Abstract
Data augmentation using generative adversarial networks (GANs) is vital in the creation of new instances that include imaging modality tasks for improved deep learning classification. In this study, conditional generative adversarial networks (cGANs) were used on a dataset of OCT (Optical Coherence Tomography)-acquired images of coronary atrial plaques for synthetic data creation for the first time, and further validated using deep learning architecture. A new OCT images dataset of 51 patients marked by three professionals was created and programmed. We used cGANs to synthetically populate the coronary aerial plaques dataset by factors of 5×, 10×, 50× and 100× from a limited original dataset to enhance its volume and diversification. The loss functions for the generator and the discriminator were set up to generate perfect aliases. The augmented OCT dataset was then used in the training phase of the leading AlexNet architecture. We used cGANs to create synthetic images and envisaged the impact of the ratio of real data to synthetic data on classification accuracy. We illustrated through experiments that augmenting real images with synthetic images by a factor of 50× during training helped improve the test accuracy of the classification architecture for label prediction by 15.8%. Further, we performed training time assessments against a number of iterations to identify optimum time efficiency. Automated plaques detection was found to be in conformity with clinical results using our proposed class conditioning GAN architecture.
1. Introduction
Deep neural networks (DNNs) have emerged as a promising solution in medical imaging classification tasks. Deep learning algorithms model high level abstractions by making use of immense neural networks with multiple layers of execution units. These advanced computing networks realize ameliorated training techniques to unfold complex data patterns in big data problems. DNNs discern features incrementally and build feature sets independently, and thus do not require any supervision. Broadly, they are classified as multi-layer perceptrons (MLPs), recurrent neural networks (RNNs) and convolutional neural networks (CNNs) [1]. Generally, CNNs are preferred over other modalities for medical imaging problems and classification purposes.
Data augmentation (DA) techniques assist with the reliable characterization of limited medical image datasets processed through DNNs [2,3]. Limited training data in deep models can be handled by applying DA techniques to scale up the dataset in terms of size and diversity [4,5]. To reduce the generalization error of the classification model, augmented data should focus primarily on generic features to mitigate overfitting [6,7]. Aggregating different augmentations assists with significant data augmentation, but leads to overfitting for limited medical imaging datasets, including OCT-acquired coronary plaque images [8,9]. A new classification algorithm suitable for real-time clinical assessment of coronary arterial plaques has been reported [10,11]; however, the real impact of synthetic data creation has not been thoroughly elaborated. Feature space augmentation is generally not preferred for medical imaging datasets due to interpretation, time and space complexities. Using GANs, we can synthetically generate new medical images, including OCT images, from limited available samples by setting up the generator and the discriminator in competition. This helps resolve the prevalent issues of data volume, diversification and class balance for coronary plaques images acquired through OCT [12]. The availability of public datasets has allowed clinical professionals to draw inter-comparisons between different GANs [13,14]. Standard GANs are being replaced by different variants to achieve better classification results and parameters optimization [15]. These include WGAN, Super GAN, LS-GAN, Bi-GAN and StyleGAN for medical imaging datasets of Alzheimer’s and brain and liver tumors, and high-resolution synthesis of sclerosis [16,17,18,19,20,21,22]. Generally, cGANs are favored for creating synthetic images related to cardiac imaging modalities, as they leverage additional information in the form of labelled data [23,24,25,26,27,28,29]. Further, they avoid modal collapse [30] and result in faster convergence by conditioning class labels to both the discriminator and the generator.
To the best of the authors’ knowledge, cGANs have not been exploited for the augmentation of coronary images acquired via OCT imaging for the classification of coronary atrial plaques. The resolve of this study was to perform data augmentation on a newly created OCT dataset using cGANs. The created images were then appended to an original OCT dataset and validated using leading AlexNet classification architecture. Further, we aimed to establish the impact of augmentation on the deep learning classification of coronary atrial plaques. To achieve this, the objective functions of the generator and the discriminator were set in competition against each other. Using the gradient descent method for optimization ensured the creation of new instances that exactly replicated the original images. Transfer learning was exploited for training purposes. We froze the initial layers of our model and fine-tuned them accordingly. To establish ground-truth, three professional clinicians determined the plaque type present in an A-scan. We envisaged the impact of the ratio of real data to synthetic data on classification accuracy. Using assessments, we illustrated how populating real images with created instances during the training phase increased our confidence in the reliable label prediction of coronary plaques. Using cGANs-created images and varying the ratio between created images and real data within each sub-batch resulted in up to a 15.8% improvement in classification accuracy.
2. Methods
A newly created dataset of 88 atrial stenoses in 51 patients was developed using the commercially available OCT system. Our target vessels were those with stenosis. However, for the sake of simplicity, serial stenosis and by-pass graft stenosis were not taken into consideration. The study was approved by Galway’s Clinical Research Ethics Committee (GCREC) with informed consents from the patients. A general flow of the algorithm implemented in the study is presented in Figure 1.
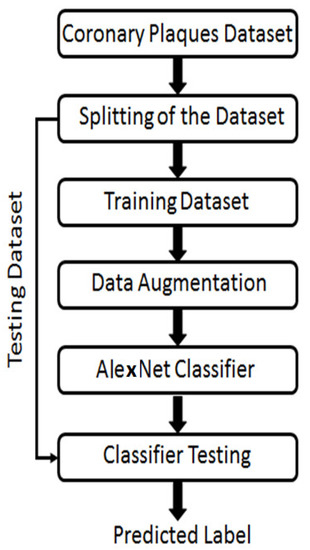
Figure 1.
An executional flow of the phases performed to achieve classification of different coronary atrial plaques using cGANs.
All three clinicians individually labelled the OCT acquired images, but each label was finally decided on the basis of mutual consensus. We set up a classification problem by setting up a specific label against the rest. Preprocessing steps were applied to raw OCT acquired images before feeding them further into our model. Vulnerable plaques that mapped with the established fibrous cap thickness criterion were excluded from this study. Measuring the signal intensity to lumen helped characterize them as either lipid (low signal intensity) or calcified (high signal intensity) plaques. In the original dataset, 27% of images were labeled as “calcified”, 23% as “lipid plaque”, 21% as “mixed plaque” and 29% as “no plaque”.
2.1. Medical Images Augmentation Using GANs
Conditional GAN transformed image data from lower to higher dimensional space using a random noise vector [31,32]. Adding a vector of features derived from an OCT image, along with the noise input, the discriminator’s task was to find the similarity index between the real and fake images and assign false images to its input labels. Conditioning was accomplished by feeding one hot vector into the input of the generator [33]. A high-level exhibit of our cGAN is presented in Figure 2. The generator and the discriminator operated with multiple convolution layers, batch normalization layers and ReLU functions. Class encoding was one hot vector with a length equivalent to the number of classes (4 in our case), and the position of the nth class was one, with the rest set to zero. In our cGAN network, we parsed in OCT images along with the class information to which each image belonged. We approximated an unfamiliar data pattern through a generator that attempted to fit samples to a known prior distribution. The generator (G) competed against the discriminator (D) to compute the desired convergence point.
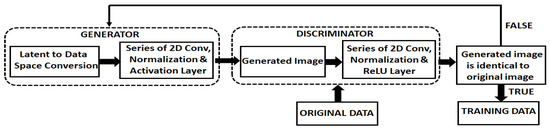
Figure 2.
An overview of the scheme used to implement the cGAN to create new instances.
The generator created accurate new instances using the algorithm presented in Figure 3. The discriminator’s task was to differentiate between the real and real-like created instances, as illustrated in Figure 2. If the cGANs-produced image was identical to the original image, then it was appended to the labelled class of images.
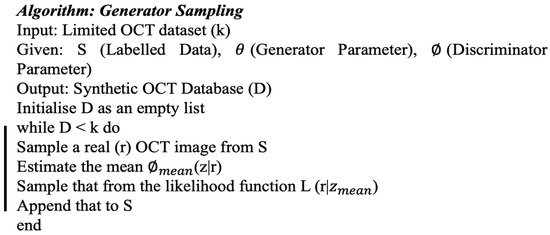
Figure 3.
Algorithm used for the generator sampling.
Our discriminator correctly labeled the incident real and false images into their specific classes. Our loss function for the discriminator was the sum of false and real image loss. Our aim was to diminish the error of forecasting real images of the dataset and the generated fake images [34,35].
The discriminator’s training flow is presented in Figure 4. In this Figure, G is the generator, D represents the discriminator, X indicates the fake images conditioned on Y, Y is the hot label for real data images and Y is the hot label for fake images. Random noise was fed to the generator along with class encoded hot label Y to foster fake images conditioned upon Y. In this cycle, gradients were not passed down the generator. The generator training process was similar to the discriminator training cycle, as illustrated in Figure 4. Both the discriminator and the generator were proficient enough to foster data close to the original data. During this training phase, discriminator gradients were conceded by the generator.
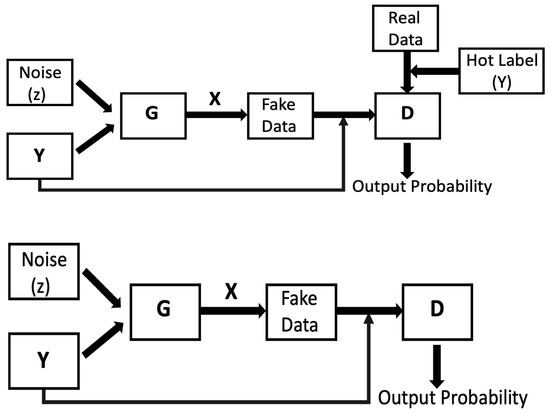
Figure 4.
A schematic illustration of the process involved in training the discriminator (top) and the generator (bottom).
Generator and discriminator loss functions are already reported in the literature [36,37] and are expressed in Equations (1) and (2).
As illustrated in Equations (1) and (2), we were dealing with two neural networks in which the generator began with a random data distribution and attempted to foster exact replicas of the real images. The discriminator network improved at distinguishing between real and generated images through successive training iterations. Both networks work against each other to generate perfect aliases of the original images.
2.2. Deep Learning Classifier
Figure 5 represents the cutting edge CNN algorithm (AlexNet), in which each chunk signifies the input and output characteristics [38]. AlexNet architecture consists of eight layers with five convolutional and three fully-connected layers. AlexNet was preferred to validate of our synthetically created images due to its competitive edge in training the cGANs. Dropout was functional on the first and second fully-connected layers. The input image dimensions were 227 × 227 × 3. For down-sampling, a max-pooling operation was implemented for a stride of two between the adjacent frames. In the dropout phase, a neuron with a probability of ½ was disregarded. This means that the neuron had not contributed to the forward propagation or the back propagation loss. This ensured that every iteration captured a diverse sample of the model’s parameters without being over fitted.
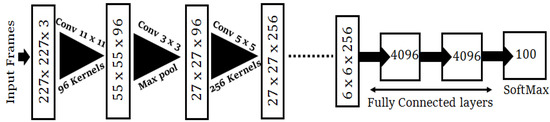
Figure 5.
The AlexNet classification model used for the validation of our created dataset.
To achieve transfer learning, we detached the last layer and used AlexNet as our pre-prepared model. For the second pass of the training cycle, we release the 4th and 5th convolutional layers with our first three layers jammed. Fine-tuning was completed by eliminating the fully linked nodes and inserting new layers. We regularized our classification forecasts via cross-entropy loss amid the factual label distribution and the projected label.
3. Results and Discussion
We performed simulations on the newly created OCT dataset, which contained 146 original images belonging to four classes that were then augmented using cGAN with multiplication factors of 5×, 10×, 50× and 100×. The × indicates images acquired via OCT and factor indicates images generated using augmentation. We used 70:30 factors for training and testing purposes. We used TensorFlow, an open-source framework (v2.10.0), to build the model and to implement it. We used CUDA to accelerate training a batch size of 64. For model training, a momentum of 0.9 was used with an initial learning rate of Lr = 10−4. This initial value of momentum helped accelerate training and converge the optimization cycle at the end of training. The learning drop factor was 0.1 with a learning drop rate period of five. The Adam optimizer was used to update network weights. Our model distinguished the coronary arterial plaques in a time-competitive fashion. The results were considered using the testing dataset, whereas assorted hyper-parameters were performed using the validation set. Figure 6 exhibits GANs-generated sample images of our four classes; namely, normal, calcified, lipid and mixed atrial plaques.
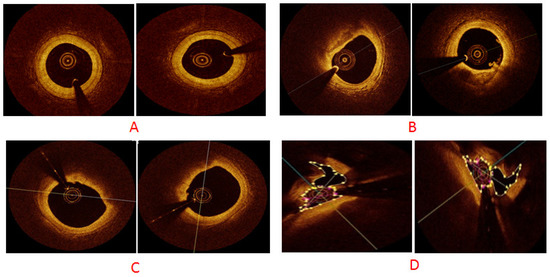
Figure 6.
cGAN-generated synthetic sample images for different classes: (A) normal plaque, (B) calcium plaque, (C) lipid plaque and (D) mixed plaque.
Next, cGAN-generated images were validated using the leading AlexNet architecture, in which each layer performed multi-level feature extraction and local features alongside. The composition of our fully connected layer helped reduced the dimensionality of training parameters. During the discriminator’s training, the same hot label was transformed to a tensor and combined with a fake image to feed it. The discriminator also received real data images along with the respective hot label transformed tensors. The discriminator determined the input was a real image based on binary cross-entropy loss. We abated the cross-entropy loss L in our algorithm using Equation (3) for multiclass classification. As we already had target probability distribution for an input class label, our aim was to predict the target distribution with reasonable confidence.
where m denotes the number of classes, y is the ground truth label and Y represents the softmax normalized model prediction.
We minimized cross-entropy across our training dataset by averaging out the cross-entropy for all training images. Our classification problem had three classes; any sample belonged to one of these three classes. The discrete probability distribution had a value of one when a sample belonged to a specific class, and zero for the rest of the classes.
Table 1 indicates the validation accuracy with and without the augmented data. It is clear that without synthetic data the classification accuracy of our model was 82.9%; it reached 98.7% after the synthetic data was merged with the original data. This resulted in reduced overfitting and improved the model’s generality. The number of iterations and its impact on the validation accuracy is highlighted in Table 1.

Table 1.
Percentage accuracy against the number of iterations with and without data augmentation.
The relationship between the iterations and training time is exhibited in Figure 7 for different augmentation scaling. We performed experiments for 5×, 10×, 50× and 100× and recorded the training time in minutes for each set of experiments. These multiplicative factors were chosen for simplicity and by observing the diversity and imbalance present in our original dataset. To observe the pronounced effect of augmentation, we scaled for 50× and 100×. This helped determine the extent to which data could be augmented and enhanced our classification accuracy. It can be inferred from Figure 7 that 5× was the optimum augmentation scheme for our created dataset. This is attributed to fact that our original OCT dataset was limited in size; therefore, 5× and 10× did not significantly improvise in terms of the samples’ diversification. Similarly, 100× led to overfitting our sample space along with adding an additional computational burden. An interesting point that can be deduced from the results presented in Figure 7 is that the increase in the training data was not exactly proportional to the increase in the performance. When a specific threshold was achieved, no further improvement in the performance was observed. This is attributed to the fact that the diversity of data could not be further improved.
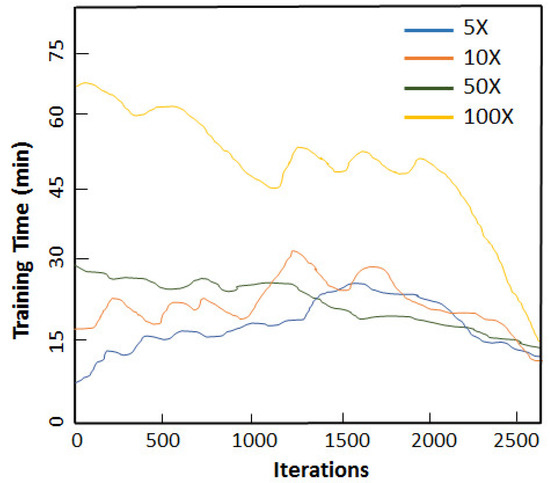
Figure 7.
Relationship between the number of iterations and training time for different augmentation scaling.
We further envisaged the effect of the augmented-to-real ratio on the increase in the classifier’s performance, as illustrated in Figure 8. The dotted line indicates the non-augmented baseline. As we increased the augmented-to-real ratio, the dataset inflated in terms of size and diversity, allowing the classifier to train more rigorously. This diversification of samples ameliorated the classifier’s ability to more correctly ascertain the labels. The classifier’s accuracy increased until our augmented-to-real ratio reached 50, as indicated in Figure 8. It then flattened due to similar and repeated samples being created. This filled the dataset, but could not further boost the generality of the model. The overall increase in the classifier’s performance due to augmented data was 15.8%. In Figure 8, the height of the bar indicates variance and the outer stubbles mark the maximums and minimums.
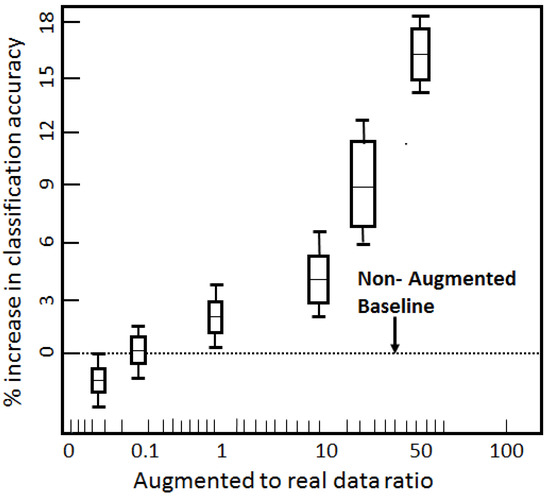
Figure 8.
Impact of augmented-to-real data ratio on the classifier’s performance.
Fundamental augmentation methods generate synthetic medical images relatively easily, whereas DNNs employ cross domains to create new images that achieve more diversity in the creation of new data. The choice of augmentation technique is contingent upon the available dataset and the ultimate objective involved. To the best of the authors’ knowledge, there is no reported published research in which augmentation has been applied directly to coronary atrial plaques data. For the sake of assessment, our proposed augmentation technique was compared to other datasets for an inter-comparison, as illustrated in Table 2. In Table 2, we present the dataset length, type of dataset and achieved accuracies both without and with augmentation techniques applied. Only fundamental data creation techniques, including random cropping, distortion, blurring and random erasing, were performed on certain datasets indicated in Table 2. However, Table 2 also includes more sophisticated data creation techniques, such as the one we proposed, to validate the efficacy of artificial data creation via deep learning coupled augmentation.

Table 2.
An inter-comparison of our proposed algorithm with data augmentation techniques applied on similar and other datasets, along with respective overall improved accuracies.
4. Conclusions and Future Work
We presented an in-depth consideration of using cGANs to generate synthetic coronary atrial plaques images. Data augmentation was proven particularly advantageous to avoid overfitting in our case and in other scenarios where there is limited training data available. The observed percentage increase in classification accuracy upholds the importance of exploiting cGANs for data augmentation in medical imaging datasets, including ours. However, the ratio of synthetic data to real data to populate the real OCT dataset was found to be crucial in terms of model overfitting, time competitiveness and confident assessment of the model’s performance.
Building a deep learning algorithm for real-time clinical assessments with data privacy intact and aggregating cGANs with other augmentation meta-learning architectures, such as neural style transfers, are imperative areas for future work. We also envisage the possibility of increasing GANs’ training speed via concurrent networks for large-scale medical imaging datasets. Furthermore, the practical coupling of data augmentation algorithms into software development tools and the optimization of applications also offer blue sky research avenues to unleash the real potential of data augmentation in automated medical imaging.
Author Contributions
Conceptualization, H.Z. and J.Z.; methodology, H.Z., J.Z. and F.S.; software, J.Z.; validation, H.Z. and J.Z.; formal analysis, H.Z., J.Z. and F.S.; investigation, H.Z. and J.Z.; resources, H.Z. and F.S.; data curation, F.S.; writing—original draft preparation, H.Z., J.Z. and F.S.; writing—review and editing, H.Z., J.Z. and F.S.; visualization, J.Z. and F.S.; supervision, H.Z. and F.S.; project administration, H.Z.; funding acquisition, H.Z. and J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Galway Clinical Research Ethics Committee.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to basic character of research.
Acknowledgments
Faisal Sharif is supported by an SFI Research Infrastructure Grant (grant number: 17/RI/5353).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Qayyum, A.; Anwar, S.M.; Awais, M.; Majid, M. Medical image retrieval using deep convolutional neural network. Neurocomputing 2017, 266, 8–20. [Google Scholar] [CrossRef]
- Fabio, G.; Alessio, S.; Lamberti, L.M. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152, 106391. [Google Scholar]
- Majdoubi, J.; Iyer, A.S.; Ashique, A.M.; Perumal, D.A.; Mahrous, Y.M.; Rahimi-Gorji, M.; Issakhov, A. Estimation of tumor parameters using neural networks for inverse bioheat problem. Comput Methods Programs Biomed. 2021, 205, 106092. [Google Scholar] [CrossRef]
- Huang, P.; Liu, X.; Huang, Y. Data Augmentation for Medical MR Image Using Generative Adversarial Networks. arXiv 2021, arXiv:2111.14297. [Google Scholar] [CrossRef]
- He, W.; Liu, M.; Tang, Y.; Liu, Q.; Wang, Y. Differentiable Automatic Data Augmentation by Proximal Update for Medical Image Segmentation. IEEE/CAA J. Autom. Sin. 2022, 9, 1315–1318. [Google Scholar] [CrossRef]
- Adar, M.F.; Diamant, I.; Klang, E. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 2018, 321–331. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Hu, B. Minimal GAN: Diverse medical image synthesis for data augmentation using minimal training data. Appl. Intell. 2022, 53, 3899–3916. [Google Scholar] [CrossRef]
- Muhammad Hussain, N.; Rehman, A.U.; Othman, M.T.B.; Zafar, J.; Zafar, H.; Hamam, H. Accessing Artificial Intelligence for Fetus Health Status Using Hybrid Deep Learning Algorithm (AlexNet-SVM) on Cardiotocographic Data. Sensors 2022, 22, 5103. [Google Scholar] [CrossRef]
- Shahwar, T.; Zafar, J.; Almogren, A.; Zafar, H.; Rehman, A.U.; Shafiq, M.; Hamam, H. Automated Detection of Alzheimer’s via Hybrid Classical Quantum Neural Networks. Electronics 2022, 11, 721. [Google Scholar] [CrossRef]
- Zafar, H.; Zafar, J.; Sharif, F.; Zafar, H.; Zafar, J.; Sharif, F. Automated Clinical Decision Support for Coronary Plaques Characterization from Optical Coherence Tomography Imaging with Fused Neural Networks. Optics 2022, 3, 8–18. [Google Scholar] [CrossRef]
- Nanni, L.; Paci, M.; Brahnam, S.; Lumini, A. Comparison of Different Image Data Augmentation Approaches. J. Imaging 2021, 7, 254. [Google Scholar] [CrossRef]
- Gong, M.; Chen, S.; Chen, Q.; Zeng, Y.; Zhang, Y. Generative Adversarial Networks in Medical Image Processing. Curr. Pharm. Des. 2021, 27, 1856–1868. [Google Scholar] [CrossRef]
- Khosla, C.; Saini, B.S. Enhancing Performance of Deep Learning Models with different Data Augmentation Techniques: A Survey. In Proceedings of the International Conference on Intelligent Engineering and Management (ICIEM), London, UK, 17–19 June 2020; pp. 79–85. [Google Scholar] [CrossRef]
- Tang, S. Lessons Learned from the Training of GANs on Artificial Datasets. IEEE Access 2020, 8, 165044–165055. [Google Scholar] [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative Adversarial Network in Medical Imaging: A Review. Med. Image Anal. 2018, 58, 101552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Pan, B.; Shao, P.; Liu, P.; Shen, S.; Yao, P.; Xu, R.X. Alzheimer’s Disease Neuroimaging Initiative; Australian Imaging Biomarkers Lifestyle flagship study of ageing. A Single Model Deep Learning Approach for Alzheimer’s Disease Diagnosis. Neuroscience 2022, 491, 200–214. [Google Scholar] [CrossRef] [PubMed]
- de Farias, E.C.; di Noia, C.; Han, C.; Sala, E.; Castelli, M.; Rundo, L. Impact of GAN-based lesion-focused medical image super-resolution on the robustness of radiomic features. Sci. Rep. 2021, 11, 21361. [Google Scholar] [CrossRef]
- Delannoy, Q.; Pham, C.H.; Cazorla, C.; Tor-Díez, C.; Dollé, G.; Meunier, H.; Bednarek, N.; Fablet, R.; Passat, N.; Rousseau, F. SegSRGAN: Super-resolution and segmentation using generative adversarial networks- Application to neonatal brain MRI. Comput. Biol. Med. 2020, 120, 103755. [Google Scholar] [CrossRef]
- Chen, X.; Lian, C.; Wang, L.; Deng, H.; Kuang, T.; Fung, S.H.; Gateno, J.; Shen, D.; Xia, J.J.; Yap, P.T. Diverse data augmentation for learning image segmentation with cross-modality annotations. Med. Image Anal. 2021, 71, 102060. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumour classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Fetty, L.; Bylund, M.; Kuess, P. Latent space manipulation for high-resolution medical image synthesis via the StyleGAN. Z. Med. Phys. 2020, 30, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Barile, B.; Marzullo, A.; Stamile, C.; Durand-Dubief, F.; Sappey-Marinier, D. Data augmentation using generative adversarial neural networks on brain structural connectivity in multiple sclerosis. Comput. Methods Programs Biomed. 2021, 206, 106113. [Google Scholar] [CrossRef]
- Zyuzin, V.; Komleva, J.; Porshnev, S. Generation of echocardiographic 2D images of the heart using cGAN. J. Phys. Conf. Ser. 2021, 1727, 2013. [Google Scholar] [CrossRef]
- Diller, G.P.; Vahle, J.; Radke, R.; Vidal, M.L.; Fischer, A.J.; Bauer, U.M.; Sarikouch, S.; Berger, F.; Beerbaum, P.; Baumgartner, H.; et al. Utility of deep learning networks for the generation of artificial cardiac magnetic resonance images in congenital heart disease. BMC Med. Imaging 2020, 20, 113. [Google Scholar] [CrossRef]
- Skandarani, Y.; Lalande, A.; Afilalo, J.; Jodoin, P.M. Generative Adversarial Networks in Cardiology. Can. J. Cardiol. 2022, 38, 196–203. [Google Scholar] [CrossRef]
- Uzunova, H.; Ehrhardt, J.; Handels, H. Memory-efficient GAN-based domain translation of high resolution 3D medical images. Comput. Med. Imaging Graph. 2020, 86, 101801. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, M.; Yang, H.; Harmuth, K. Conditional Generative Adversarial Refinement Networks for Unbalanced Medical Image Semantic Segmentation. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 7–11 January 2019; pp. 1836–1845. [Google Scholar] [CrossRef]
- Han, C.; Kitamura, Y.; Kudo, A.; Ichinose, A.; Rundo, L.; Furukawa, Y.; Umemoto, K.; Li, Y.; Nakayama, H. Synthesizing Diverse Lung Nodules Wherever Massively: 3D Multi-Conditional GAN-Based CT Image Augmentation for Object Detection. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Quebec City, QC, Canada, 16–19 September 2019; pp. 729–737. [Google Scholar] [CrossRef]
- Naglaha, A.; Khalifaa, F.; El-Baza, A. Conditional GANs based system for fibrosis detection and quantification in Hematoxylin and Eosin whole slide images. Med. Image Anal. 2022, 81, 102537. [Google Scholar] [CrossRef]
- Thanh-Tung, H.; Tran, T. Catastrophic forgetting and mode collapse in GANs. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–10. [Google Scholar] [CrossRef]
- Dar, S.U.; Yurt, M.; Karacan, L.; Erdem, A.; Erdem, E.; Cukur, T. Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Trans. Med. Imaging 2019, 38, 2375–2388. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Xu, H.; Jiang, J.; Mei, X.; Zhang, X.P. DDcGAN: A Dual-Discriminator Conditional Generative Adversarial Network for Multi-Resolution Image Fusion. IEEE Trans. Image Process. 2020, 29, 4980–4995. [Google Scholar] [CrossRef] [PubMed]
- Celard, P.; Iglesias, E.L.; Sorribes-Fdez, J.M.; Romero, R.; Vieira, A.S.; Borrajo, L. A survey on deep learning applied to medical images: From simple artificial neural networks to generative models. Neural Comput. Applic. 2023, 35, 2291–2323. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar] [CrossRef]
- Douzas, G.; Bacao, F. Effective data generation for imbalanced learning using conditional generative adversarial networks. Expert Syst. Appl. 2018, 91, 464–471. [Google Scholar] [CrossRef]
- Strelcenia, E.; Prakoonwit, S. A Survey on GAN Techniques for Data Augmentation to Address the Imbalanced Data Issues in Credit Card Fraud Detection. Mach. Learn. Knowl. Extr. 2023, 5, 304–329. [Google Scholar] [CrossRef]
- Lucas, A.; Tapia, S.L.; Molina, R.; Katsaggelo, A.K. Generative Adversarial Networks and Perceptual Losses for Video Super-Resolution. Available online: https://arxiv.org/abs/1806.05764 (accessed on 14 February 2023).
- Lu, S.; Lu, Z.; Zhang, Y.D. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Perez, F.; Vasconcelos, C.; Avila, S.; Valle, E. Data Augmentation for Skin Lesion Analysis. Available online: https://arxiv.org/abs/1809.01442 (accessed on 14 February 2023).
- Muramatsu, C.; Nishio, M.; Goto, T.; Oiwa, M.; Morita, T.; Yakami, M.; Kubo, T.; Togashi, K.; Fujita, H. Improving breast mass classification by shared data with domain transformation using a generative adversarial network. Comput. Biol. Med. 2020, 119, 103698. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.; Hong, H.; Bae, H.; Lim, J.S.; Kim, J. Classification of focal liver lesions in CT images using convolutional neural networks with lesion information augmented patches and synthetic data augmentation. Med. Phys. 2021, 48, 5029–5046. [Google Scholar] [CrossRef]
- Uemura, T.; Näppi, J.J.; Ryu, Y.; Watari, C.; Kamiya, T.; Yoshida, H. A generative flow-based model for volumetric data augmentation in 3D deep learning for computed tomographic colonography. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.R.; Elshamy, M.R.; Taha, T.E.; El-Fishawy, A.S.; Abd El-Samie, F.E. Efficient deep learning models for brain tumor detection with segmentation and data augmentation techniques. Concurr. Comput. Pract. Exp. 2022, 34, e7031. [Google Scholar] [CrossRef]
- Tasci, E.; Uluturk, C.; Ugur, A. A voting-based ensemble deep learning method focusing on image augmentation and preprocessing variations for tuberculosis detection. Neural Comput. Applic. 2021, 33, 15541–15555. [Google Scholar] [CrossRef] [PubMed]
- Ioannis, D.; Apostolopoulos, N.; Spyridonidis, T.; Apostolopoulos, D.J. Automatic characterization of myocardial perfusion imaging polar maps employing deep learning and data augmentation. Hell. J. Nucl. Med. 2020, 23, 125–132. [Google Scholar]
- He, C.; Wang, J.; Yin, Y.; Li, Z. Automated classification of coronary plaque calcification in OCT pullbacks with 3D deep neural networks. J. Biomed. Opt. 2020, 25, 095003. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; He, C.; Xu, B.; Li, Z. Coronary Plaque Characterization from Optical Coherence Tomography Imaging with a Two-Pathway Cascade Convolutional Neural Network Architecture. Front. Cardiovasc. Med. 2021, 8, 670502. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).