Urinary Mitochondrial DNA Is Related to Allograft Function in Living Donor Kidney Transplantation—An Observational Study of the VAPOR-1 Cohort
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Outcome
2.3. Timepoints
2.4. Nuclear and Mitochondrial DNA Analysis
2.5. mtDNA Copy Number Analysis
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics
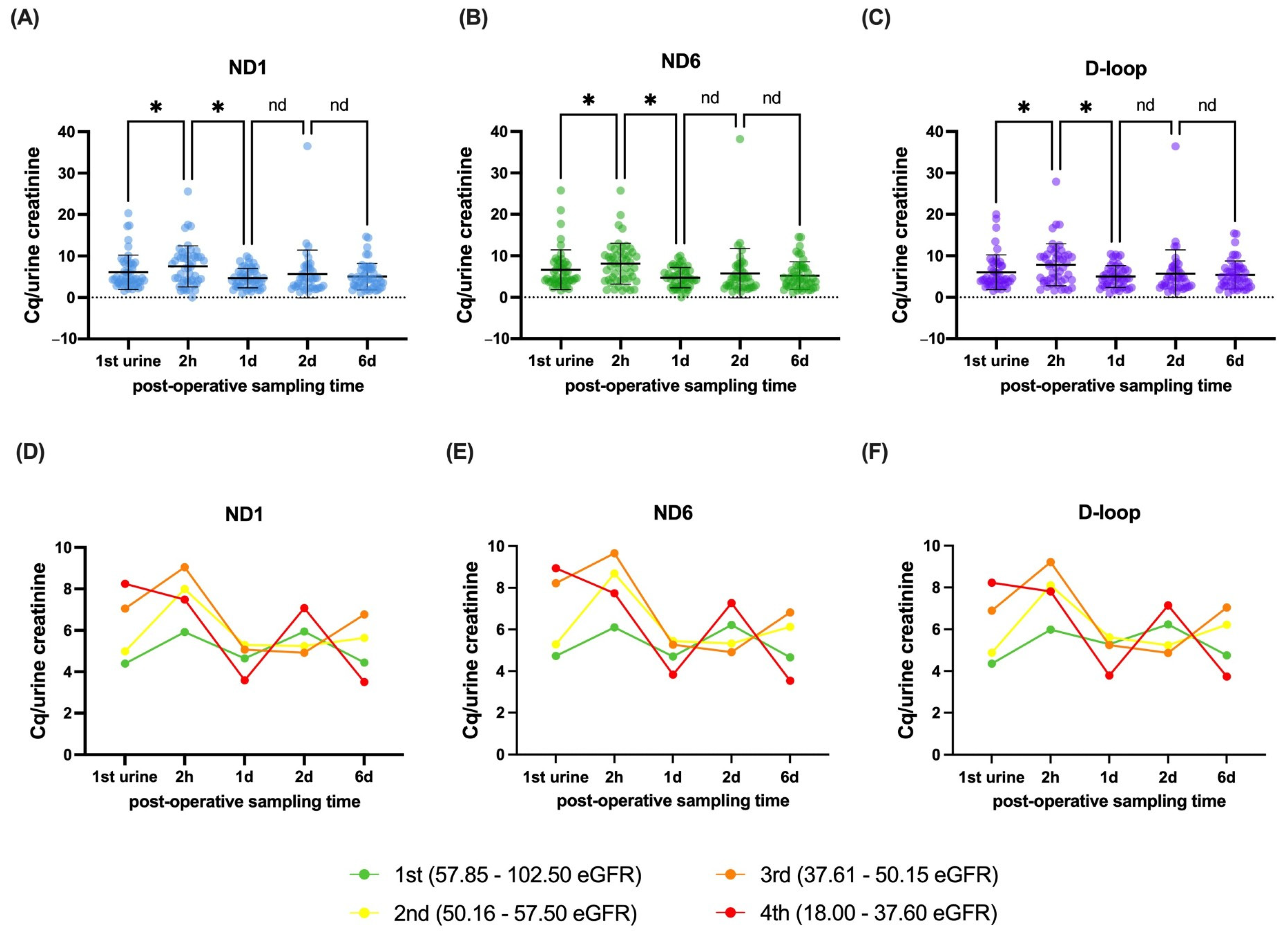
3.2. Dynamics and Stratification of mtDNA Genes in Recipients After Transplantation
3.3. The Additional Value of mtDNA Genes to Models for Renal Outcome
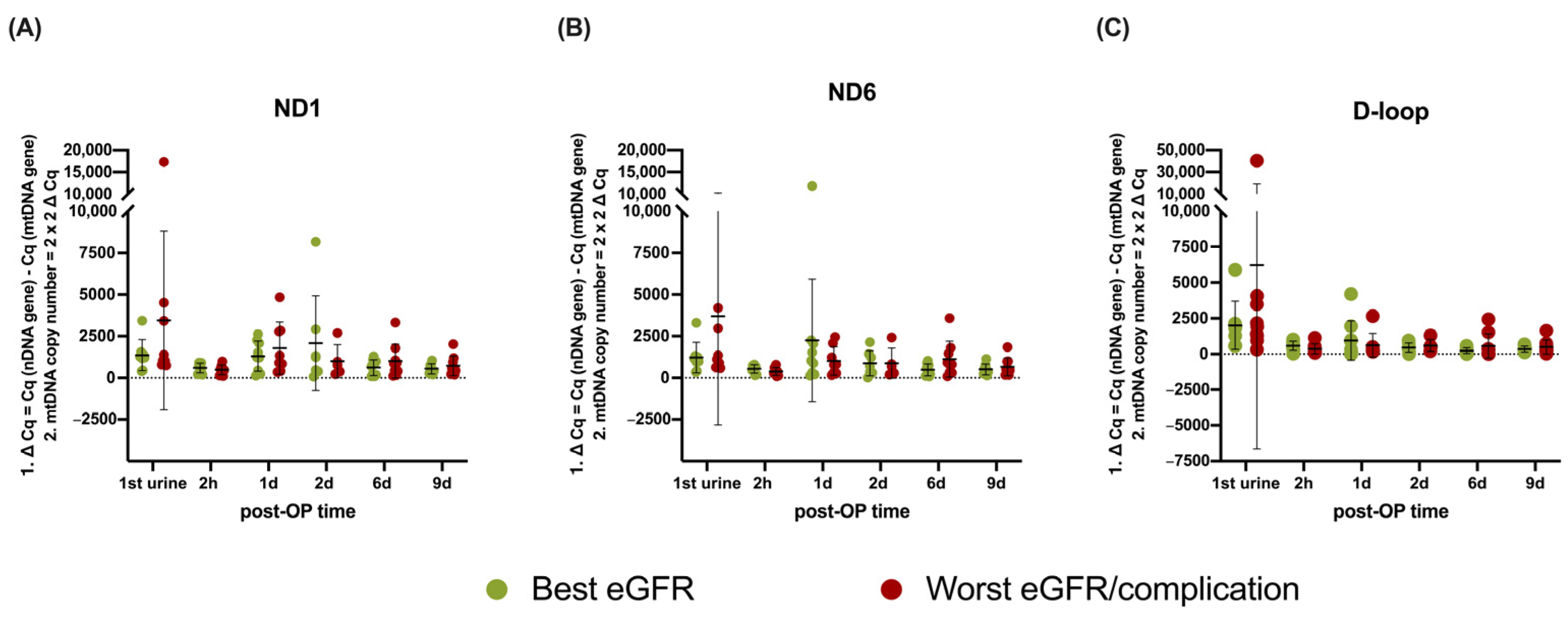
3.4. mtDNA Copy Number Analysis
4. Discussion
5. Future Perspectives and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKI | acute kidney injury |
| ATP | adenosine triphosphate |
| β2M | beta-2 microglobulin |
| BMI | body mass index |
| cGAS | cyclic GMP–AMP synthase |
| cGAMP | cyclic GMP-AMP |
| CI | confidence interval |
| CIT | cold ischemia time |
| CKD | chronic kidney disease |
| Cq | quantification cycle |
| D | day |
| DAMP | damage-associated molecular pattern |
| DGF | delayed graft function |
| D-loop | displacement-loop |
| eGFR | estimated glomerular filtration rate |
| ETC | electron transport chain |
| HLA | human leukocyte antigens |
| H-strand | heavy strand |
| IFTA | interstitial fibrosis and tubular atrophy |
| IQR | interquartile range |
| IRI | ischemia–reperfusion injury |
| Kg | kilogram |
| KIM-1 | kidney injury molecule-1 |
| LDKT | living donor kidney transplantation |
| L-strand | light strand |
| m2 | square meter |
| mGFR | measured glomerular filtration rate |
| Min | minute |
| Ml | milliliter |
| mtDNA | mitochondrial DNA |
| N | number |
| NAG | n-acetyl-β-D-glucosaminidase |
| nDNA | nuclear DNA |
| ND1 | NADH ubiquinone oxidoreductase subunit 1 |
| ND6 | NADH ubiquinone oxidoreductase subunit 6 |
| NGAL | neutrophil gelatinase-associated Lipocalin |
| NFκB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| PNF | primary non-function |
| post-OP | post-operative |
| PPIs | proton pump inhibitors |
| PRA | panel-specific antibodies |
| PRR | pattern recognition receptor |
| qPCR | quantitative polymerase chain reaction |
| REML | restricted maximum likelihood |
| ROS | reactive oxygen species |
| SD | standard deviation |
| STING | stimulator interferon genes |
| TLR | toll-like receptor |
| VAPOR-1 | Volatile Anesthetic Protection of Renal Transplants-1 |
| WIT1 | first warm ischemia time |
| WIT2 | second warm ischemia time |
| Y | years |
Appendix A
Appendix A.1. Urinary Creatinine Does Not Explain mtDNA Associations
| 1-Month eGFR | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| Creatinine 1st urine | 0.971 | 0.981 | −0.032 (−1.793–1.728) |
| Creatinine 2 h | 0.784 | 0.905 | −0.181 (−1.498–1.136) |
| 12-Month eGFR | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| Creatinine 1st urine | 0.032 | 0.083 | 1.764 (0.154–3.374) |
| Creatinine 2 h | 0.395 | 0.532 | 0.535 (−0.716–1.787) |
| 24-Month eGFR | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| Creatinine 1st urine | 0.067 | 0.108 | 1.894 (−0.140–3.928) |
| Creatinine 2 h | 0.007 | 0.057 | 2.337 (0.656–4.019) |
| ΔeGFR 1–24 Months | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| Creatinine 1st urine | 0.041 | 0.083 | 1.740 (0.076–3.405) |
| Creatinine 2 h | 0.026 | 0.083 | 1.628 (0.200–3.056) |
References
- Salvadori, M.; Rosso, G.; Bertoni, E. Update on ischemia-reperfusion injury in kidney transplantation: Pathogenesis and treatment. World J. Transplant. 2015, 5, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Ponticelli, C. Ischaemia-reperfusion injury: A major protagonist in kidney transplantation. Nephrol. Dial. Transplant. 2014, 29, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Jahn, N.; Sack, U.; Stehr, S.; Vöelker, M.T.; Laudi, S.; Seehofer, D.; Atay, S.; Zgoura, P.; Viebahn, R.; Boldt, A.; et al. The role of innate immune cells in the prediction of early renal allograft injury following kidney transplantation. J. Clin. Med. 2022, 11, 6148. [Google Scholar] [CrossRef] [PubMed]
- Yarlagadda, S.G.; Coca, S.G.; Formica, R.N.; Poggio, E.D.; Parikh, C.R. Association between delayed graft function and allograft and patient survival: A systematic review and meta-analysis. Nephrol. Dial. Transplant. 2009, 24, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuijs-Moeke, G.J.; Pischke, S.E.; Berger, S.P.; Sanders, J.S.F.; Pol, R.A.; Struys, M.M.R.F.; Ploeg, R.J.; Leuvenink, H.G.D. Ischemia and reperfusion injury in kidney transplantation: Relevant mechanisms in injury and repair. J. Clin. Med. 2020, 9, 253. [Google Scholar] [CrossRef]
- Krysko, D.V.; Agostinis, P.; Krysko, O.; Garg, A.D.; Bachert, C.; Lambrecht, B.N.; Vandenabeele, P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011, 32, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Riley, J.S.; Tait, S.W. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020, 21, e49799. [Google Scholar] [CrossRef]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef]
- Zanini, G.; Selleri, V.; Lopez Domenech, S.; Malerba, M.; Nasi, M.; Mattioli, A.V.; Pinti, M. Mitochondrial DNA as inflammatory DAMP: A warning of an aging immune system? Biochem. Soc. Trans. 2023, 51, 735–745. [Google Scholar] [CrossRef]
- Kim, K.; Moon, H.; Lee, Y.H.; Seo, J.-W.; Kim, Y.G.; Moon, J.-Y.; Kim, J.S.; Jeong, K.H.; Lee, T.W.; Ihm, C.G.; et al. Clinical relevance of cell-free mitochondrial DNA during the early postoperative period in kidney transplant recipients. Sci. Rep. 2019, 9, 18607. [Google Scholar] [CrossRef]
- Jansen, M.P.B.; Pulskens, W.P.C.; Uil, M.; Claessen, N.; Nieuwenhuizen, G.; Standaar, D.; Hau, C.M.; Nieuwland, R.; Florquin, S.; Bemelman, F.J.; et al. Urinary mitochondrial DNA associates with delayed graft function following renal transplantation. Nephrol. Dial. Transplant. 2020, 35, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Haines, R.W.; Fowler, A.J.; Liang, K.; Pearse, R.M.; Larsson, A.O.; Puthucheary, Z.; Prowle, J.R. Comparison of Cystatin C and Creatinine in the Assessment of Measured Kidney Function during Critical Illness. Clin. J. Am. Soc. Nephrol. 2023, 18, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.; Binjie, H.; Min, L.; Lipei, F.; Yanli, N.; Junwen, Z.; Xianghua, S. A meta-analysis on diagnostic value of serum cystatin C and creatinine for the evaluation of glomerular filtration function in renal transplant patients. Afr. Health Sci. 2014, 14, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Waikar, S.S.; Betensky, R.A.; Bonventre, J.V. Creatinine as the gold standard for kidney injury biomarker studies? Nephrol. Dial. Transplant. 2009, 24, 3263–3265. [Google Scholar] [CrossRef] [PubMed]
- Waikar, S.S.; Betensky, R.A.; Emerson, S.C.; Bonventre, J.V. Imperfect gold standards for kidney injury biomarker evaluation. J. Am. Soc. Nephrol. 2012, 23, 13–21. [Google Scholar] [CrossRef]
- Harman, G.; Akbari, A.; Hiremath, S.; White, C.A.; Ramsay, T.; Kokolo, M.B.; Craig, J.; Knoll, G.A. Accuracy of cystatin C-based estimates of glomerular filtration rate in kidney transplant recipients: A systematic review. Nephrol. Dial. Transplant. 2013, 28, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Ricci, Z.; Cruz, D.; Ronco, C. The RIFLE criteria and mortality in acute kidney injury: A systematic review. Kidney Int. 2008, 73, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Malyszko, J.; Lukaszyk, E.; Glowinska, I.; Durlik, M. Biomarkers of delayed graft function as a form of acute kidney injury in kidney transplantation. Sci. Rep. 2015, 5, 11684. [Google Scholar] [CrossRef] [PubMed]
- Halawa, A. The early diagnosis of acute renal graft dysfunction: A challenge we face. The role of novel biomarkers. Ann. Transplant. 2011, 16, 90–98. [Google Scholar] [PubMed]
- Huisman, G.J.J.; Spraakman, N.A.; Koomen, J.V.; Talsma, A.M.; Pol, R.A.; Berger, S.P.; Leuvenink, H.G.D.; Struys, M.M.R.F.; Nieuwenhuijs-Moeke, G.J. Urinary Biomarkers in a Living Donor Kidney Transplantation Cohort-Predictive Value on Graft Function. Int. J. Mol. Sci. 2023, 24, 5649. [Google Scholar] [CrossRef] [PubMed]
- Vanmassenhove, J.; Vanholder, R.; Nagler, E.; Van Biesen, W. Urinary and serum biomarkers for the diagnosis of acute kidney injury: An in-depth review of the literature. Nephrol. Dial. Transplant. 2013, 28, 254–273. [Google Scholar] [CrossRef] [PubMed]
- Beker, B.M.; Corleto, M.G.; Fieiras, C.; Musso, C.G. Novel acute kidney injury biomarkers: Their characteristics, utility and concerns. Int. Urol. Nephrol. 2018, 50, 705–713. [Google Scholar] [CrossRef]
- Obermüller, N.; Geiger, H.; Weipert, C.; Urbschat, A. Current developments in early diagnosis of acute kidney injury. Int. Urol. Nephrol. 2014, 46, 1–7. [Google Scholar] [CrossRef]
- Han, F.; Wan, S.; Sun, Q.; Chen, N.; Li, H.; Zheng, L.; Zhang, N.; Huang, Z.; Hong, L.; Qiquan, S. Donor Plasma Mitochondrial DNA Is Correlated with Posttransplant Renal Allograft Function. Transplantation 2019, 103, 2347–2358. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Sun, Q.; Huang, Z.; Li, H.; Ma, M.; Liao, T.; Luo, Z.; Zheng, L.; Zhang, N.; Chen, N.; et al. Donor plasma mitochondrial DNA is associated with antibody-mediated rejection in renal allograft recipients. Aging 2021, 13, 8440–8453. [Google Scholar] [CrossRef]
- Kroneisl, M.; Spraakman, N.A.; Koomen, J.V.; Hijazi, Z.; Hoogstra-Berends, F.H.; Leuvenink, H.G.D.; Struys, M.M.R.F.; Henning, R.H.; Nieuwenhuijs-Moeke, G.J. Peri-Operative Kinetics of Plasma Mitochondrial DNA Levels during Living Donor Kidney Transplantation. Int. J. Mol. Sci. 2023, 24, 13579. [Google Scholar] [CrossRef]
- Jansen, M.P.B.; Pulskens, W.P.; Butter, L.M.; Florquin, S.; Juffermans, N.P.; Roelofs, J.J.T.H.; Leemans, J.C. Mitochondrial DNA is released in urine of SIRS patients with acute kidney injury and correlates with severity of renal dysfunction. Shock 2018, 49, 301–310. [Google Scholar] [CrossRef]
- Liu, J.; Jia, Z.; Gong, W. Circulating mitochondrial DNA stimulates innate immune signaling pathways to mediate acute kidney injury. Front. Immunol. 2021, 12, 680648. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuijs-Moeke, G.J.; Nieuwenhuijs, V.B.; Seelen, M.A.J.; Berger, S.P.; van den Heuvel, M.C.; Burgerhof, J.G.M.; Ottens, P.J.; Ploeg, R.J.; Leuvenink, H.G.D.; Struys, M.M.R.F. Propofol-based anaesthesia versus sevoflurane-based anaesthesia for living donor kidney transplantation: Results of the VAPOR-1 randomized controlled trial. Br. J. Anaesth. 2017, 118, 720–732. [Google Scholar] [CrossRef]
- Simmons, J.D.; Lee, Y.-L.; Mulekar, S.; Kuck, J.L.; Brevard, S.B.; Gonzalez, R.P.; Gillespie, M.N.; Richards, W.O. Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann. Surg. 2013, 258, 591–596; discussion 596. [Google Scholar] [CrossRef]
- Quiros, P.M.; Goyal, A.; Jha, P.; Auwerx, J. Analysis of mtDNA/nDNA Ratio in Mice. Curr. Protoc. Mouse Biol. 2017, 7, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, A.V.; Javadov, S.; Margreiter, R.; Grimm, M.; Hagenbuchner, J.; Ausserlechner, M.J. The Role of Mitochondria in the Mechanisms of Cardiac Ischemia-Reperfusion Injury. Antioxidants 2019, 8, 454. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ren, J.; Wu, J.; Li, G.; Wu, X.; Liu, S.; Wang, G.; Gu, G.; Ren, H.; Hong, Z.; et al. Urinary mitochondrial DNA levels identify acute kidney injury in surgical critical illness patients. Shock 2017, 48, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ren, J.; Ren, H.; Wu, J.; Wu, X.; Liu, S.; Wang, G.; Gu, G.; Guo, K.; Li, J. Urinary Mitochondrial DNA Identifies Renal Dysfunction and Mitochondrial Damage in Sepsis-Induced Acute Kidney Injury. Oxid. Med. Cell. Longev. 2018, 2018, 8074936. [Google Scholar] [CrossRef]
- Ho, P.W.-L.; Pang, W.-F.; Luk, C.C.-W.; Ng, J.K.-C.; Chow, K.-M.; Kwan, B.C.-H.; Li, P.K.-T.; Szeto, C.-C. Urinary mitochondrial DNA level as a biomarker of acute kidney injury severity. Kidney Dis. 2017, 3, 78–83. [Google Scholar] [CrossRef]
- Cao, H.; Wu, J.; Luo, J.; Chen, X.; Yang, J.; Fang, L. Urinary mitochondrial DNA: A potential early biomarker of diabetic nephropathy. Diabetes Metab. Res. Rev. 2019, 35, e3131. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.Z.; Kwan, B.C.-H.; Chow, K.M.; Cheng, P.M.-S.; Luk, C.C.-W.; Li, P.K.-T.; Szeto, C.C. Urinary mitochondrial DNA level is an indicator of intra-renal mitochondrial depletion and renal scarring in diabetic nephropathy. Nephrol. Dial. Transplant. 2018, 33, 784–788. [Google Scholar] [CrossRef]
- Yu, B.C.; Cho, N.-J.; Park, S.; Kim, H.; Gil, H.-W.; Lee, E.Y.; Kwon, S.H.; Jeon, J.S.; Noh, H.; Han, D.C.; et al. Minor Glomerular Abnormalities are Associated with Deterioration of Long-Term Kidney Function and Mitochondrial Injury. J. Clin. Med. 2019, 9, 33. [Google Scholar] [CrossRef]
- Wei, P.Z.; Kwan, B.C.-H.; Chow, K.M.; Cheng, P.M.-S.; Luk, C.C.-W.; Lai, K.-B.; Li, P.K.-T.; Szeto, C.C. Urinary mitochondrial DNA level in non-diabetic chronic kidney diseases. Clin. Chim. Acta 2018, 484, 36–39. [Google Scholar] [CrossRef]
- Ye, J.; Hu, X.; Wang, Z.; Li, R.; Gan, L.; Zhang, M.; Wang, T. The role of mtDAMPs in the trauma-induced systemic inflammatory response syndrome. Front. Immunol. 2023, 14, 1164187. [Google Scholar] [CrossRef]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef]
- Bullen, A.; Liu, Z.Z.; Hepokoski, M.; Li, Y.; Singh, P. Renal oxygenation and hemodynamics in kidney injury. Nephron 2017, 137, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Nørgård, M.Ø.; Svenningsen, P. Acute kidney injury by ischemia/reperfusion and extracellular vesicles. Int. J. Mol. Sci. 2023, 24, 15312. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Rangel, W.Á.; García-Valdés, L.; Méndez-Del Villar, M.; Castañeda-Arellano, R.; Totsuka-Sutto, S.E.; García-Benavides, L. Therapeutic Targets for Regulating Oxidative Damage Induced by Ischemia-Reperfusion Injury: A Study from a Pharmacological Perspective. Oxid. Med. Cell. Longev. 2022, 2022, 8624318. [Google Scholar] [CrossRef]
- Nadalutti, C.A.; Ayala-Peña, S.; Santos, J.H. Mitochondrial DNA damage as driver of cellular outcomes. Am. J. Physiol. Cell Physiol. 2022, 322, C136–C150. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, Z.; Liang, W.; Wei, Z.; Ding, G. Roles of mitochondrial DNA damage in kidney diseases: A new biomarker. Int. J. Mol. Sci. 2022, 23, 15166. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-M.; Liu, N.; Qin, Z.-H.; Wang, Y. Mitochondrial-derived damage-associated molecular patterns amplify neuroinflammation in neurodegenerative diseases. Acta Pharmacol. Sin. 2022, 43, 2439–2447. [Google Scholar] [CrossRef]
- Lira Chavez, F.M.; Gartzke, L.P.; van Beuningen, F.E.; Wink, S.E.; Henning, R.H.; Krenning, G.; Bouma, H.R. Restoring the infected powerhouse: Mitochondrial quality control in sepsis. Redox Biol. 2023, 68, 102968. [Google Scholar] [CrossRef] [PubMed]
- Brealey, D.; Brand, M.; Hargreaves, I.; Heales, S.; Land, J.; Smolenski, R.; Davies, N.A.; Cooper, C.E.; Singer, M. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet 2002, 360, 219–223. [Google Scholar] [CrossRef]
- Huang, W.; Wang, X.; Zhang, H.; Wang, G.; Xie, F.; Liu, D. Serum Mitochondrial Quality Control Related Biomarker Levels are Associated with Organ Dysfunction in Septic Patients. Shock 2021, 56, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Yu, B.; Armando, I.; Han, F. Mitochondrial DNA-Mediated Inflammation in Acute Kidney Injury and Chronic Kidney Disease. Oxid. Med. Cell. Longev. 2021, 2021, 9985603. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; Sanchez-Niño, M.D.; Ramos, A.M.; Ortiz, A. Regulated cell death pathways in kidney disease. Nat. Rev. Nephrol. 2023, 19, 281–299. [Google Scholar] [CrossRef] [PubMed]
- Tummers, B.; Green, D.R. The evolution of regulated cell death pathways in animals and their evasion by pathogens. Physiol. Rev. 2022, 102, 411–454. [Google Scholar] [CrossRef]
- Linkermann, A.; Chen, G.; Dong, G.; Kunzendorf, U.; Krautwald, S.; Dong, Z. Regulated cell death in AKI. J. Am. Soc. Nephrol. 2014, 25, 2689–2701. [Google Scholar] [CrossRef]
- Guerrero-Mauvecin, J.; Villar-Gómez, N.; Rayego-Mateos, S.; Ramos, A.M.; Ruiz-Ortega, M.; Ortiz, A.; Sanz, A.B. Regulated necrosis role in inflammation and repair in acute kidney injury. Front. Immunol. 2023, 14, 1324996. [Google Scholar] [CrossRef]
- Guo, R.; Duan, J.; Pan, S.; Cheng, F.; Qiao, Y.; Feng, Q.; Liu, D.; Liu, Z. The Road from AKI to CKD: Molecular Mechanisms and Therapeutic Targets of Ferroptosis. Cell Death Dis. 2023, 14, 426. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed]
- Marchi, S.; Guilbaud, E.; Tait, S.W.G.; Yamazaki, T.; Galluzzi, L. Mitochondrial control of inflammation. Nat. Rev. Immunol. 2023, 23, 159–173. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Iwai, K. Roles of the NF-κB Pathway in B-Lymphocyte Biology. Curr. Top. Microbiol. Immunol. 2016, 393, 177–209. [Google Scholar] [CrossRef]
- He, W.J.; Li, C.; Huang, Z.; Geng, S.; Rao, V.S.; Kelly, T.N.; Hamm, L.L.; Grams, M.E.; Arking, D.E.; Appel, L.J.; et al. Association of Mitochondrial DNA Copy Number with Risk of Progression of Kidney Disease. Clin. J. Am. Soc. Nephrol. 2022, 17, 966–975. [Google Scholar] [CrossRef]
- Chang, C.-C.; Chiu, P.-F.; Wu, C.-L.; Kuo, C.-L.; Huang, C.-S.; Liu, C.-S.; Huang, C.-H. Urinary cell-free mitochondrial and nuclear deoxyribonucleic acid correlates with the prognosis of chronic kidney diseases. BMC Nephrol. 2019, 20, 391. [Google Scholar] [CrossRef]
- Irazabal, M.V.; Chade, A.R.; Eirin, A. Renal mitochondrial injury in the pathogenesis of CKD: mtDNA and mitomiRs. Clin. Sci. 2022, 136, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Thurairajah, K.; Briggs, G.D.; Balogh, Z.J. The source of cell-free mitochondrial DNA in trauma and potential therapeutic strategies. Eur. J. Trauma Emerg. Surg. 2018, 44, 325–334. [Google Scholar] [CrossRef]
- Torp, M.-K.; Stensløkken, K.-O.; Vaage, J. When our best friend becomes our worst enemy: The mitochondrion in trauma, surgery, and critical illness. J. Intensive Care Med. 2024, 40, 8850666241237715. [Google Scholar] [CrossRef]
| Primer | Sequence |
|---|---|
| ND1, ETC complex 1 | Forward: TGGCTCCTTTAACCTCTCCA Reverse: GGTTCGGTTGGTCTCTGCTA |
| ND6, ETC complex 1 | Forward: TGATTGTTAGCGGTGTGGTC Reverse: CCTCAATAGCCATCGCTGTA |
| D-loop, regulative region | Forward: AACCTACCCACCCTTAACAG Reverse: CACTCTTGTGCGGGATATTG |
| β2M, nuclear housekeeping gene | Forward: CTGGTAGCTCTAAACAATGTATTCA Reverse: CATGTACTCAAATGTCTAAAATGGT |
| Donor | n = 57 |
| Age [y] | 52.3 (±11.0) |
| Male [n (%)] | 26 (45.6%) |
| BMI [kg/m2] | 27 (±3.2) |
| Active smokers [n (%)] | 16 (28.1%) |
| Cardiovascular comorbidity [n (%)] Medication use [n (%)] Antihypertensive therapy Statins PPIs | 17 (29.8%) 15 (26.3%) 7 (12.3%) 9 (15.8%) |
| Pre-donation mGFR [mL/min] | 116 (97–134) |
| Recipient | n = 57 |
| Age [y] | 51.2 (45.0–58.5) |
| Male [n (%)] | 27 (47.4%) |
| BMI [kg/m2] | 25.4 (22.5–28.3) |
| Cardiovascular comorbidity [n (%)] Medication use [n (%)] Antihypertensive therapy Phosphate binders Statins | 39 (68.4%) 52 (91.2%) 32 (56.1%) 28 (49.1%) |
| Unrelated donor [n (%)] | 29 (50.9%) |
| Pre-emptive transplantation [n (%)] | 28 (49.1%) |
| Re-transplantation [n (%)] | 7 (12.3%) |
| ≥3 HLA mismatches [n (%)] | 35 (61.4%) |
| Positive PRA [n (%)] | 7 (12.3%) |
| Ischemia times [min] WIT1 CIT WIT2 | 4 (3–4) 175.5 (156.0–187.0) 43.0 (±7.3) |
| Kidney and patient outcomes | n = 57 |
| DGF [n (%)] | 3 (5.4%) |
| eGFR 1 month post-transplantation [mL/min/1.73 m2] | 50.8 (±14.9) |
| eGFR 3 months post-transplantation [mL/min/1.73 m2] | 49.6 (38.8–58.2) |
| eGFR 6 months post-transplantation [mL/min/1.73 m2] | 50.4 (38.8–61.1) |
| eGFR 12 months post-transplantation [mL/min/1.73 m2] | 50.2 (±14.2) |
| eGFR 24 months post-transplantation [mL/min/1.73 m2] | 51.4 (±17.6) |
| Acute rejection 2 years [n (%)] | 9 (16.1%) |
| Graft loss [n (%)] | 2 (3.5%) |
| Mortality [n (%)] | 1 (1.8%) |
| 1-Month eGFR | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| ND1 1st urine | 0.742 | 0.515 | 0.183 (−0.929–1.294) |
| ND1 2 h | 0.479 | 0.391 | 0.335 (−0.609–1.278) |
| ND6 1st urine | 0.752 | 0.515 | 0.154 (−0.823–1.132) |
| ND6 2 h | 0.585 | 0.439 | 0.250 (−0.666–1.167) |
| D-loop 1st urine | 0.952 | 0.625 | 0.032 (−1.046–1.110) |
| D-loop 2 h | 0.496 | 0.391 | 0.309 (−0.598–1.216) |
| 12-Month eGFR | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| ND1 1st urine | 0.008 | 0.030 * | −1.303 (−2.257–−0.350) |
| ND1 2 h | 0.250 | 0.219 | −0.512 (−1.398–0.373) |
| ND6 1st urine | 0.016 | 0.030 * | −1.050 (−1.895–−0.204) |
| ND6 2 h | 0.227 | 0.210 | −0.520 (−1.374–0.334) |
| D-loop 1st urine | 0.011 | 0.030 * | −1.259 (−2.213–−0.306) |
| D-loop 2 h | 0.217 | 0.210 | −0.528 (−1.377–0.321) |
| 24-Month eGFR | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| ND1 1st urine | 0.036 | 0.052 | −1.311 (−2.533–−0.089) |
| ND1 2 h | 0.064 | 0.067 | −1.029 (−2.120–0.062) |
| ND6 1st urine | 0.046 | 0.060 | −1.098 (−2.176–−0.020) |
| ND6 2 h | 0.064 | 0.067 | −0.997 (−2.056–0.062) |
| D-loop 1st urine | 0.036 | 0.052 | −1.326 (−2.563–−0.089) |
| D-loop 2 h | 0.054 | 0.065 | −1.025 (−2.069–0.019) |
| ΔGFR 1–24 Months | |||
| Variable | p-value | q-value | Estimate (95% CI) |
| ND1 1st urine | 0.009 | 0.030 * | −1.372 (−2.388–−0.356) |
| ND1 2 h | 0.007 | 0.030 * | −1.188 (−2.031–−0.345) |
| ND6 1st urine | 0.015 | 0.030 * | −1.141 (−2.049–−0.234) |
| ND6 2 h | 0.014 | 0.030 * | −1.066 (−1.910–−0.222) |
| D-loop 1st urine | 0.017 | 0.030 * | −1.231 (−2.228–−0.233) |
| D-loop 2 h | 0.005 | 0.030 * | −1.168 (−1.974–−0.362) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gartzke, L.; Huisman, J.; Spraakman, N.; Lira Chavez, F.; Struys, M.; Leuvenink, H.; Henning, R.; Nieuwenhuijs-Moeke, G. Urinary Mitochondrial DNA Is Related to Allograft Function in Living Donor Kidney Transplantation—An Observational Study of the VAPOR-1 Cohort. Transplantology 2025, 6, 20. https://doi.org/10.3390/transplantology6030020
Gartzke L, Huisman J, Spraakman N, Lira Chavez F, Struys M, Leuvenink H, Henning R, Nieuwenhuijs-Moeke G. Urinary Mitochondrial DNA Is Related to Allograft Function in Living Donor Kidney Transplantation—An Observational Study of the VAPOR-1 Cohort. Transplantology. 2025; 6(3):20. https://doi.org/10.3390/transplantology6030020
Chicago/Turabian StyleGartzke, Lucas, Julia Huisman, Nora Spraakman, Fernanda Lira Chavez, Michel Struys, Henri Leuvenink, Robert Henning, and Gertrude Nieuwenhuijs-Moeke. 2025. "Urinary Mitochondrial DNA Is Related to Allograft Function in Living Donor Kidney Transplantation—An Observational Study of the VAPOR-1 Cohort" Transplantology 6, no. 3: 20. https://doi.org/10.3390/transplantology6030020
APA StyleGartzke, L., Huisman, J., Spraakman, N., Lira Chavez, F., Struys, M., Leuvenink, H., Henning, R., & Nieuwenhuijs-Moeke, G. (2025). Urinary Mitochondrial DNA Is Related to Allograft Function in Living Donor Kidney Transplantation—An Observational Study of the VAPOR-1 Cohort. Transplantology, 6(3), 20. https://doi.org/10.3390/transplantology6030020







