion imaging for EVAR with mobile c-arm. Ann. Vasc. Surg. 2018, 55. [Google Scholar] [CrossRef]
- Gindre, J.; Bel-Brunon, A.; Kaladji, A.; Duménil, A.; Rochette, M.; Lucas, A.; Haigron, P.; Combescure, A. Finite element simulation of the insertion of guidewires during an EVAR procedure: Example of a complex patient case, a first step toward patient-specific parameterized models. Int. J. Numer. Methods Biomed. Eng. 2015, 31. [Google Scholar] [CrossRef] [PubMed]
- Gindre, J.; Bel-Brunon, A.; Rochette, M.; Lucas, A.; Kaladji, A.; Haigron, P.; Combescure, A. Patient-Specific Finite-Element Simulation of the Insertion of Guidewire During an EVAR Procedure: Guidewire Position Prediction Validation on 28 Cases. IEEE Trans. Biomed. Eng. 2017, 64, 1057–1066. [Google Scholar] [CrossRef]
- Mohammadi, H.; Lessard, S.; Therasse, E.; Mongrain, R.; Soulez, G. A Numerical Preoperative Planning Model to Predict Arterial Deformations in Endovascular Aortic Aneurysm Repair. Ann. Biomed. Eng. 2018, 46, 2148–2161. [Google Scholar] [CrossRef]
- Liao, R.; Tan, Y.; Sundar, H.; Pfister, M.; Kamen, A. An Efficient Graph-Based Deformable 2D/3D Registration Algorithm with Applications for Abdominal Aortic Aneurysm Interventions. In Proceedings of the MIAR 2010: Medical Imaging and Augmented Reality, Beijing, China, 19–20 September 2010. [Google Scholar]
- Toth, D.; Pfister, M.; Maier, A.; Kowarschik, M.; Hornegger, J. Adaption of 3D models to 2D x-ray images during endovascular abdominal aneurysm repair. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Raheem, A.; Carrell, T.; Modarai, B.; Penney, G. Non-rigid 2D-3D image registration for use in Endovascular repair of Abdominal Aortic Aneurysms. Med. Image Underst. Anal. 2010, 15. [Google Scholar]
- Groher, M.; Zikic, D.; Navab, N. Deformable 2D-3D Registration of Vascular Structures in a One View Scenario. IEEE Trans. Med. Imaging 2009, 28. [Google Scholar] [CrossRef]
- Zheng, J.Q.; Zhou, X.Y.; Riga, C.; Yang, G.Z. Towards 3d path planning from a single 2d fluoroscopic image for robot assisted fenestrated endovascular aortic repair. Int. Conf. Robot. Autom. (Icra) 2019, 8747–8753. [Google Scholar] [CrossRef]
- The Vascular Modeling Toolkit. Available online: www.vmtk.org (accessed on 24 October 2018).
- Bergel, D.H. The static elastic properties of the arterial wall. J. Physiol. 1961, 445–457. [Google Scholar] [CrossRef]
- Veer, M.V.; Buth, J.; Merkx, M.; Tonino, P.; van den Bosch, H.; Pijls, N.; van de Vosse, F. Biomechanical properties of abdominal aortic aneurysms assessed by simultaneously measured pressure and changes in human. J. Vasc. Surg. 2008, 1401–1407. [Google Scholar] [CrossRef] [Green Version]
- Derycke, L.; Sénémaud, J.; Perrin, D.; Avril, S.; Desgranges, P.; Albertini, J.N.; Haulon, S. Patient Specific Computer Modelling for Automated Sizing of Fenestrated Stent Grafts. Eur. J. Vasc. Endovasc. Surg. 2020, 59, 237–246. [Google Scholar] [CrossRef]
- Derycke, L.; Perrin, D.; Cochennec, F.; Albertini, J.N.; Avril, S. Predictive numerical simulations of double branch stent-graft deployment in an aortic arch aneurysm. Ann. Biomed. Eng. 2019, 47, 1051–1062. [Google Scholar] [CrossRef] [Green Version]
- Santamaría, V.A.A.; Daniel, G.; Perrin, D.; Albertini, J.N.; Rosset, E.; Avril, S. Model reduction methodology for computational simulations of endovascular repair. Comput. Methods Biomech. Biomed. Eng. 2018, 21, 139–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrin, D.; Badel, P.; Orgéas, L.; Geindreau, C.; Dumenil, A.; Albertini, J.N.; Avril, S. Patient-specific numerical simulation of stent-graft deployment: Validation on three clinical cases. J. Biomech. 2015, 48, 1868–1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandwell, D.T. Biharmonic spline interpolation of GEOS-3 and SEASAT altimeter data. Geophys. Res. Lett. 1987. [Google Scholar] [CrossRef] [Green Version]
- Duriez, C. Real-time haptic simulation of medical procedures involving deformations and device-tissue interactions. Ph.D. Thesis, Université des Sciences et Technologie de Lille–Lille I, Villeneuve-d’Ascq, France, 2013. [Google Scholar]


| Patient | Sex | Age at CT Scan 1 | CT Scans Intervals (Months) |
|---|---|---|---|
| 1 | Female | 59 | 8 |
| 2 | Male | 78 | 6 |
| 3 | Male | 66 | 9 |
| 4 | Male | 68 | 4 |
| 5 | Male | 77 | 3 |
| 6 | Male | 54 | 5 |
| 7 | Male | 59 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pionteck, A.; Pierrat, B.; Gorges, S.; Albertini, J.-N.; Avril, S. Finite-Element Based Image Registration for Endovascular Aortic Aneurysm Repair. Modelling 2020, 1, 22-38. https://doi.org/10.3390/modelling1010002
Pionteck A, Pierrat B, Gorges S, Albertini J-N, Avril S. Finite-Element Based Image Registration for Endovascular Aortic Aneurysm Repair. Modelling. 2020; 1(1):22-38. https://doi.org/10.3390/modelling1010002
Chicago/Turabian StylePionteck, Aymeric, Baptiste Pierrat, Sébastien Gorges, Jean-Noël Albertini, and Stéphane Avril. 2020. "Finite-Element Based Image Registration for Endovascular Aortic Aneurysm Repair" Modelling 1, no. 1: 22-38. https://doi.org/10.3390/modelling1010002
APA StylePionteck, A., Pierrat, B., Gorges, S., Albertini, J.-N., & Avril, S. (2020). Finite-Element Based Image Registration for Endovascular Aortic Aneurysm Repair. Modelling, 1(1), 22-38. https://doi.org/10.3390/modelling1010002






