Epitope Specificity of HLA Class I Alloantibodies in Indian Renal Transplant Patients: A Single-Center Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Sample Collection and Processing
2.3. Antibody Detection
2.4. Epitope Analysis
2.5. Data Compilation and Categorization
3. Results
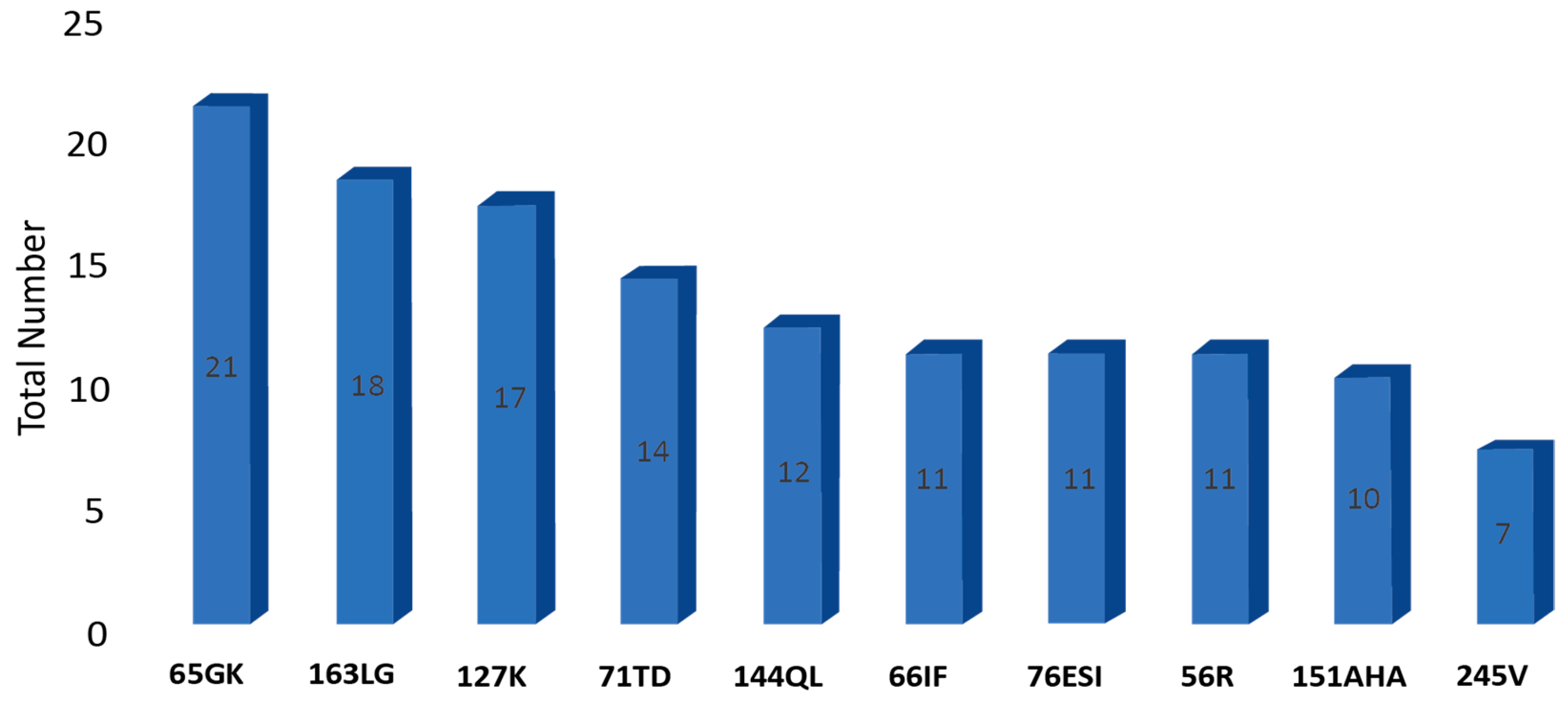
3.1. Overall Epitope Distribution
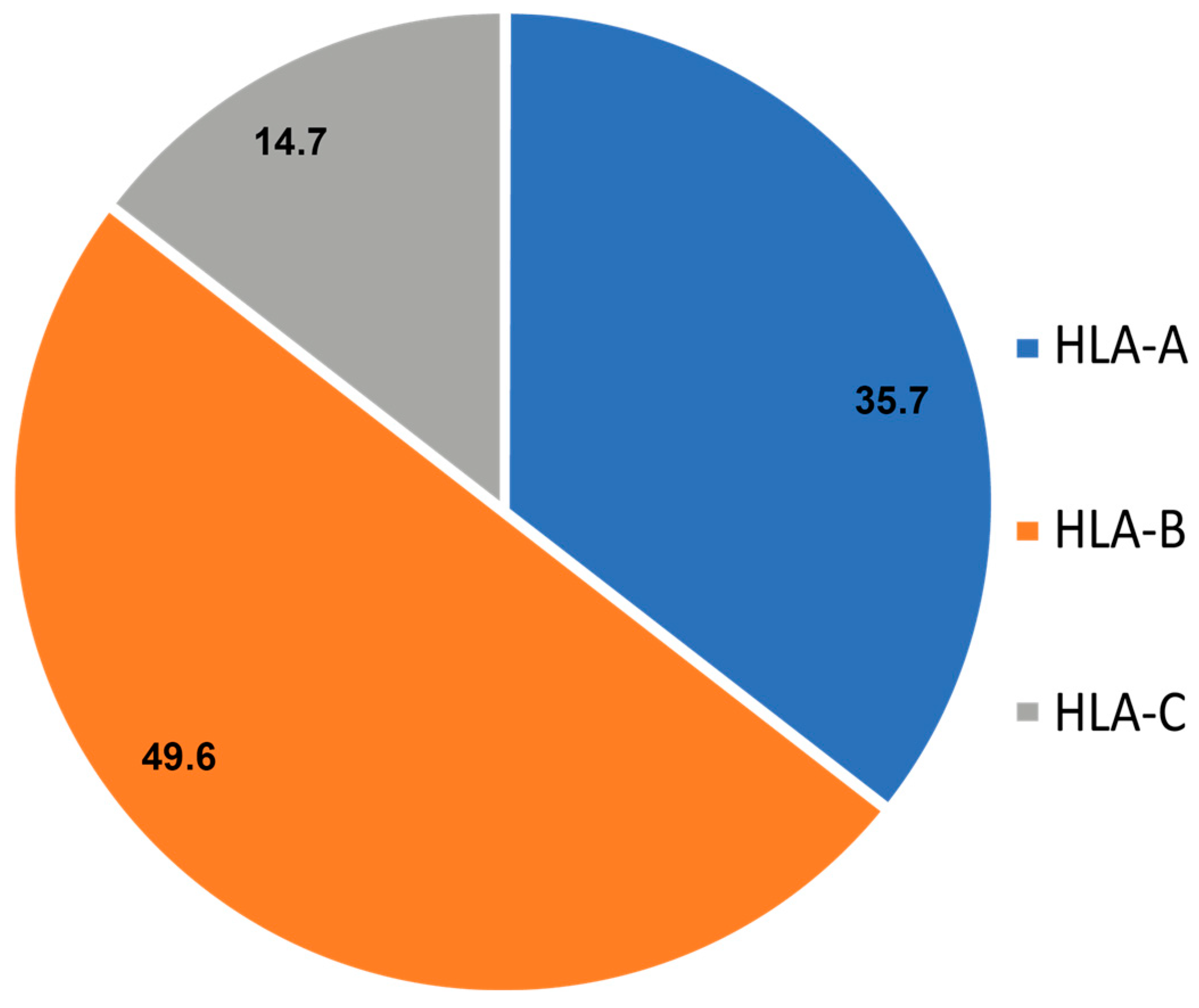
3.2. Locus-Wise Distribution
3.3. Sex-Wise Comparison
4. Discussion
5. Conclusions
6. Limitation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSA | Donor-Specific Antibodies |
| HLA | Human Leukocyte Antigen |
| MFI | Mean Fluorescence Intensity |
| NGS | Next Generation Sequencing |
| SAB | Single Antigen Bead |
| TCRs | T-cell Receptors |
References
- Renaldo, A.; Roa-Bautista, A.; González-López, E.; López-Hoyos, M.; San Segundo, D. Epitope-Level matching-a review of the novel concept of eplets in transplant histocompatibility. Transplantology 2021, 2, 336–347. [Google Scholar] [CrossRef]
- Yip, V.L.; Marson, A.G.; Jorgensen, A.L.; Pirmohamed, M.; Alfirevic, A.H.L.A. HLA genotype and carbamazepine—induced cutaneous adverse drug reactions: A systematic review. Clin. Pharmacol. Ther. 2012, 92, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Larkins, N.G.; Wong, G.; Taverniti, A.; Lim, W.H. Epitope matching in kidney transplantation: Recent advances and current limitations. Curr. Opin. Organ Transplant. 2019, 24, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Oguz, F.S.; Oguz, S.R.; Ogret, Y.; Karadeniz, T.S.; Ciftci, H.S.; Karatas, S.; Kivanc, D.; Aydin, F. Distribution of HLA epitope frequencies in Turkish population. Turk. J. Biochem. 2022, 47, 289–295. [Google Scholar] [CrossRef]
- Rodey, G.E.; Neylan, J.F.; Whelchel, J.D.; Revels, K.W.; Bray, R.A. Epitope specificity of HLA class I alloantibodies I. Frequency analysis of antibodies to private versus public specificities in potential transplant recipients. Hum. Immunol. 1994, 39, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Tambur, A.R.; Das, R. Can we use eplets (or molecular) mismatch load analysis to improve organ allocation? The hope and the hype. Transplantation 2023, 107, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Duquesnoy, R.J. HLA epitope based matching for transplantation. Transpl. Immunol. 2014, 31, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Duquesnoy, R.J. Reflections on HLA epitope-based matching for transplantation. Front. Immunol. 2016, 7, 469. [Google Scholar] [CrossRef] [PubMed]
- El-Awar, N. HLA epitopes–Empirically defined as conformational amino acids sequences of the HLA antigen and are likely to be part of the binding sites of anti-HLA antibodies. Hum. Immunol. 2022, 83, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Mangiola, M.; Ellison, M.; Marrari, M.; Xu, Q.; Mankowski, M.; Sese, D.; Lonze, B.E.; Montgomery, R.A.; Zeevi, A. HLA EPLET Frequencies Are Similar in Six Population Groups and Are Expressed by the Most Common HLA Alleles. HLA 2024, 104, e70000. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Prasad, N.; Kushwaha, R.S.; Patel, M.; Bhadauria, D.S.; Kaul, A. Prevalence of Human Leukocyte Antigen Alleles Polymorphism in North Indian Population. Indian J. Nephrol. 2025, 35, 536. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.C.; Chandra, D.; Raina, A.; Sharma, R.; Raina, V. Diversity of HLA Class I and II Genes in the North Indian Population. GenoMed Connect 2024, 1, 1–10. [Google Scholar] [CrossRef]
- Kumar, M.; Chakroborty, S.; Raina, V.; Kandpal, U.; Kumar, M. Analysis of distribution of HLA class I antigens in population from six north Indian states. Apollo Med. 2007, 4, 29–31. [Google Scholar] [CrossRef]
- Mishra, V.C.; Chandra, D.; Singh, P.; Deshpande, T.; Dorwal, P.; Raina, V. Prevalence and specificity of anti-HLA antibodies in Indian patients–single-centre data! ISBT Sci. Ser. 2019, 14, 374–378. [Google Scholar] [CrossRef]
- Chauhan, R.; Tiwari, A.K.; Rajvanshi, C.; Mehra, S.; Saini, A.; Aggarwal, G.; Bansal, S.B.; Kher, V.; Nandi, S.P. Prevalence of clinically significant anti-HLA antibodies in renal transplant patients: Single-center report from North India. Indian J. Nephrol. 2021, 31, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.C.; Raina, V. Enhancing Precision of the Single-antigen Bead (SAB) Assay: Considerations and Challenges. J. Clin. Transl. Pathol. 2024, 4, 12–17. [Google Scholar] [CrossRef]
- Mishra, V.C.; Chandra, D.; Raina, V. Histocompatibility Testing: A Fundamental Aspect of Renal Transplant Workup. Transplantology 2024, 5, 85–97. [Google Scholar] [CrossRef]
| S. No | Epitope | Associated HLA Locus | Total Number | FEMALES (N = 89) | MALES (N = 129) |
|---|---|---|---|---|---|
| 1 | 9D | BC | 2 | 1 | 1 |
| 2 | 9F | AC | 1 | 1 | 0 |
| 3 | 9T | A | 1 | 0 | 1 |
| 4 | 9Y | ABC | 1 | 0 | 1 |
| 5 | 11AV | BC | 1 | 0 | 1 |
| 6 | 12M | B | 3 | 1 | 2 |
| 7 | 14W | C | 1 | 0 | 1 |
| 8 | 16S | C | 2 | 1 | 1 |
| 9 | 19K | A | 5 | 2 | 3 |
| 10 | 24T | B | 2 | 1 | 1 |
| 11 | 30G | B | 6 | 4 | 2 |
| 12 | 32L | B | 1 | 1 | 0 |
| 13 | 41T | B | 2 | 1 | 1 |
| 14 | 43R | A | 6 | 3 | 3 |
| 15 | 44KM | A | 7 | 2 | 5 |
| 16 | 44RM | AB | 2 | 0 | 2 |
| 17 | 44RMA | B | 2 | 1 | 1 |
| 18 | 44RME | A | 2 | 1 | 1 |
| 19 | 44RT | B | 6 | 4 | 2 |
| 20 | 45EE | B | 2 | 0 | 2 |
| 21 | 56R | A | 11 | 2 | 9 |
| 22 | 62EE | A | 1 | 1 | 0 |
| 23 | 62GE | AB | 9 | 6 | 3 |
| 24 | 62GK | A | 5 | 2 | 3 |
| 25 | 62GRN | AB | 5 | 3 | 2 |
| 26 | 62LQ | A | 9 | 4 | 5 |
| 27 | 62QE | A | 3 | 2 | 1 |
| 28 | 62RN | AB | 2 | 1 | 1 |
| 29 | 62RR | AB | 6 | 4 | 2 |
| 30 | 63NI | B | 1 | 1 | 0 |
| 31 | 65GK | A | 21 | 5 | 16 |
| 32 | 65QIA | B | 9 | 9 | 0 |
| 33 | 65RA | AB | 3 | 0 | 3 |
| 34 | 65RNA | AB | 2 | 2 | 0 |
| 35 | 66I | B | 3 | 3 | 0 |
| 36 | 66IC | B | 1 | 1 | 0 |
| 37 | 66IF | B | 11 | 5 | 6 |
| 38 | 66IS | B | 1 | 1 | 0 |
| 39 | 66IY | B | 1 | 1 | 0 |
| 40 | 66KA | A | 1 | 1 | 0 |
| 41 | 66NM | AB | 1 | 0 | 1 |
| 42 | 66NV | A | 2 | 0 | 2 |
| 43 | 69AA | B | 1 | 0 | 1 |
| 44 | 69RA | C | 1 | 0 | 1 |
| 45 | 69TNT | B | 2 | 0 | 2 |
| 46 | 70IAQ | B | 3 | 2 | 1 |
| 47 | 71ATD | B | 4 | 3 | 1 |
| 48 | 71QS | A | 2 | 1 | 1 |
| 49 | 71SA | B | 4 | 2 | 2 |
| 50 | 71TD | B | 14 | 7 | 7 |
| 51 | 71TN | B | 2 | 2 | 0 |
| 52 | 71TTS | B | 1 | 1 | 0 |
| 53 | 73AN | C | 1 | 0 | 1 |
| 54 | 73AS | C | 1 | 1 | 0 |
| 55 | 73ID | A | 7 | 3 | 4 |
| 56 | 76ANT | A | 1 | 0 | 1 |
| 57 | 76ED | B | 2 | 1 | 1 |
| 58 | 76ESI | A | 11 | 7 | 4 |
| 59 | 76ESN | B | 4 | 2 | 2 |
| 60 | 76ET | AB | 1 | 1 | 0 |
| 61 | 76VRN | BC | 1 | 1 | 0 |
| 62 | 77S | ABC | 1 | 1 | 0 |
| 63 | 80I | AB | 5 | 4 | 1 |
| 64 | 82LR | AB | 8 | 4 | 4 |
| 65 | 90D | AC | 2 | 2 | 0 |
| 66 | 91R | C | 1 | 1 | 0 |
| 67 | 95V | A | 1 | 0 | 1 |
| 68 | 97N | B | 11 | 6 | 5 |
| 69 | 97S | B | 1 | 1 | 0 |
| 70 | 97V | B | 7 | 3 | 4 |
| 71 | 99Y | ABC | 1 | 0 | 1 |
| 72 | 103M | 2 | 1 | 1 | |
| 73 | 105S | A | 2 | 0 | 2 |
| 74 | 107W | A | 2 | 0 | 2 |
| 75 | 109F | AB | 2 | 0 | 2 |
| 76 | 113HD | BC | 1 | 0 | 1 |
| 77 | 113HN | B | 2 | 1 | 1 |
| 78 | 114H | AB | 2 | 2 | 0 |
| 79 | 114Q | A | 1 | 1 | 0 |
| 80 | 114R | A | 1 | 1 | 0 |
| 81 | 116L | BC | 1 | 1 | 0 |
| 82 | 116Y | ABC | 1 | 0 | 1 |
| 83 | 127K | A | 17 | 12 | 5 |
| 84 | 131S | B | 3 | 2 | 1 |
| 85 | 138K | C | 2 | 0 | 2 |
| 86 | 138MI | A | 1 | 1 | 0 |
| 87 | 144K | A | 2 | 0 | 2 |
| 88 | 144KR | A | 1 | 1 | 0 |
| 89 | 144QL | B | 12 | 3 | 9 |
| 90 | 144TKH | A | 10 | 4 | 6 |
| 91 | 145HT | A | 2 | 0 | 2 |
| 92 | 145RT | A | 7 | 3 | 4 |
| 93 | 147L | BC | 2 | 0 | 2 |
| 94 | 149TAH | A | 1 | 1 | 0 |
| 95 | 150AAH | A | 1 | 1 | 0 |
| 96 | 150AH | A | 4 | 1 | 3 |
| 97 | 151AHA | A | 10 | 6 | 4 |
| 98 | 151AHV | A | 3 | 2 | 1 |
| 99 | 151ARV | AB | 1 | 0 | 1 |
| 100 | 151H | A | 1 | 1 | 0 |
| 101 | 152RA | C | 1 | 0 | 1 |
| 102 | 152RR | A | 4 | 1 | 3 |
| 103 | 152T | C | 1 | 0 | 1 |
| 104 | 152V | AB | 1 | 0 | 1 |
| 105 | 152W | A | 1 | 0 | 1 |
| 106 | 156DA | BC | 3 | 3 | 0 |
| 107 | 156QA | AC | 1 | 0 | 1 |
| 108 | 156WA | ABC | 5 | 0 | 5 |
| 109 | 158T | B | 5 | 3 | 2 |
| 110 | 161D | A | 4 | 1 | 3 |
| 111 | 162GLS | B | 10 | 2 | 8 |
| 112 | 163EW | ABC | 5 | 2 | 3 |
| 113 | 163L | BC | 4 | 1 | 3 |
| 114 | 163LG | B | 18 | 4 | 14 |
| 115 | 163LS/G | B | 10 | 4 | 6 |
| 116 | 163LW | BC | 8 | 6 | 2 |
| 117 | 163RG | A | 4 | 1 | 3 |
| 118 | 163RW | A | 2 | 0 | 2 |
| 119 | 166DG | A | 6 | 2 | 4 |
| 120 | 170RH | AB | 4 | 4 | 0 |
| 121 | 173K | C | 5 | 2 | 3 |
| 122 | 177DK | B | 5 | 3 | 2 |
| 123 | 177DT | B | 2 | 2 | 0 |
| 124 | 177KT | C | 2 | 0 | 2 |
| 125 | 180E | B | 3 | 3 | 0 |
| 126 | 184A | A | 1 | 0 | 1 |
| 127 | 184R | C | 1 | 0 | 1 |
| 128 | 193AV | A | 2 | 1 | 1 |
| 129 | 193LV | C | 1 | 0 | 1 |
| 130 | 193PI | AB | 2 | 1 | 1 |
| 131 | 193PL | C | 4 | 2 | 2 |
| 132 | 199V | B | 4 | 2 | 2 |
| 133 | 211T | C | 2 | 1 | 1 |
| 134 | 245TA | B | 2 | 1 | 1 |
| 135 | 245V | A | 7 | 4 | 3 |
| 136 | 253Q | AC | 2 | 2 | 0 |
| 137 | 275EL | A | 1 | 0 | 1 |
| Total | 504 | 237 | 267 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, V.C.; Chandra, D.; Sharma, R.; Dhuliya, D.; Raina, V. Epitope Specificity of HLA Class I Alloantibodies in Indian Renal Transplant Patients: A Single-Center Study. Transplantology 2025, 6, 34. https://doi.org/10.3390/transplantology6040034
Mishra VC, Chandra D, Sharma R, Dhuliya D, Raina V. Epitope Specificity of HLA Class I Alloantibodies in Indian Renal Transplant Patients: A Single-Center Study. Transplantology. 2025; 6(4):34. https://doi.org/10.3390/transplantology6040034
Chicago/Turabian StyleMishra, Vikash Chandra, Dinesh Chandra, Ritu Sharma, Diksha Dhuliya, and Vimarsh Raina. 2025. "Epitope Specificity of HLA Class I Alloantibodies in Indian Renal Transplant Patients: A Single-Center Study" Transplantology 6, no. 4: 34. https://doi.org/10.3390/transplantology6040034
APA StyleMishra, V. C., Chandra, D., Sharma, R., Dhuliya, D., & Raina, V. (2025). Epitope Specificity of HLA Class I Alloantibodies in Indian Renal Transplant Patients: A Single-Center Study. Transplantology, 6(4), 34. https://doi.org/10.3390/transplantology6040034







