Abstract
Background: Single cell transcriptomics offers an avenue for predicting, with improved accuracy, the gene networks that are involved in the establishment of the first direct cell–cell interactions between the blastocyst and the maternal luminal epithelium. We hypothesised that in silico modelling of the maternal–embryonic interface may provide a causal model of these interactions, leading to the identification of genes associated with a successful initiation of implantation. Methods: Bulk and single cell RNA-sequencing of endometrial epithelium and scRNAseq of day 6 and 7 trophectoderm (TE) were used to model the initial encounter between the blastocyst and the maternal uterine lining epithelium in silico. In silico modelling of the maternal–embryonic interface was performed using hypernetwork (HN) analysis of genes mediating endometrial–TE interactions and the wider endometrial epithelial transcriptome. A hypernetwork analysis identifies genes that co-ordinate the expression of many other genes to derive a higher order interaction likely to be causally linked to the function. Potential interactions of TE with non-ciliated luminal cells, ciliated cells, and glandular cells were examined. Results: Prominent epithelial activities include secretion, endocytosis, ion transport, adhesion, and immune modulation. Three highly correlated clusters of 25, 22 and 26 TE-interacting epithelial surface genes were identified, each with distinct properties. Genes in both ciliated and non-ciliated luminal epithelial cells and glandular cells exhibit significant functional associations. Ciliated cells are predicted to bind to TE via galectin–glycan interaction. Day 6 and day 7 embryonic–epithelial interactomes are largely similar. The removal of aneuploid TE-derived mRNA invoked only subtle differences. No direct interaction with the maternal gland epithelial cell surface is predicted. These functional differences validate the in silico segregation of phenotypes. Single cell analysis of the epithelium revealed significant change with the cycle phase, but differences in the cell phenotype between individual donors were also present. Conclusions: A hypernetwork analysis can identify epithelial gene clusters that show correlated change during the menstrual cycle and can be interfaced with TE genes to predict pathways and processes occurring during the initiation of embryo–epithelial interaction in the mid-secretory phase. The data are on a scale that is realistic for functional dissection using current ex vivo human implantation models. A focus on luminal epithelial cells may allow a resolution to the current bottleneck of endometrial receptivity testing based on tissue lysates, which is confounded by noise from multiple diverse cell populations.
1. Introduction
The first direct intercellular interaction at implantation is between embryonic trophectoderm (TE) and endometrial luminal epithelium (LE) [1]. The acquisition of receptivity may therefore be, at least initially, an epithelially-centred transition. There is evidence that the apical glycocalyx of the epithelium forms a protective barrier to be overcome by the embryo for stable attachment to be achieved [2,3]. Further evidence suggests that changes in TE trophoblast gene expression required for the acquisition of invasive characteristics are dependent on direct contact with LE [4,5]. Not only do apposition and attachment define the site of implantation, but they are also required to initiate the first phase of a gene expression program in the embryo that will allow it to progress to invade the stroma and develop a placenta. Some aneuploid embryos develop to become morphologically normal blastocysts but cannot be traced in products of conception [6], and early morphological, metabolic and hCG monitoring shows that failure to progress may be seen broadly at any stage from fertilisation to blastocyst transfer and beyond [7,8,9]. Though the numbers remain controversial [10,11], it seems clear that some IVF embryos with the potential to develop into healthy offspring do not do so because of endometrial factors. Impaired attachment could be a cause of implantation failure, and so there is good reason to investigate the molecular mechanisms involved.
Transcriptomic evaluation of endometrial status has been posited as a way of characterising the program of maternal gene expression that leads to receptivity and predicting (and even personalising) the optimal time to replace embryos created with IVF [12]. So far, however, the results have not been sufficiently convincing in terms of improved outcomes to persuade regulatory bodies to support its widespread use, despite the availability and active marketing of testing kits [13,14]. Amongst a number of issues, the use of tissue biopsies containing multiple cell types stands out as a source of noise in the transcriptome, arising from variation in proportions of both resident and colonial cell types, contamination with blood cells as well as natural variation in timing within the ovarian-uterine endocrine axis. Single cell RNAseq offers a potential avenue for characterizing changes in specific cell subpopulations including the minorities.
We have previously used in silico approaches to model the maternal–embryo interface, focusing on TE responses to interactions with endometrial epithelial cells (EEC) [5]. Here, we aimed to identify gene networks that are activated at implantation by deriving an in silico interactome between endometrial epithelium and day 6/7 human trophectoderm. We then used hypernetwork (HN) analysis [15,16], an approach that enumerates the number of genes with shared correlated expression (Figure 1) to delineate maternal downstream gene networks that might become involved in the response to an embryo. Specifically, bulk (aggregated) as well as scRNAseq data from primary endometrial epithelial cells (EEC) were used to examine EEC gene networks coupled to the TE interactome. The data reveal the distinct interaction characteristics of LE, ciliated (CE), and glandular epithelial (GE) cells and show them all changing with the cycle stage. The relatively small scale of the resultant data means that functional evaluation using current in vitro models [5,17] should be achievable. We also detect inter-individual differences that might confound attempts to define a conserved state of receptivity.

Figure 1.
Generating and summarising a hypernetwork model of co-correlated gene expression as a causal model. (A). A bipartite network model of correlated gene expression between differentially expressed genes (circles) and the rest of the expressed genes in the genome (triangles). Strong correlations are defined as by the upper and lower standard deviations of the distribution of correlation r-values. (B). The number of shared strong correlations between each differentially expressed gene and genes in the genome is calculated and expressed in a square matrix form (symmetrical)—this is the adjacency matrix of the hypernetwork. (C). Genes with high numbers of shared correlations are identified by clustering using the Euclidean metric.
2. Materials and Methods
2.1. Bulk RNA-Seq Data
Endometrial epithelial bulk RNA-seq data [18], represented as read counts, were filtered and normalized using the edgeR R package with the filterByExpr function [19]. The resulting data matrix consisted of a total of 9352 genes and 9 samples (4 proliferative and 5 mid-secretory samples). Differentially expressed genes were determined by comparing gene expression between the proliferative (n = 4) and mid-secretory (n = 5) samples. This was performed using the “DGELIST” function. The p values for differentially expressed genes were calculated using the “topTags” function. Differentially expressed genes were defined by a cut-off of an adjusted p < 0.01. The p values were adjusted with the Benjamini-Hochberg method [20].Transcriptomic differences between samples were visualised with principal component analysis (PCA).
2.2. ScRNA-Seq Data
Endometrial epithelial scRNA-seq data [21] were represented as mapped read counts. Proliferative, early secretory, early mid-secretory, and mid-secretory samples were extracted from the data. Endometrial epithelial cells (EEC) defined as luminal, glandular, or ciliated were further refined from the original data matrix.
Genes expressed in fewer than 3 cells, and cells expressing fewer than 200 genes were removed, as were cells with more than 5% mitochondrial gene transcripts to minimise doublets and low-quality (broken or damaged) cells, respectively. The remaining total comprised 20,669 genes and 2493 cells. Data were normalised using the “LogNormalize” method in the Seurat R package [22]. The gene expression measurement is normalised for each cell by multiplying the total expression by a scale factor (10,000 by default) and log-transforms the resulting variable genes. Differential gene analysis was performed via the “FindMarkers” function in Seurat R. Differentially expressed genes (DEGs) were determined by comparing gene expression between proliferative and secretory phases for ciliated, luminal, and glandular epithelial subtypes separately.
DEGs were defined by the cut-off adjusted p < 0.01. The p values were adjusted with the Benjamini-Hochberg method [20]. The single epithelial cell differential transcriptomes were visualised using Uniform Manifold Approximation and Projection (UMAP) to cluster subtypes.
We analysed single-cell RNA-seq of day 6 and day 7 TE [23]. To identify outliers’ data was visualised using violin plots. Genes expressed in fewer than 3 cells, and cells expressing fewer than 7500 genes were removed. We identified aneuploid cells within the TE dataset as described [24].
2.3. In Silico Modelling of the TE-EEC Interface
Maternal cell surface genes were refined from the list of DEGs from the scRNA-seq endometrial data. Cell surface genes were identified by the Database for Annotation, Visualisation, and Integrated Discovery (DAVID) [25]. Genes encode proteins localised to any of ‘extracellular space’, ‘extracellular matrix’, ‘proteinaceous extracellular matrix’, or ‘cell surface’ were considered to be cell surface. Putative protein binding partners were identified using the Biological General Repository for Interaction Datasets (BioGRID) [26]. Binding ligands were refined to those present in the TE single-cell RNA transcriptomes.
2.4. Hypernetworks
HN were utilised to identify clusters of highly correlated genes in endometrial epithelium. This can be represented by a hypergraph, . The vertices, represent the genes present at the EEC surface and each edge represents a subset of called hyperedges. In our hypergraph, the set represents correlations of paired genes, whereas the set represents genes within the EEC network.
To calculate the correlation of endometrial surface genes to the rest of the transcriptome, EEC bulk transcriptomes were used. The trimmed mean of m-values (TMM) were extracted from the count matrix. Hypernetworks are calculated by multiplying the incidence matrix of the hypergraph against the incidence matrix of the dual hypergraph to forma an adjacency matrix. To binarise the resulting matrix, values within one standard deviation of the correlation r-value were given the value 0 and all other correlation values become 1. 0 indicates no relationship between a gene pair whereas 1 indicates a relationship between a gene pair. Hypernetwork calculations were repeated with scRNA-seq dataset for each endometrial epithelial subtype: glandular, ciliated, and luminal.
Data are presented as a heatmap, and hierarchical clustering was applied to cluster genes with shared connections within the transcriptome. Genes were clustered based on a dendrogram representing the Euclidean distance between samples as a measure of similarity. The Galois correspondence of highly co-ordinated genes was defined in the original incidence matrix to identify the set of genes being influenced by higher order interactions and, therefore, the networks of genes functioning at the maternal-fetal interface.
2.5. Gene Network Construction and Visualisation
The gene–gene interaction network from HN was constructed from the adjacency matrix. Cytoscape was used for the visualisation of the clusters [27]. The adjacency matrix was exported into Cytoscape via the aMatReader plugin [28]. The network was illustrated in the form of nodes and edges, where each node represents a single gene, and each edge represents the gene-gene interactions. The Perfuse force-directed layout of the Cytoscape application was used to display the gene–gene network.
2.6. Enrichment Analysis
To identify functions associated with each indirect action set extracted from the hypernetwork, we performed a Gene Ontology (GO) enrichment analysis on “WEB-based Gene SeT AnaLysis Toolkit”(WebGestalt) [29]. GO annotates genes to molecular function, cellular component and biological processes (BP) terms, and Weighted Set Cover was used to identify overarching BP terms.
2.7. Data Availability and Materials
All analysis was performed in R version 3.4.2 (R Foundation for Statistical Computing), and the code has been deposited at https://github.com/Aikibloke/Taqua-Project (accessed on 15 July 2021). All the data used come from publicly available repositories and data portals.
Raw sequencing data of individual cells from 88 human preimplantation embryos [23] are available from Array Express under E-MTAB-392957. Day 6 and day 7 single TE cell data were extracted.
Endometrial epithelial bulk RNA sequencing (RNAseq) transcriptomes from 9 endometrial samples were extracted through the Gene Expression Omnibus (GEO) under the accession number GSE13271151. Endometrial epithelium scRNA-seq data were downloaded through the web portal www.reproductivecellatlas.org (accessed on 15 March 2021).
3. Results
3.1. Secretory Phase EEC Genes Used to Model the Maternal-TE Interface
Principal component analysis was carried out on EEC transcriptomes from 4 proliferative phase and 9 mid secretory phase samples. As expected [30], they resolved into two distinct clusters representing the cycle phases pre- and post-ovulation (Supplementary Figure S1) with 440 differentially expressed genes (DEG) at p < 0.01, 115 of which were either associated with the cell surface or secreted. Genes differentially expressed between proliferative and secretory phases were considered as candidates for the regulation of implantation, including either up- or down-regulation. Figure 2 demonstrates the molecular function GO terms for EEC cycle-responsive cell surface and secreted genes. In agreement with the most prominent terms, growth factor, chemokine, cytokine, and adhesion/ECM receptors are candidates in current work on embryo interaction with receptive phase endometrium, as summarised in a recent review [31].

Figure 2.
Gene Ontology (GO) enrichment analysis. Molecular function of differential expressed endometrial epithelial surface genes were identified using the over-representation analysis tool. The X axis reports gene enrichment in the GO term and the Y axis the statistical significance of over-representation (negative log10 false discovery rate). The size of dot reflects the number of genes in each GO term and the colour reflects the number of genes in the sample matching the term.
3.2. Networks Functioning at the EEC-TE Interface Examined Using Hypernetwork Analysis
Single-cell transcriptomic datasets from blastocyst day 7 TE were interfaced to the EEC data to identify gene networks functioning at the TE-EEC interface. A hypernetwork (HN) is a graphical model that, by mathematical capture of changes of expression correlated in the response to a variable (Figure 1), identifies higher-order interactions, in this case the multiple simultaneous ones that occur between cellular macromolecules. Hypernetworks cluster genes based on co-expression within the transcriptome, which relates to connectivity in functional gene networks. 63% of genes present at the EEC surface with cognate cell surface genes in day 7 TE were highly correlated to gene networks functioning within the EEC transcriptome. Three highly correlated clusters of 25, 22 and 26 TE interacting EEC surface genes were identified (Figure 3, Table S1) as is most readily evident in the dendrogram to the left of the figure.
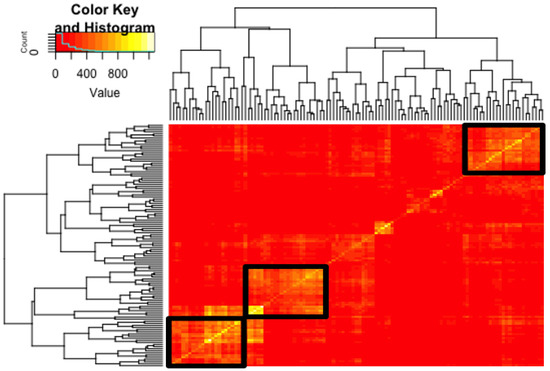
Figure 3.
Hypernetwork analysis of day 7 TE interacting endometrial epithelial genes (EEC). Heatmap demonstrating the correlations between EEC genes identified as possessing cognate cell-surface genes in day 7 TE, against the rest of the EEC transcriptome. The same set of genes (n = 115) is present on the columns and rows, thus generating a symmetrical matrix in which the colour scale represents the number of shared correlations between each gene pair. Hierarchical clustering via the dendrogram identified three gene clusters that share highly connected correlations (1, 2 and 3 from bottom left to top right). Red in the heatmap depicts fewer correlations whereas yellow depicts highly correlated genes. The black boxes highlight the most connected clusters.
The TE-interacting EEC genes and those strongly correlated to them in the hypernetwork were extracted; cluster 1 n = 158, cluster 2 n = 206, and cluster 3 n = 240. The ontology associated with each cluster was calculated using overrepresentation analysis (Figure 4). No significantly enriched biological process terms were identified in the first cluster. Twenty-six biological process terms were significantly over-represented in the second cluster (Figure 4A), and these could be consolidated to 9 overarching terms including “immune response”, “regulation of response to stress”, and “regulation of programmed cell death”, among others. The third cluster contained many more significantly enriched biological process (Figure 4B), 64 in total that consolidated to 10 umbrella terms including the related terms secretion, vesicle-mediated transport and regulation of transport, and regulation of cell proliferation, cell migration, and cell adhesion. The data emphasise the importance of the epithelial-immune cell dialogue at peri-implantation [32].
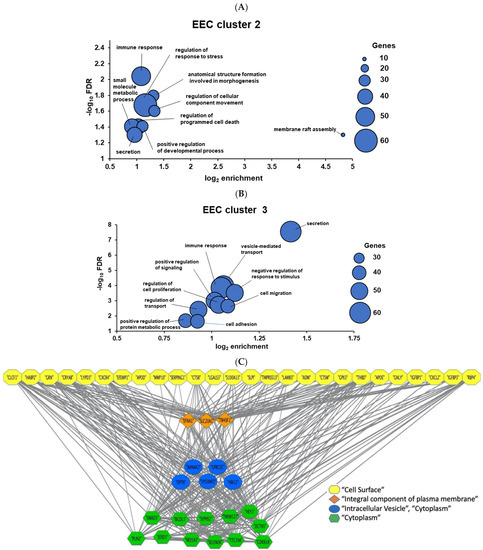
Figure 4.
Gene Ontology (GO) of endometrial epithelium networks reported via indirect action analysis of hypernetwork clusters 2 and 3 ((A,B), respectively). The X axis reports gene enrichment in the GO term and the Y axis the statistical significance of over-representation (negative log10 false discovery rate). Dot size reflects the number of genes in the sample matching the GO term. (C) Cystoscope network visualisation of genes in cluster 3. Edges with weight above a threshold of 0.9 are displayed. Yellow nodes denote cell surface genes that interact with TE. Downstream genes are depicted as orange nodes (integral component of plasma membrane), blue nodes (intracellular vesicle) and green nodes (cytoplasm).
As cluster 3 was by far the most enriched for biological process terms, the gene networks in this cluster were visualised in the form of nodes (genes) and edges (gene-gene interactions) on Cytoscape (Figure 4C). A high-confidence network of 46 nodes and 442 edges was identified, where a group of 5 “intracellular vesicle” genes downstream of the interface genes were found alongside “cytoplasm” genes. This reflects the secretory GO terms identified for this cluster, including 3/12 cytoplasmic genes involved in vesicular trafficking (DCTN5, BICDL1, and TMEM127). The results thus suggest the epithelial surface is highly dynamic and capable of rapid change.
3.3. Epithelial Heterogeneity
A fundamental problem with bulk sequencing is the averaging of gene expression over the cell types in a full tissue. To refine the in silico model and discern epithelial cell type-specific interactions at the interface with TE, scRNA-seq data of 10,729 endometrial cells from 15 individuals were extracted from a publicly accessible repository. In keeping with our focus on interactions at embryo attachment, data were filtered to include only epithelial (luminal, ciliated, and glandular) subtypes (Figure 5), based on markers identified by Garcia-Alonso et al. [21] and previous literature.
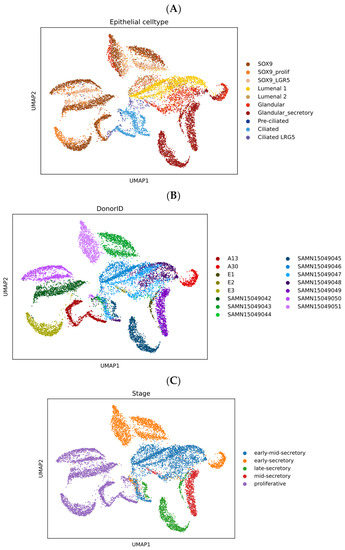
Figure 5.
Uniform Manifold Approximation and Projection (UMAP) clustering of the scRNA-seq dataset (n = 10,729 cells with unadjusted sampling, so showing different cell numbers per donor) representing endometrial epithelial subsets (A), donor individuals (B) and cycle stages (C) reported by Garcia-Alonso et al. [21]. The epithelial subsets bearing SOX9 and LGR5 are specific phenotypes of interest to these authors, with SOX9 being enriched in, but not exclusive to, the proliferative phase. LGR5 cells are mainly luminal and sometimes ciliated.
As expected [33], the data indicate substantial changes in gene expression from proliferative to early secretory phase (Figure 5C), and further changes are evident that continue through the secretory phase. While cells from the same histologically characterised phases of the cycle cluster nearby one another, indicating conserved phenotypic change with cycle progression, samples are also notably subclustered by individual donor (Figure 5B). Though there are insufficient mid secretory phase samples to draw any firm conclusion, the significant differences between donors in other phases of the cycle would suggest that gene expression at the implantation stage may also differ quite markedly between women.
Based on these findings, and the possibility that there may be discrete functions for luminal, ciliated, and glandular cells at implantation, their secretory phase interactomes were analysed separately.
3.4. Trophectoderm-Luminal Epithelial Gene Networks
Starting from LE cell DEGs (n = 341 at an adjusted p-value < 0.05), cell surface or secreted proteins were identified (n = 127) which might be involved most directly in the initial intercellular interaction with TE. ScRNA-seq profiles of TE at day 6 and day 7 of development were utilised to predict possible interactomes between LE and the two embryo stages. There was no significant difference between the number of LE surface genes that matched cognate binding partners on the TE surface at day 6 (83%) or day 7 (82%). HN analysis identified 23 surface genes in TE that were highly correlated to surface genes expressed by LE at day 7 (Figure 6, Table S2).
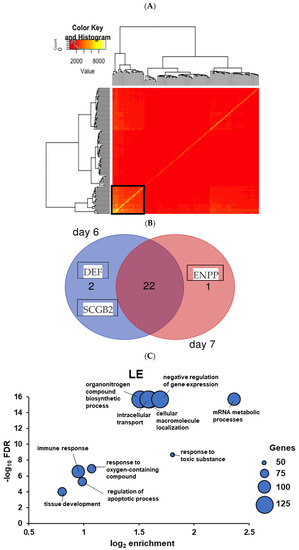
Figure 6.
(A) Hypernetwork analysis demonstrating the correlations between LE genes with cognate cell-surface genes in day 6 TE against the rest of the luminal transcriptome. Hierarchical clustering based on the dendrogram identified gene clusters (black box) that share highly connected correlations. Red within the heatmap depicts a low number of gene correlations whereas yellow depicts more highly correlated genes. (B) Venn diagram of the LE cluster genes comparing day 6 and day 7 interactomes. The overlapping number represents LE cluster components which interact with the TE surface proteins on both day 6 and day 7. Non-overlapping numbers specify the genes unique to each interactome. (C) Overarching GO terms of LE networks reported via indirect action analysis of LE hypernetwork cluster in A. The X axis shows gene enrichment in the GO term and the Y axis the statistical significance of over-representation (negative log10 false discovery rate). Dot size relates to the number of genes in the sample matching the GO term.
LE cell functions suggested by this analysis include secretion and release of soluble signals (IL6, midkine, CRISP3, SCGB2A1) and prostaglandin production (COX-1/PTGS1/prostaglandin H2 synthase), both in keeping with the literature [34]. Other LE functions—adjustment of membrane structure and composition (ACSL4; ANXA4), protection from complement-mediated damage (CD55), and ion transport (ATP1A1, SLC26A2)—are generally conserved in epithelial tissues. It would not be surprising if these were affected by an implanting embryo that crosses the barrier. Several genes in the LE interactome were also represented in the bulk sequencing data (ANXA4, CRYAB, GRN, SERPINA5, and VIM).
A proportion of aneuploid cells can often be found in the human blastocyst [35,36]. To predict the possible impact at the level of gene expression, aneuploid cells (a total of 22) were removed from the data matrix, effecting a 9% decrease on day 7 TE. Limited changes were noted: ENPP3 (Ectonucleotide pyrophosphatase) was exclusively associated with aneuploid-containing day 7 TE interactions while CRISP3 (cysteine-rich secretory protein 3) was absent from aneuploid-free day 7 interactome (data not shown). All subsequent maternal-embryo interface modelling was performed with euploid-only TE data. This analysis is consistent with the observation that, at least in some cases, implantation can proceed even when aneuploid cells are present in the blastocyst [37].
We assessed GO biological process terms associated with the highly correlated day 7 TE-interacting LE genes of the HN cluster (n = 441). Over-representation analysis established 83 significantly enriched terms, more than in the bulk EEC model. These were consolidated into 10 overarching terms, including ‘mRNA metabolic processes’, ‘intracellular transport‘, and ‘immune response’ (Figure 5), the latter reflecting what was observed in bulk EEC hypernetworks.
3.5. Trophectoderm-Ciliated Epithelium and Trophectoderm-Glandular Epithelium Gene Networks
DEG analysis of ciliated epithelial cells (CE) identified 1240 genes that differed between proliferative and secretory phases. Of these, 385 encoded cell surface proteins, and HN identified 85 interacting genes in day 7 TE that were highly correlated to the rest of the CE transcriptome (Figure 7, Table S2). Gene ontology analysis identified 127 biological processes associated with the CE cluster which were consolidated to 10 overarching pathways including ‘cellular response to stress’, ‘response to toxic substance’, apoptotic processes’, and ‘regulation of cell proliferation’.
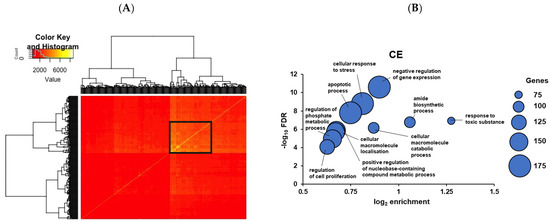
Figure 7.
(A) Hypernetwork analysis of day 6 TE-interacting endometrial ciliated epithelial genes (CE). The heatmap demonstrates correlations between CE genes and cognate cell-surface genes in day 6 TE against the rest of the CE transcriptome. Hierarchical clustering selects genes that share highly connected correlations (black box). (B) GO analysis of the indirect action analysis of CE hypernetwork cluster in A. Gene enrichment in the GO term and statistical significance of over-representation (negative log10 false discovery rate) are plotted. Dot size relates to the number of genes in the sample matching the GO term.
Analysis of the CE genes suggests processes important in this cell subset: ion transport (eg SLC26A2), membrane organization, and membrane-cytoskeletal interaction (TSPAN, CD81, EZR), and cell adhesion (LGALS1, LGALS3 (also represented in the bulk sequencing data), EPHA2).
TE may interact at implantation with cells at the necks of glands, and early cytotrophoblasts can enter gland lumena [38]. Therefore, it was relevant to profile the prospective TE-gland epithelial interactome. Changes in 340 glandular cell DEGs were detected between the proliferative and secretory phases at an adjusted p < 0.05. This list was refined to leave 77 surface or secreted gene products through DAVID utilising the gene ontology terms “extracellular space”, “extracellular matrix”, “extracellular region”, and “proteinaceous extracellular matrix”. The term “cell surface” was not associated with any of the DEGs, consistent with GE cells mainly functioning through secretory activity as implantation is initiated.
Interactions of genes encoding proteins secreted by GE with cognate TE cell-surface genes on day 7 were examined. HN analysis of the interactome defined only 3 GE genes (IGFBP4, EPB41L2, MTRNR2L1) to be highly correlated to genes in TE (Table S2). Insulin-like growth factor binding protein 4 (IGFBP4) is associated with cell growth via regulation of IGF bioavailability [39] and is an inhibitor of canonical Wnt signalling [40]. Wnt has been implicated in the development of ciliated epithelial cells [21]. GO enrichment analysis of the gene sets implicated in indirect action correlated with the cluster genes (n = 1380) shows association with 85 biological process that make up 10 overarching processes (Figure 8). However, it should be noted that the correlated dataset may not reflect genes that respond to TE interactions, as could be the case for cell surface genes in the previous analyses. Furthermore, enrichment ratios are lower, so caution is required in interpretation.
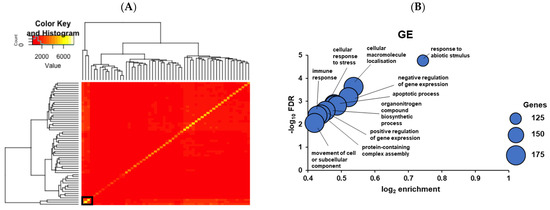
Figure 8.
(A) Hypernetwork analysis of day 6 TE-interacting glandular epithelial genes (GE). The heatmap demonstrates correlations between GE genes and cognate cell-surface genes in day 6 TE against the rest of the GE transcriptome. Hierarchical clustering selects genes that share highly connected correlations (black box). (B) Gene Ontology (GO) of GE networks identified via indirect action analysis. Enriched biological pathways were identified using over-representation analysis. The X-axis represents the log2 (fold enrichment) and the Y-axis represents the negative log10 (FDR) of each enriched biological process (GO term). Each dot demonstrates a specific pathway, and the size of the dot shows the gene set size of each enriched pathway. All GO terms were significant at an FDR < 0.01.
3.6. Comparison of Maternal Gene Networks at the Different TE Interface Models
The maternal–TE interface model determined from bulk EEC transcriptome data as expected yielded the highest number of transcriptome-correlated genes, as derived by HN analysis (Figure 9A). Amongst the epithelial subsets, the associated gene networks of the LE interface model were the most coherent in terms of enriched biological process ontology terms The CE model contained a similar number of highly correlated interface genes to the LE model but fewer associated ontology terms while the GE interface model yielded very few transcriptome-correlated interface genes and their associations were much less enriched in ontology terms. A comparison of the significance and enrichment levels of the umbrella ontology terms for each interface model shows that the LE and CE interface models are associated with much more highly significant ontology terms than any other model apart from the ‘secretory’ term in EEC cluster 3 (Figure 9B; cf Figure 4B). Thus, the results predict that both ciliated and non-ciliated cells in the luminal compartment contribute to interaction with an implanting embryo.
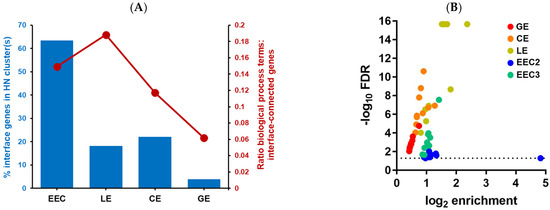
Figure 9.
Maternal gene networks at the TE interface elucidated from bulk EEC and single cell (LE, CE and GE) transcriptomes. (A) Comparison of hypernetwork (HN) cluster size and enrichment of biological process terms in connections downstream of the maternal-embryo interface between the transcriptomes. (B) Summary plot of significance and enrichment levels of overarching biological process terms identified downstream of maternal-embryo interface genes (dotted line represents FDR cut off of 0.05).
4. Discussion
The hypernetwork approach to analysis of correlated gene expression allows identification of coregulation patterns that would not be detected in pairwise analysis of gene expression, so observations may stem from actions upstream (such as a transcriptional activator or repressor) or downstream (such as an adaptor protein or solute transporter) control points that may not be visible after data filtration. Central gene clusters are associated with different degrees of pathway promiscuity, reflecting regulatory coordination of a stochastic change [41]. Correlation is non-directional; that is, co-variation and inverse variation are treated the same. Since the analysis arises from system change, in this case in cycling EEC, we cannot expect to identify all interactions that are relevant to the success of implantation; rather, genes emerge that play a part in mediating changes in the function of the cells against a background of conserved genes that may still be interactive. The results suggest that the single cell LE cluster yields the most highly causally enriched model (better than bulk EEC), and that CE modelling indicates a likely additional contribution to the interface with TE.
In the secretory phase, obtaining larger single cell data sets will be important as it is here that receptivity to the blastocyst arises, and the comparison between bulk sequencing and single cell data does not account for possible changes in the abundance of the three cell subpopulations. This applies especially to mid secretory phase. Given the presence of inter-individual variation overlaid on cycle progression, with speed of response to progesterone known to vary between individuals [42], we are not currently in possession of enough samples to see the extent of variation. However, it is likely that this combination of factors accounts for the documented problems with attempts to specify and schedule the receptive phase using transcriptomics [14].
A further limitation of the analysis is that we are considering both EEC and TE in their respective states before interaction begins, thus ignoring any effects of the spatiotemporal advance of the mutual dialogue. Indeed, we have shown [4] that contact between TE and LE generates a signal that turns on differentiation-related transcription factors within a few hours, consistent with the rapid differentiation of trophoblast required to advance implantation. Therefore, we can only consider the interactions predicted here as representing an initial phase of implantation. Day 6 and Day 7 TE behave similarly, consistent with the idea that implantation could occur on either day.
To examine the maternal-embryonic dialogue as it evolves, ex vivo models in which blastocysts or blastoids interact with 3D endometrial tissue constructs can be combined with scRNA analysis; while blastoids have already been reported interacting with endometrial cells in 2D [17], as well as embryos interacting with 3D endometrial assembloids [43], there are still technical challenges to overcome. Spatial transcriptomics of implantation sites generated ex vivo will also be informative as spatial relationships are lost in single cell analysis [44].
The substantial differences between gene expression in epithelial cells in proliferative vs secretory phase are consistent with many previous studies showing that progesterone can act directly on receptor in epithelial cells, or indirectly on receptor-bearing stromal cells, which subsequently emit paracrine signals that act on the epithelium [33]. In both instances, the effects are far reaching.
It is clear from morphology studies that both glands and luminal epithelium contain ciliated as well as non-ciliated cells [45]. Ciliated cells are thought to arise in response to estrogen stimulation in the proliferative phase [45,46]. They change phenotype in the proliferative to secretory transition, but according to the snapshot achieved here, do so less radically than other epithelial subsets. They are abundant in the LE and express the galectin LGAL3 (also detected in EEC and LE), which suggests a possible role in the initial, perhaps relatively weak, interaction with glycoprotein on TE at apposition. This could be linked to repositioning or rotation of the embryo prior to stable adhesion. The data also suggest that CE cells might act as sensors of the luminal environment, invoking metabolic pathways to respond to stressors such as nutrient deprivation, which, even if confined to the peri-implantation period, could exert far-reaching effects on the conceptus [47,48,49].
Comparisons of bulk sequencing data with the single cell analysis identifies other pathways with plausible functional significance in specific epithelial subsets. The epithelial surface is dynamic with prominent paracrine signalling, vesicular trafficking and ion transport functions. Protease activity and its inhibition appear widely, for example in the presence of SLPI in both EEC and LE. Down-regulation of this inhibitor has been implicated at implantation [50]. Ephrin A1 (EFNA1), the hyaluronan binding protein HABP2 and CXCL2 are expressed in both EEC and LE. TM4SF1, a tetraspanin associated with cell migration and Wnt inhibition [51] is present in EEC and all three single cell types. SMAD3, a transcription factor activated by TGFβ and implicated in endometrial priming for implantation [52,53,54], is found in EEC bulk sequencing, but only in the GE single cell data. The cluster analysis supports the idea that distinct functional repertoires in different epithelial subsets contribute to interactions at implantation.
In GE, little evidence was found for direct cell-cell interaction with TE, consistent with the spatial relationship with TE at early implantation. However the data are consistent with a secretory role for GE, offering a biological readout that supports the validity of this methodology. A combination of secretory at-a-distance signals with direct cell-cell interactions involving distinct epithelial subsets is likely to be the emerging picture of implantation biology. Localised activation of GE is likely at and near the implantation site to target secretory activity, and the glands show a capacity where necessary to activate immune and stromal cell populations adjacent to the embryo.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/reprodmed4010006/s1, Figure S1: Principal Component analysis (PCA) of primary endometrial epithelial cell transcriptomes; Table S1: Genes identified in the hypernetwork clusters from bulk transcriptome EEC; Table S2: Genes identified in the hypernetwork clusters from single-cell LE, CE and GE transcriptomes.
Author Contributions
A.S. conceptualised and directed the project with input from, P.T.R. and J.D.A., and they all contributed to the analytical design and choice of data. A.S. and T.G. supervised the computational work including choice of software, visualisation, debugging and validation. T.K., M.S. and T.G. downloaded the data and managed software and curation. M.S. ran the scRNAseq analysis. T.K. ran the other analysis with help from T.G., P.T.R. and A.S. as well as preparing a dissertation. J.D.A. and P.T.R. undertook data interpretation and developed a full text from T.K.’s report. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not relevant—There is no new wet experimentation here. All data were anonymised.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data and software are accessible on public databases.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aplin, J.D.; Ruane, P.T. Embryo-epithelium interactions during implantation at a glance. J. Cell Sci. 2017, 130, 15–22. [Google Scholar] [CrossRef]
- Meseguer, M.; Aplin, J.D.; Caballero-Campo, P.; O’Connor, J.E.; Martín, J.C.; Remohí, J.; Pellicer, A.; Simón, C. Human endometrial mucin MUC1 is up-regulated by progesterone and down-regulated in vitro by the human blastocyst. Biol. Reprod. 2001, 64, 590–601. [Google Scholar] [CrossRef]
- Aplin, J.D.; Meseguer, M.; Simón, C.; Ortíz, M.E.; Croxatto, H.; Jones, C.J. MUC1, glycans and the cell-surface barrier to embryo implantation. Biochem. Soc. Trans. 2001, 29 Pt 2, 153–156. [Google Scholar] [CrossRef]
- Ruane, P.T.; Berneau, S.C.; Koeck, R.; Watts, J.; Kimber, S.J.; Brison, D.R.; Westwood, M.; Aplin, J.D. Apposition to endometrial epithelial cells activates mouse blastocysts for implantation. Mol. Hum. Reprod. 2017, 23, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Ruane, P.T.; Garner, T.; Parsons, L.; Babbington, P.A.; Wangsaputra, I.; Kimber, S.J.; Stevens, A.; Westwood, M.; Brison, D.R.; Aplin, J.D. Trophectoderm differentiation to invasive syncytiotrophoblast is promoted by endometrial epithelial cells during human embryo implantation. Hum. Reprod. 2022, 37, 777–792. [Google Scholar] [CrossRef]
- Savio Figueira Rde, C.; Setti, A.S.; Braga, D.P.A.F.; Iaconelli, A., Jr.; Borges, E., Jr. Blastocyst Morphology Holds Clues Concerning The Chromosomal Status of The Embryo. Int. J. Fertil. Steril. 2015, 9, 215–220. [Google Scholar] [PubMed]
- Wilcox, A.J.; Weinberg, C.R.; O’Connor, J.F.; Baird, D.D.; Schlatterer, J.P.; Canfield, R.E.; Armstrong, E.G.; Nisula, B.C. Incidence of early loss of pregnancy. N. Engl. J. Med. 1988, 319, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, C.; Wang, L.; Chen, D.; Guang, W.; French, J. Conception, early pregnancy loss, and time to clinical pregnancy: A population-based prospective study. Fertil. Steril. 2003, 79, 577–584. [Google Scholar] [CrossRef]
- Foo, L.; Johnson, S.; Marriott, L.; Bourne, T.; Bennett, P.; Lees, C. Peri-implantation urinary hormone monitoring distinguishes between types of first-trimester spontaneous pregnancy loss. Paediatr. Perinat. Epidemiol. 2020, 34, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Franasiak, J.M.; Alecsandru, D.; Forman, E.J.; Gemmell, L.C.; Goldberg, J.M.; Llarena, N.; Margolis, C.; Laven, J.; Schoenmakers, S.; Seli, E. A review of the pathophysiology of recurrent implantation failure. Fertil. Steril. 2021, 116, 1436–1448. [Google Scholar] [CrossRef]
- Pirtea, P.; De Ziegler, D.; Tao, X.; Sun, L.; Zhan, Y.; Ayoubi, J.M.; Seli, E.; Franasiak, J.M.; Scott, R.T. Rate of true recurrent implantation failure is low: Results of three successive frozen euploid single embryo transfers. Fertil. Steril. 2021, 115, 45–53. [Google Scholar] [CrossRef]
- Díaz-Gimeno, P.; Ruiz-Alonso, M.; Sebastian-Leon, P.; Pellicer, A.; Valbuena, D.; Simón, C. Window of implantation transcriptomic stratification reveals different endometrial subsignatures associated with live birth and biochemical pregnancy. Fertil. Steril. 2017, 108, 703–710.e3. [Google Scholar] [CrossRef]
- Ben Rafael, Z. Endometrial Receptivity Analysis (ERA) test: An unproven technology. Hum. Reprod. Open 2021, 2021, hoab010. [Google Scholar] [CrossRef]
- Aplin, J.D.; Stevens, A. Use of ’omics for endometrial timing: The cycle moves on. Hum. Reprod. 2022, 37, 644–650. [Google Scholar] [CrossRef]
- Battiston, F.; Amico, E.; Barrat, A.; Bianconi, G.; de Arruda, G.F.; Franceschiello, B.; Iacopini, I.; Kéfi, S.; Latora, V.; Moreno, Y.; et al. The physics of higher-order interactions in complex systems. Nat. Phys. 2021, 17, 1093–1098. [Google Scholar] [CrossRef]
- Murgas, K.A.; Saucan, E.; Sandhu, R. Hypergraph geometry reflects higher-order dynamics in protein interaction networks. Sci. Rep. 2022, 12, 20879. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, H.; Javali, A.; Khoei, H.H.; Sommer, T.M.; Sestini, G.; Novatchkova, M.; Reimer, Y.S.O.; Castel, G.; Bruneau, A.; Maenhoudt, N.; et al. Human blastoids model blastocyst development and implantation. Nature 2021, 601, 600–605. [Google Scholar] [CrossRef]
- Chi, R.-P.A.; Wang, T.; Adams, N.; Wu, S.-P.; Young, S.L.; Spencer, T.E.; DeMayo, F. Human Endometrial Transcriptome and Progesterone Receptor Cistrome Reveal Important Pathways and Epithelial Regulators. J. Clin. Endocrinol. Metab. 2020, 105, e1419-39. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple hypothesis testing. J. R Stat. Soc. B. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Handfield, L.-F.; Roberts, K.; Nikolakopoulou, K.; Fernando, R.C.; Gardner, L.; Woodhams, B.; Arutyunyan, A.; Polanski, K.; Hoo, R.; et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet. 2021, 53, 1698–1711. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587 e29. [Google Scholar] [CrossRef]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Reyes, A.P.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef]
- Starostik, M.R.; Sosina, O.A.; McCoy, R.C. Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism. Genome Res. 2020, 30, 814–825. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Zheng, X.; Yang, J.; Imamichi, T.; Stephens, R.; Lempicki, R.A. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinform. 2009, 27, 13.11.1–13.11.13. [Google Scholar] [CrossRef]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Settle, B.; Otasek, D.; Morris, J.H.; Demchak, B. aMatReader: Importing adjacency matrices via Cytoscape Automation. F1000Research 2018, 7, 823. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wang, W.; Vilella, F.; Alama, P.; Moreno, I.; Mignardi, M.; Isakova, A.; Pan, W.; Simon, C.; Quake, S.R. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat. Med. 2020, 26, 1644–1653. [Google Scholar] [CrossRef]
- Deng, W.; Wang, H. Efficient cell chatting between embryo and uterus ensures embryo implantationdagger. Biol. Reprod. 2022, 107, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.A.; Moldenhauer, L.M.; Green, E.S.; Care, A.S.; Hull, M.L. Immune determinants of endometrial receptivity: A biological perspective. Fertil. Steril. 2022, 117, 1107–1120. [Google Scholar] [CrossRef]
- Aplin, J. Uterus-Endometrium, in Encyclopedia of Reproduction; Skinner, M.K., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2018; pp. 326–332. [Google Scholar]
- Haller-Kikkatalo, K.; Tagoma, A.; Uibo, R.; Salumets, A.; Altmäe, S. Autoimmune activation toward embryo implantation is rare in immune-privileged human endometrium. Semin. Reprod. Med. 2014, 32, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Fragouli, E.; Alfarawati, S.; Spath, K.; Babariya, D.; Tarozzi, N.; Borini, A.; Wells, D. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum. Genet. 2017, 136, 805–819. [Google Scholar] [CrossRef]
- Liu, X.; Tan, J.P.; Schröder, J.; Aberkane, A.; Ouyang, J.F.; Mohenska, M.; Lim, S.M.; Sun, Y.B.Y.; Chen, J.; Sun, G.; et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021, 591, 627–632. [Google Scholar] [CrossRef]
- Yang, M.; Rito, T.; Metzger, J.; Naftaly, J.; Soman, R.; Hu, J.; Albertini, D.F.; Barad, D.H.; Brivanlou, A.H.; Gleicher, N. Depletion of aneuploid cells in human embryos and gastruloids. Nat. Cell Biol. 2021, 23, 314–321. [Google Scholar] [CrossRef]
- Moser, G.; Windsperger, K.; Pollheimer, J.; de Sousa Lopes, S.C.; Huppertz, B. Human trophoblast invasion: New and unexpected routes and functions. Histochem. Cell Biol. 2018, 150, 361–370. [Google Scholar] [CrossRef]
- Hjortebjerg, R. IGFBP-4 and PAPP-A in normal physiology and disease. Growth Horm. IGF Res. 2018, 41, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Shiojima, I.; Ito, Y.; Li, Z.; Ikeda, H.; Yoshida, M.; Naito, A.T.; Nishi, J.-I.; Ueno, H.; Umezawa, A.; et al. IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature 2008, 454, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Allègre, N.; Chauveau, S.; Dennis, C.; Renaud, Y.; Meistermann, D.; Estrella, L.V.; Pouchin, P.; Cohen-Tannoudji, M.; David, L.; Chazaud, C. NANOG initiates epiblast fate through the coordination of pluripotency genes expression. Nat. Commun. 2022, 13, 3550. [Google Scholar] [CrossRef]
- Lipecki, J.; Mitchell, A.; Muter, J.; Lucas, E.; Makwana, K.; Fishwick, K.; Odendaal, J.; Hawkes, A.; Vrljicak, P.; Brosens, J.; et al. EndoTime: Non-categorical timing estimates for luteal endometrium. Hum. Reprod. 2022, 37, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, T.M.; Makwana, K.; Taylor, D.M.; Molè, M.A.; Fishwick, K.J.; Tryfonos, M.; Odendaal, J.; Hawkes, A.; Zernicka-Goetz, M.; Hartshorne, G.M.; et al. Modelling the impact of decidual senescence on embryo implantation in human endometrial assembloids. Elife 2021, 10, e69603. [Google Scholar] [CrossRef] [PubMed]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polański, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Ferenczy, A.; Richart, R.M.; Agate, F.J.; Purkerson, M.L.; Dempsey, E.W. Scanning electron microscopy of the human endometrial surface epithelium. Fertil. Steril. 1972, 23, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Gamperl, M.; Burkard, T.R.; Kunihs, V.; Kaindl, U.; Junttila, S.; Fiala, C.; Schmidt, K.; Mendjan, S.; Knöfler, M.; et al. Estrogen Signaling Drives Ciliogenesis in Human Endometrial Organoids. Endocrinology 2019, 160, 2282–2297. [Google Scholar] [CrossRef]
- Ruane, P.T.; Tan, C.M.J.; Adlam, D.J.; Kimber, S.J.; Brison, D.R.; Aplin, J.D.; Westwood, M. Protein O-GlcNAcylation Promotes Trophoblast Differentiation at Implantation. Cells 2020, 9, 2246. [Google Scholar] [CrossRef]
- Ruane, P.T.; Koeck, R.; Berneau, S.; Kimber, S.J.; Westwood, M.; Brison, D.R.; Aplin, J. Osmotic stress induces JNK-dependent embryo invasion in a model of implantation. Reproduction 2018, 156, 421–428. [Google Scholar] [CrossRef]
- Fleming, T.P.; Eckert, J.J.; Denisenko, O. The Role of Maternal Nutrition During the Periconceptional Period and Its Effect on Offspring Phenotype. Adv. Exp. Med. Biol. 2017, 1014, 87–105. [Google Scholar]
- Sharma, N.; Kaur, J.; Xu, H.; Nieden, N.Z.; Rancourt, D. Characterization of secretory leukocyte protease inhibitor as an inhibitor of implantation serine proteinases. Mol. Reprod. Dev. 2008, 75, 1136–1142. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, J.; Di, Z.; Yuan, W.; Zhou, Z.; Liu, Z.; Han, S.; Liu, Y.; Ying, G.; Shu, X.; et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/beta-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 232. [Google Scholar] [CrossRef]
- Xu, H.; Li, J.; Jin, L.; Zhang, D.; Chen, B.; Liu, X.; Lin, X.; Huang, Y.; Ke, Z.; Liu, J.; et al. Intrauterine hyperglycemia impairs endometrial receptivity via up-regulating SGK1 in diabetes. Sci. China Life Sci. 2022, 65, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, X.-Y.; Yang, P.-W.; Yang, S.-L.; Hu, D.; Zhang, H.-W.; Sui, C. Activation of Uterine Smad3 Pathway Is Crucial for Embryo Implantation. Curr. Med. Sci. 2019, 39, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Kriseman, M.; Monsivais, D.; Agno, J.; Masand, R.P.; Creighton, C.J.; Matzuk, M.M. Uterine double-conditional inactivation of Smad2 and Smad3 in mice causes endometrial dysregulation, infertility, and uterine cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 3873–3882. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).