1. Introduction
Colorectal cancer (CRC) continues to rank among the most prevalent and lethal cancers globally, with a notable and rapid increase in early-onset cases—defined as diagnoses occurring before age 50—over the past several decades [
1,
2,
3,
4,
5]. This upward trend is particularly pronounced in high-risk groups, including Hispanic/Latino (H/L) individuals [
6,
7,
8,
9,
10]. Although multiple oncogenic pathways contribute to CRC development and progression, the transforming growth factor-beta (TGF-β) signaling pathway plays a pivotal role by facilitating processes such as epithelial-to-mesenchymal transition (EMT), immune system evasion, and metastatic spread [
11,
12,
13]. However, efforts to comprehensively characterize TGF-β dysregulation in EOCRC—especially among the underrepresented populations—have been constrained by the underrepresentation of diverse patient cohorts in genomic databases and the absence of tools capable of linking clinical and molecular data in an integrative manner [
6,
7,
14,
15].
The TGF-β pathway is frequently altered in CRC through mutations in key components such as SMAD4, BMPR1A, and TGFBR2, which are associated with poor prognosis, therapy resistance, and aggressive tumor phenotypes [
16,
17,
18,
19]. Recent studies suggest that these mutations may present with distinct patterns in EOCRC compared to late-onset CRC (LOCRC) and may vary by ethnicity—highlighting a need for population-specific investigation [
6,
7,
19,
20]. For example, alterations in BMPR1A and BMP7 have been identified in H/L patients with EOCRC, suggesting unique mechanisms of TGF-β dysregulation in this group [
6,
7]. However, few tools exist that can efficiently integrate and stratify these genomic insights alongside clinical factors, such as age, tumor stage, treatment response, and microsatellite instability (MSI) status.
Although public platforms such as The Cancer Genome Atlas (TCGA) and AACR GENIE provide rich datasets for CRC research, existing analysis tools—like cBio Cancer Genomics Portal (cBioPortal) [
21] and the University of California Santa Cruz (UCSC) Xena [
22]—require multi-step workflows and offer limited functionality for pathway-specific, population-disaggregated, or treatment-contextualized exploration in investigating CRC TGF-β dysregulation. These constraints disproportionately affect non-computational researchers, impeding precision oncology efforts in real-world and subpopulation contexts.
The emergence of artificial intelligence (AI), especially advancements in large language models (LLMs), has paved the way for conversational tools that convert natural language input into executable data analysis pipelines [
23,
24]. Although these technologies have demonstrated potential in streamlining multiomic investigations [
25,
26,
27,
28,
29,
30], there remains a lack of platforms specifically designed to target signaling pathways or to support integrative clinical–genomic research with an emphasis on hypothesis generation and health disparity considerations.
AI-HOPE-TGFbeta (Artificial Intelligence agent for High-Optimization and Precision Medicine focused on TGF-β) was developed to directly address the lack of tools capable of pathway-specific integrative analysis in CRC. This conversational AI system enables users to pose natural language questions that are translated into executable workflows, facilitating real-time synthesis of harmonized genomic and clinical data. With built-in automation for statistical tasks—including Kaplan–Meier survival analysis and odds ratio estimation—the platform streamlines both validation studies and exploratory investigations across large datasets. In the present study, we (1) created AI-HOPE-TGFbeta to enable a user-friendly, pathway-centered interrogation of CRC cohorts; (2) assessed its performance by replicating well-established clinical–genomic associations in EOCRC; and (3) applied the system to reveal novel links between TGF-β mutations, MSI, tumor staging, and population-level variables. These results highlight AI-HOPE-TGFbeta as an innovative and inclusive solution to support scalable, translational TGF-β pathway research in precision oncology.
To guide the reader through the manuscript, the remainder of the paper is organized as follows: In
Section 2, we describe the architecture of the AI-HOPE-TGFbeta platform, including its system components, data sources, conversational query handling, statistical framework, and validation strategy.
Section 3 presents the results of validation and exploratory analyses, highlighting both recapitulated findings and novel associations between TGF-β pathway alterations and clinical–genomic features in colorectal cancer. In
Section 4, we discuss the translational significance of the findings, implications for health equity, and technical advantages of the platform, while also outlining limitations and future directions. Finally,
Section 5 provides our concluding remarks on the broader impact of AI-HOPE-TGFbeta for precision oncology and population-focused cancer research.
This study contributes several novel methodological advancements at the intersection of AI and clinical–genomic research. AI-HOPE-TGFbeta leverages a fine-tuned biomedical variant of the Large Language Model Meta AI 3 (LLaMA 3) for the semantic interpretation of natural language queries. It incorporates a natural language-to-code interpreter that translates user inputs into executable Python 3.12-based statistical workflows. These AI components enable real-time cohort stratification, hypothesis testing, and interpretation of TGF-β pathway alterations in CRC—without requiring programming expertise. These technical innovations distinguish AI-HOPE-TGFbeta as a scalable and user-friendly system for translational bioinformatics and precision oncology.
2. Materials and Methods
2.1. System Architecture and Workflow of AI-HOPE-TGFbeta
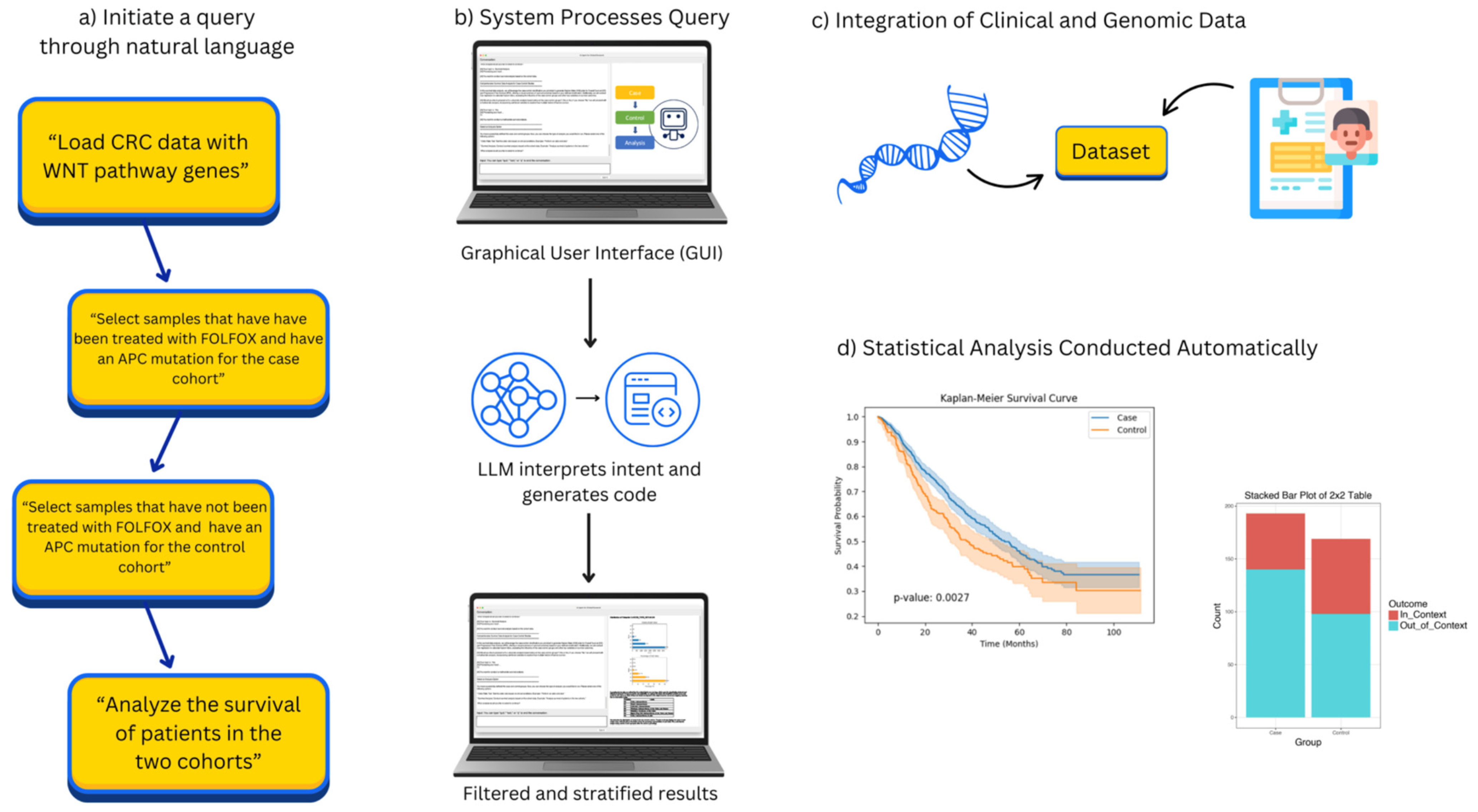
AI-HOPE-TGFbeta is a natural language-enabled AI system engineered to explore CRC with a specific focus on alterations within the TGF-β signaling pathway. The platform is built on a layered, modular framework that integrates three core components: a built-in LLM based on the LLaMA 3 architecture for semantic query interpretation, a translation layer that converts user prompts into executable code, and a statistical backend designed to automate case generation, analytical processing, and result visualization. When a user submits a question in plain English, the system identifies the analytical intent, applies relevant filters to harmonized clinical–genomic datasets, and generates a suite of outputs—including survival analyses, mutation frequencies, odds ratios, and explanatory text summaries tailored to the context of the query (
Figure 1).
The choice of the LLaMA 3 architecture as the core language model for AI-HOPE-TGFbeta was based on its strong performance in biomedical natural language processing tasks, its open-source accessibility, and its adaptability for domain-specific fine-tuning. Compared to other available LLMs, LLaMA 3 offers an optimal balance between computational efficiency and contextual accuracy, making it well-suited for real-time, query-driven clinical–genomic analysis. The model’s architecture supports long-context comprehension and precise semantic parsing, both essential for translating complex natural language queries into executable bioinformatics workflows. Furthermore, its open-access licensing facilitates reproducibility and broader deployment across research environments without commercial restrictions, aligning with the goal of democratizing precision oncology tools.
2.2. Data Sources and Preparation for AI-HOPE-TGFbeta
To power its analyses, AI-HOPE-TGFbeta draws from harmonized CRC datasets derived from public repositories such as TCGA and cBioPortal, with a targeted focus on genes implicated in TGF-β signaling. Key genomic features include alterations in SMAD4, TGFBR1, TGFBR2, BMPR1A, BMPR2, ACVR1B, and BMP7. The accompanying clinical metadata encompass a broad array of attributes—patient age, disease stage, treatment history (including FOLFOX—fluorouracil, leucovorin, and oxaliplatin—exposure), MSI classification, ethnicity, tumor tissue origin (primary versus metastatic), and overall survival metrics. Raw data underwent extensive preprocessing to ensure analytical compatibility: files were converted into standardized, tab-delimited matrices with harmonized sample identifiers, and ontology-based frameworks, such as OncoTree and the Disease Ontology, were applied to unify clinical annotations. Mutation data were cross-validated across sources, and all TGF-β pathway gene sets were curated using publicly available knowledge bases to ensure accuracy and biological relevance.
2.3. Conversational Query Handling and Cohort Definition in AI-HOPE-TGFbeta
AI-HOPE-TGFbeta enables users to initiate complex clinical–genomic analyses using natural language inputs. Queries such as “Compare survival outcomes for SMAD4-mutated versus wild-type EOCRC patients treated with FOLFOX” or “Evaluate TGFBR1 mutation prevalence between H/L and non-Hispanic White (NHW) individuals” are interpreted by a built-in LLM based on the LLaMA 3 architecture. This LLM translates conversational prompts into executable code that filters datasets, defines cohorts, and launches the appropriate statistical analyses. When necessary, the system prompts the user for clarification to resolve ambiguity and ensure accurate query interpretation. AI-HOPE-TGFbeta accommodates a wide range of stratification parameters, including genetic mutation status, MSI, tumor stage, race/ethnicity, and chemotherapy exposure, allowing users to flexibly define custom subgroups for targeted analysis.
To ensure data integrity and analytical reliability, all colorectal cancer datasets were extracted from cBioPortal, which provides harmonized and quality-controlled clinical–genomic data. For this study, we selected only those cases with complete clinical variables—including tumor stage, MSI status, treatment history, and survival data—and comprehensive genomic profiles covering key TGF-β pathway genes. The dataset underwent preprocessing to remove any entries with missing values across variables critical for cohort stratification and statistical analysis. Additionally, class balance was assessed across primary stratification variables (e.g., mutation status, MSI phenotype, ethnicity), and cohort sizes were adjusted where necessary to maintain representativeness and minimize analytical bias. These measures ensured that the final dataset was both complete and sufficiently balanced for robust statistical evaluation.
2.4. Analytical Framework and Statistical Methods in AI-HOPE-TGFbeta
AI-HOPE-TGFbeta’s analytical engine is powered by a Python-based bioinformatics workflow that supports a comprehensive suite of statistical methods tailored for clinical–genomic analysis. Categorical variables are evaluated using either chi-square or Fisher’s exact tests, with odds ratios and corresponding 95% confidence intervals calculated to quantify associations. For survival-related outcomes, the system implements Kaplan–Meier estimations and log-rank tests to compare groups, while multivariable Cox proportional hazards regression is available to adjust for confounding variables in time-to-event analyses. The platform also includes specialized modules for examining TGF-β pathway enrichment, identifying co-mutation patterns, and performing stratified survival analysis. Users can conduct subgroup comparisons across a variety of dimensions, including age groups (<50 vs. ≥50 years), racial and ethnic backgrounds (e.g., H/L vs. NHW), tumor sample type (primary vs. metastatic), and MSI status.
The selection of statistical models integrated into AI-HOPE-TGFbeta was guided by standard practices in clinical–genomic research and the specific analytical needs of CRC studies. Chi-square and Fisher’s exact tests were employed for categorical variable comparisons due to their robustness in evaluating associations across stratified groups, particularly with small or imbalanced sample sizes. Odds ratios with 95% confidence intervals were used to quantify the strength of associations between genomic alterations and clinical characteristics. Kaplan–Meier survival analysis and log-rank tests were selected for time-to-event outcomes, given their widespread application and interpretability in oncology. To account for potential confounding variables, multivariable Cox proportional hazards regression was incorporated for more complex survival modeling. These models were chosen for their proven reliability, reproducibility, and suitability for the types of queries AI-HOPE-TGFbeta is designed to support—including population-level comparisons, mutation enrichment analyses, and treatment-specific outcome evaluations.
2.5. Platform Design and Validation Strategy
AI-HOPE-TGFbeta was engineered with a strong emphasis on analytical rigor and reproducibility. At its core, the platform integrates a retrieval-augmented generation (RAG) mechanism that continuously references a structured biomedical knowledge base to enhance the contextual accuracy of its outputs and reduce the likelihood of AI-generated errors or hallucinations. The system applies schema-guided prompting to standardize how queries are interpreted and how results are formatted, ensuring consistency across diverse analyses. To assess the reliability of the platform, we validated its performance by successfully reproducing known clinical–genomic relationships from prior studies involving SMAD4, BMPR1A, and TGFBR2 in EOCRC [
6,
7], including stage-specific survival outcomes and mutation frequency disparities across patient populations.
2.6. Usability Evaluation and Comparative Benchmarking
To assess the usability and performance of AI-HOPE-TGFbeta, we conducted a comparative analysis against established platforms, including cBioPortal and UCSC Xena. Evaluation criteria focused on speed of task execution, consistency of analytical output, and flexibility in constructing complex, stratified cohort queries. Benchmark tasks included identifying EOCRC patients with SMAD4 mutations stratified by treatment status, generating Kaplan–Meier survival curves based on MSI subtypes, and analyzing TGFBR1 mutation frequency across racial and ethnic groups. Across all scenarios, AI-HOPE-TGFbeta outperformed existing tools in both response time and user interaction efficiency—particularly in handling intersectional analyses that required integration of clinical, genomic, and demographic filters.
2.7. Visualization Capabilities and Exportable Results
Upon completion of each analysis, AI-HOPE-TGFbeta produces a suite of high-quality visual and tabular outputs designed for immediate interpretation and downstream application. These include Kaplan–Meier survival curves, forest plots, mutation heatmaps, and summary data tables—each rendered using backend libraries such as Matplotlib 3 and Plotly 4 to ensure visual clarity and consistency. In addition to graphical elements, the system generates narrative summaries that interpret statistical findings within the context of existing TGF-β pathway literature. All outputs can be downloaded in formats suitable for publication, presentation, or integration into clinical decision-making workflows.
3. Results
By converting user-generated natural language inputs into fully automated clinical–genomic workflows, AI-HOPE-TGFbeta enables the on-demand analysis of TGF-β signaling disruptions in CRC. Its interactive conversational design allows users to define custom cohorts based on variables such as age, tumor stage, MSI classification, mutational status, treatment exposure, and racial or ethnic background. The system then performs statistical evaluations—generating Kaplan–Meier survival curves, odds ratio calculations, and corresponding visual outputs—with no additional coding required. In both validation and discovery-focused queries, AI-HOPE-TGFbeta consistently reproduced the established associations and revealed new insights related to EOCRC, treatment efficacy, and pathway-specific biomarker patterns.
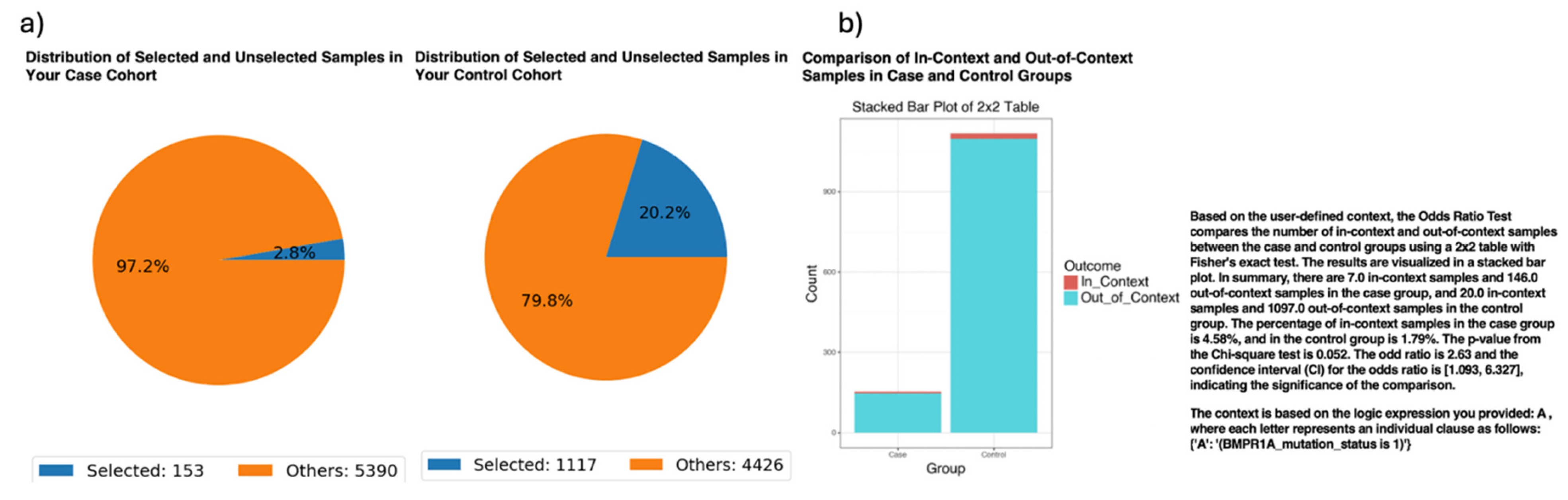
While validating the ancestry-stratified analyses, AI-HOPE-TGFbeta identified a potential disparity in the frequency of BMPR1A mutations among EOCRC patients across ethnic groups (
Figure 2). Specifically, the platform revealed that 4.58% of EOCRC H/L patients harbored BMPR1A mutations, compared to 1.79% of EOCRC NHW patients. This difference translated to an odds ratio of 2.63 (95% CI: [1.093, 6.327];
p = 0.052), suggesting that BMPR1A mutations were more than twice as likely to occur in the H/L EOCRC population. Although the association narrowly missed conventional thresholds for statistical significance, these findings underscore the potential of AI-HOPE-TGFbeta to uncover emerging ancestry-linked molecular patterns that may otherwise be overlooked. Importantly, this analysis highlights the need for a broader inclusion of racially and ethnically diverse populations in genomic studies to validate and extend these observations.
In ancestry-specific survival analyses, AI-HOPE-TGFbeta evaluated the prognostic impact of TGF-β pathway alterations in EOCRC among H/L patients (
Figure S1). The platform stratified EOCRC HL cases by mutation status in key TGF-β signaling genes, including SMAD4, TGFBR2, and BMPR1A. The case cohort consisted of 48 patients with TGF-β pathway alterations (0.9% of the dataset), while the control cohort included 105 patients without such mutations (1.9%). Kaplan–Meier survival analysis revealed no statistically significant difference in the overall survival between the two groups (
p = 0.8631), suggesting that TGF-β pathway mutations may not independently influence the prognosis in EOCRC HL under current sample sizes. Despite the lack of significance, this result underscores the value of AI-HOPE-TGFbeta in enabling fine-grained subgroup analyses and generating hypotheses about context-dependent effects. The findings also highlight the need for larger, ancestry-specific datasets to more definitively assess the clinical relevance of TGF-β signaling alterations in diverse EOCRC populations.
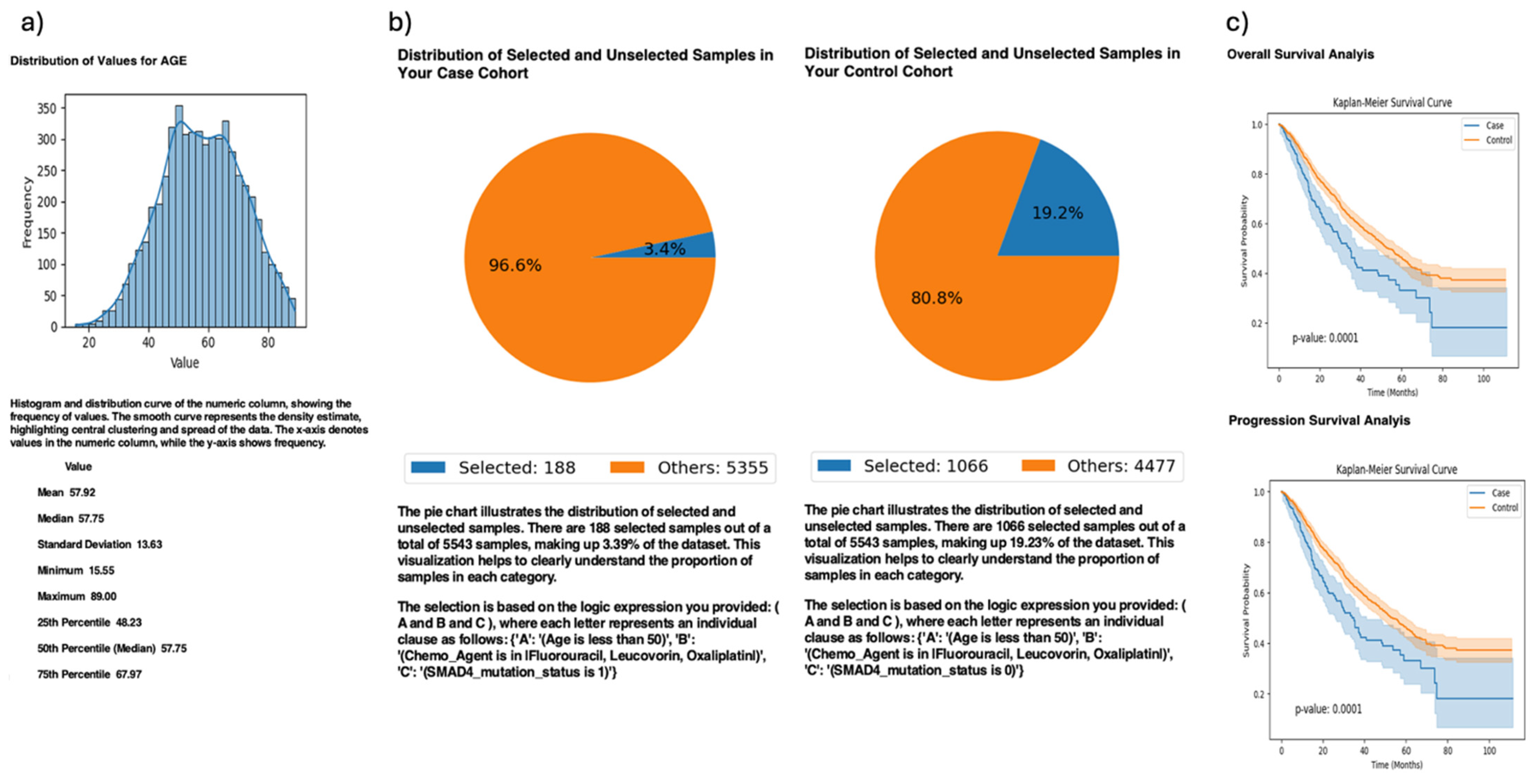
In exploratory analyses, AI-HOPE-TGFbeta reproduced a key finding from the published TGF-β literature regarding the prognostic relevance of SMAD4 mutations in EOCRC patients treated with FOLFOX chemotherapy (
Figure 3). Using a natural language query, the system stratified EOCRC patients (<50 years old) by SMAD4 mutation status and assessed treatment outcomes following FOLFOX (fluorouracil, leucovorin, and oxaliplatin) administration. The case cohort included 188 SMAD4-mutated patients (3.4% of the dataset), while the control cohort included 1066 SMAD4 wild-type patients (19.2%). Kaplan–Meier survival analyses revealed that SMAD4-mutated patients exhibited significantly worse overall and progression-free survival compared to wild-type cases (
p = 0.0001 for both), consistent with prior reports linking SMAD4 loss to chemoresistance and aggressive tumor biology in EOCRC. These results highlight the ability of AI-HOPE-TGFbeta to recapitulate known genotype–treatment–outcome relationships and underscore the clinical importance of SMAD4 as a biomarker for poor prognosis in young CRC patients undergoing standard chemotherapy.
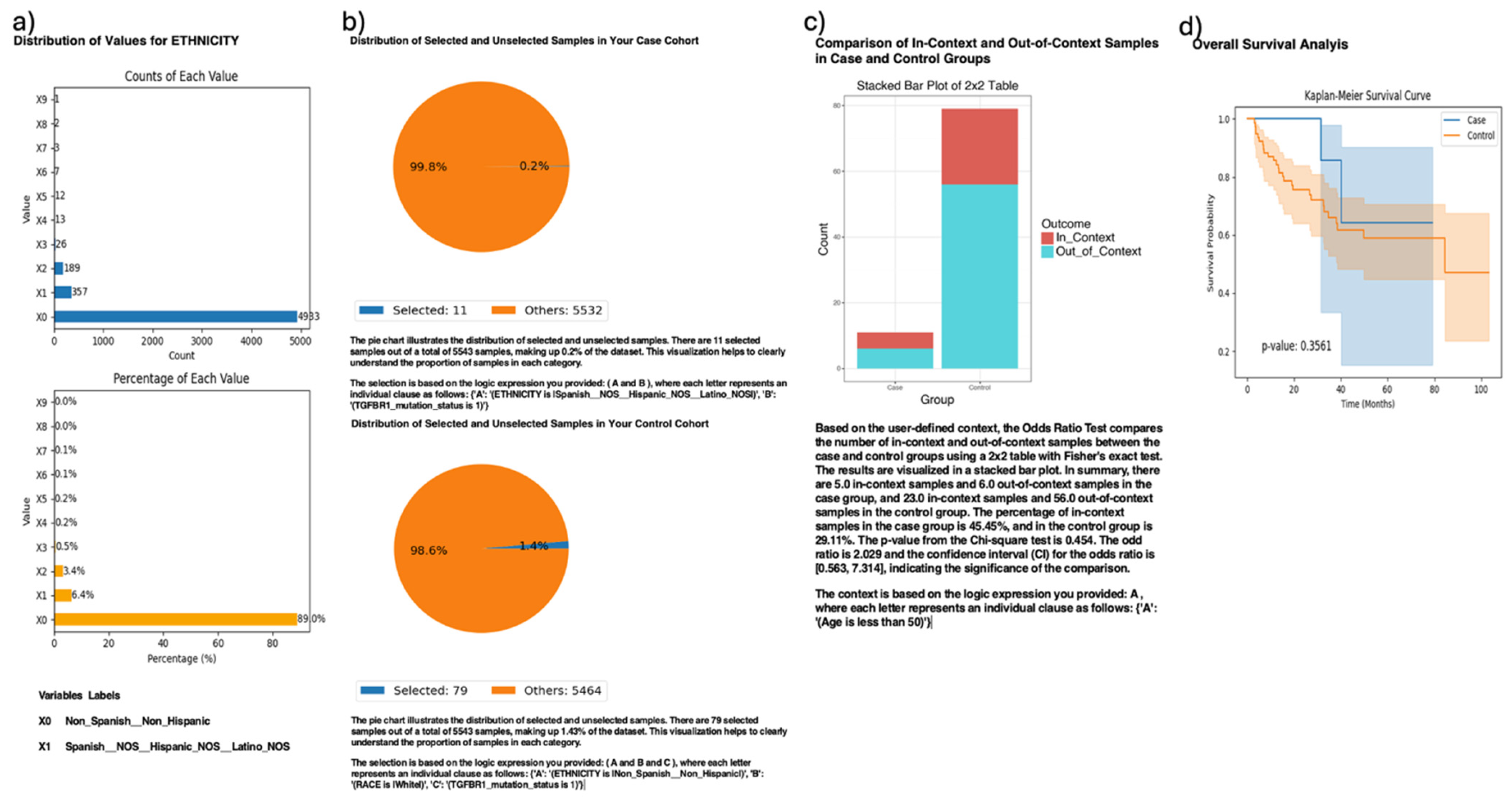
AI-HOPE-TGFbeta also enabled exploratory ethnicity-specific analysis of TGFBR1 mutation patterns in CRC patients (
Figure 4). This analysis compared TGFBR1-mutated H/L patients (case cohort,
n = 11) to TGFBR1-mutated NHW patients (control cohort, n = 79), highlighting the platform’s capacity to support disaggregated investigations despite sample imbalances. Although H/L individuals accounted for only 6.4% of the full dataset, AI-HOPE-TGFbeta successfully isolated a sufficient number of cases to perform comparative statistical analyses. Odds ratio testing, stratified by early-onset age (<50 years), yielded a value of 1.029 (95% CI: [0.563, 7.134],
p = 0.454), suggesting no significant difference in the mutation enrichment context across ethnic groups. The Kaplan–Meier survival curves similarly showed no statistically significant difference in overall survival (
p = 0.3561), despite apparent visual divergence between curves. These findings reinforce the underrepresentation of H/L populations in the existing genomic datasets and underscore the potential of AI-HOPE-TGFbeta to enable focused population-level queries that can guide future efforts toward equity-driven precision oncology.
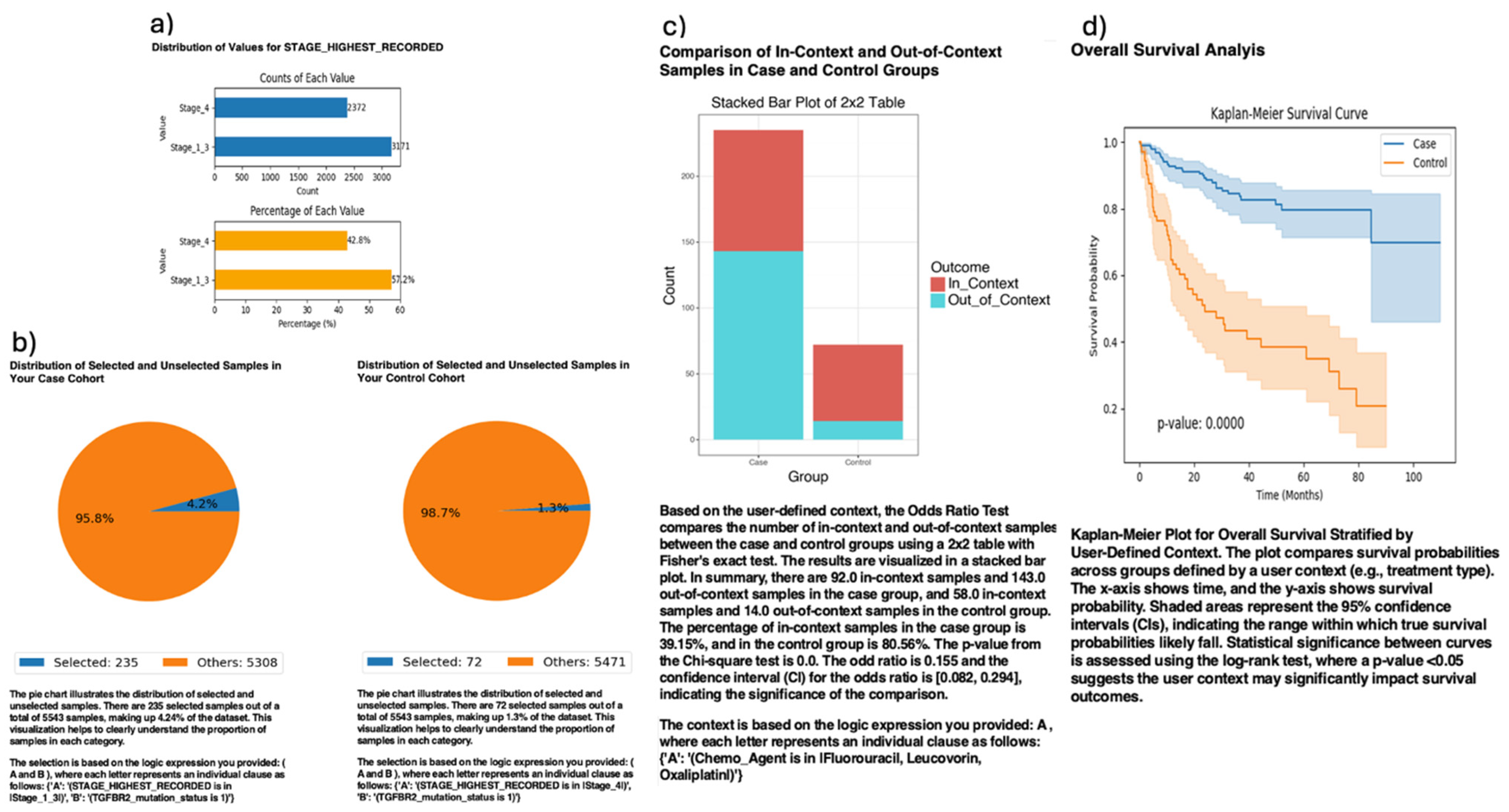
Further analysis using AI-HOPE-TGFbeta revealed clinically significant associations between TGFBR2 mutation status and tumor stage in CRC patients (
Figure 5). The platform stratified TGFBR2-mutated cases into early-stage (Stages I–III) and late-stage (Stage IV) cohorts to assess the prognostic impact of disease stage in the context of TGF-β pathway disruption. Among the 307 TGFBR2-mutated patients analyzed, those with early-stage disease (n = 235) exhibited markedly improved overall survival compared to their late-stage counterparts (n = 72), with Kaplan–Meier analysis yielding a highly significant
p-value (
p = 0.0000). Additionally, a 2×2 odds ratio analysis evaluating FOLFOX chemotherapy exposure revealed that early-stage patients were significantly more likely to have received standard treatment (OR = 0.155, 95% CI: [0.082, 0.294],
p = 0.000), suggesting potential treatment-related differences contributing to improved outcomes. These findings underscore the prognostic relevance of tumor stage among TGFBR2-mutated CRC patients and highlight AI-HOPE-TGFbeta’s capacity to integrate clinical and genomic variables for nuanced outcome stratification, supporting its use in guiding precision medicine strategies.
AI-HOPE-TGFbeta was used to assess the prognostic significance of MSI status among the SMAD4-mutated CRC patients (
Figure S2). In this analysis, patients were stratified by MSI phenotype, comparing those with MSI-high (Instable) tumors to those with MSI-stable counterparts. The case cohort included 78 SMAD4-mutated patients with MSI-Instable tumors (1.4% of the dataset), while the control cohort comprised 710 SMAD4-mutated patients with MSI-Stable tumors (12.8%). The Kaplan–Meier survival analysis revealed that MSI-Instable patients had significantly better overall survival than MSI-Stable patients (
p = 0.00001), with clearly divergent survival curves and non-overlapping 95% confidence intervals. This finding suggests a potential protective interaction between MSI-associated immunogenicity and SMAD4 pathway disruption, supporting the clinical relevance of combining genomic and molecular features in CRC prognosis. Moreover, the result highlights the utility of AI-HOPE-TGFbeta in uncovering context-dependent biomarker interactions that may inform immunotherapy stratification and precision treatment strategies.
Finally, AI-HOPE-TGFbeta was employed to evaluate the prognostic relevance of tumor sample type in CRC patients harboring SMAD2 mutations (
Figure S3). The platform stratified patients by tumor origin—primary versus metastatic—within the SMAD2-mutant cohort to assess survival differences across disease progression stages. The case cohort included 209 patients with SMAD2-mutant primary tumors (3.8% of the dataset), while the control cohort consisted of 48 patients with SMAD2-mutant metastatic tumors (0.9%). The Kaplan–Meier survival analysis revealed significantly better overall survival in patients with primary tumors compared to those with metastatic lesions (
p = 0.0010), with a clear separation of survival curves and non-overlapping 95% confidence intervals. This result supports prior evidence linking TGF-β signaling dysregulation to metastatic progression and underscores the clinical importance of tumor origin in prognostic modeling. Notably, this analysis highlights the strength of AI-HOPE-TGFbeta in dissecting context-specific molecular subgroups and advancing precision oncology through AI-enabled stratification.
Together, these findings demonstrate the versatility and analytical power of AI-HOPE-TGFbeta in uncovering both validated and novel insights into TGF-β pathway alterations across CRC subtypes and populations. By translating natural language prompts into executable clinical–genomic workflows, the platform enabled real-time, interpretable analyses incorporating key variables such as age, tumor stage, MSI status, mutation profiles, treatment exposure, and race/ethnicity. AI-HOPE-TGFbeta consistently recapitulated known prognostic associations—such as SMAD4-driven chemoresistance and stage-specific TGFBR2 outcomes—while also identifying emerging patterns, including ancestry-linked BMPR1A disparities and context-dependent survival modifiers, like MSI status and tumor origin. These results highlight the potential of conversational AI to democratize integrative bioinformatics, support equity-driven investigations, and accelerate precision medicine through scalable, dynamic cohort interrogation and hypothesis generation.
4. Discussion
AI-HOPE-TGFbeta represents a paradigm shift in precision oncology, offering a novel conversational AI platform that enables real-time, natural language-driven interrogation of TGF-β signaling dysregulation in CRC. By translating user-defined prompts into rigorous, reproducible analyses that integrate genomic and clinical variables, the system addresses longstanding limitations in accessibility, usability, and stratified data exploration. Unlike conventional bioinformatics platforms that often require complex scripting or multi-step workflows, AI-HOPE-TGFbeta streamlines the analytical process, allowing researchers and clinicians—even those without programming expertise—to formulate and execute pathway-centric, population-specific hypotheses with minimal friction.
The TGF-β signaling pathway is a central regulator of CRC progression, influencing processes such as epithelial-to-mesenchymal transition (EMT), immune evasion, and metastasis. Mutations in the TGF-β pathway genes—such as SMAD4, TGFBR2, and BMPR1A—are well-documented markers of poor prognosis and therapeutic resistance, particularly in EOCRC, which is rising at alarming rates in young adults and underserved populations. Despite this clinical importance, integrative analysis of TGF-β alterations has been hindered by the fragmentation of clinical–genomic data, underrepresentation of diverse populations, and the technical inaccessibility of traditional analysis pipelines. AI-HOPE-TGFbeta was developed to close these gaps, empowering users to interrogate TGF-β dysregulation across molecular and demographic contexts with unprecedented ease.
A core strength of AI-HOPE-TGFbeta lies in its ability to validate known associations while surfacing novel insights. In this study, the platform successfully recapitulated key findings from the TGF-β literature. These included the significantly worse overall and progression-free survival observed in SMAD4-mutated EOCRC patients treated with FOLFOX chemotherapy and the markedly better outcomes among the early-stage TGFBR2-mutated patients compared to their late-stage counterparts. These results not only confirmed the accuracy of AI-HOPE-TGFbeta’s analytic engine but also highlighted its potential for reinforcing known clinical–genomic relationships in treatment stratification and prognosis modeling.
AI-HOPE-TGFbeta also enabled hypothesis-driven, population-disaggregated analyses that are critical for advancing health equity in cancer genomics. Through natural language queries, the platform identified a potential disparity in the frequency of BMPR1A mutations among H/L EOCRC patients relative to their NHW counterparts—an observation that approached statistical significance and may reflect unique molecular etiologies in underrepresented groups. Similarly, the system allowed for survival analysis of TGFBR1-mutated CRC patients by ethnicity, demonstrating the platform’s flexibility in handling small cohort comparisons and highlighting the persistent underrepresentation of minority populations in genomic datasets. These findings underscore the urgent need for inclusive datasets and the value of AI systems like AI-HOPE-TGFbeta that support such investigations despite cohort-size limitations.
Beyond ancestry and ethnicity, the platform uncovered clinically actionable interactions between the TGF-β pathway genes and molecular or histopathological features. For example, among SMAD4-mutated tumors, those with MSI-high status exhibited significantly better survival than MSI-stable counterparts. This suggests a potentially protective immunologic interaction between MSI and TGF-β pathway disruption—an observation that may have implications for immunotherapy response prediction in CRC. Likewise, AI-HOPE-TGFbeta identified a strong prognostic benefit for patients with SMAD2-mutant primary tumors compared to those with metastatic lesions, reinforcing the relevance of tumor origin in outcome prediction and supporting the growing recognition of spatial tumor context in clinical decision-making.
From a technical perspective, AI-HOPE-TGFbeta offers a uniquely powerful platform built on LLMs, a RAG engine, and harmonized clinical–genomic data. The system integrates structured biomedical ontologies to ensure accurate cohort definitions and interpretable outputs. Benchmarking results revealed that AI-HOPE-TGFbeta outperforms widely used tools such as cBioPortal and UCSC Xena in execution speed, subgroup flexibility, and multidimensional filtering—particularly for complex queries involving intersectional factors like age, race/ethnicity, MSI subtype, tumor stage, and treatment exposure. These performance advantages position the platform as a scalable, next-generation solution for precision oncology.
Looking ahead, future iterations of AI-HOPE-TGFbeta will incorporate additional statistical metrics to further refine its survival analysis capabilities. While the current version focuses on Kaplan–Meier and log-rank methods—consistent with prior TGF-β pathway studies—we recognize the value of including complementary metrics, such as the concordance index. This measure offers a more nuanced evaluation of predictive discrimination in time-to-event models and could enhance the platform’s utility in personalized prognosis estimation. Expanding the platform to support such metrics will allow users to generate more comprehensive survival insights and better evaluate the clinical relevance of molecular alterations across diverse CRC populations.
While AI-HOPE-TGFbeta represents a significant advancement in conversational AI-driven clinical–genomic analysis, several limitations should be acknowledged. First, the platform is currently limited to publicly available datasets, such as those from cBioPortal, which may not fully capture the diversity or granularity of patient populations, especially among underrepresented groups. Second, although the platform supports a wide range of analytical outputs, its current statistical capabilities do not yet include advanced machine learning models or multivariate feature selection techniques. Third, while natural language interfaces lower the barrier for non-programming users, the platform may require refinement to handle ambiguous or overly complex queries with optimal accuracy. Lastly, the generalizability of AI-HOPE-TGFbeta beyond the TGF-β pathway or colorectal cancer has not yet been formally evaluated. Future versions will aim to incorporate additional data modalities, broader pathway support, and enhanced analytical flexibility to address these limitations.
In evaluating the interpretability and communication capabilities of AI-HOPE-TGFbeta, we compared its outputs to human-generated content previously analyzed and curated in two peer-reviewed studies on colorectal cancer [
6,
7]. These prior publications provided a foundation for assessing the AI agent’s ability to generate clinically relevant and context-aware responses. However, it is important to note that this content was derived from public databases, such as cBioPortal [
21], whose primary objective is to simplify and enhance the accessibility of complex cancer genomic data for cancer biologists and clinicians. AI-HOPE-TGFbeta is intended to serve as a research-oriented AI tool that supports precision oncology investigations focused on TGF-β pathway signaling in colorectal cancer and is not a substitute for authoritative medical advice. We strongly recommend that non-professional users consult licensed healthcare providers for any clinical or treatment-related decisions. This limitation has been explicitly acknowledged to ensure transparency regarding the context of AI–human comparisons and to emphasize the importance of professional oversight when applying AI tools in healthcare settings.
An important consideration in this study is the lack of control over input complexity, response length, and answer structure across the evaluated queries. Because AI-HOPE-TGFbeta generates natural language outputs in real time based on user-defined questions, the heterogeneity in phrasing and depth of prompts may influence the comprehensiveness and formatting of the responses. This variation could introduce unintended bias in interpretability or perceived quality, particularly when outputs are assessed subjectively or in comparison with human-generated responses. While this flexibility reflects real-world user interaction with conversational AI systems, we acknowledge it as a limitation in standardized performance evaluation. Future versions of the platform will incorporate more structured benchmarking protocols to ensure consistent comparisons across different question types and complexity levels.
An acknowledged constraint of this study is that the current version of AI-HOPE-TGFbeta relies primarily on univariate statistical methods—such as Kaplan–Meier survival analysis and odds ratio testing—without formal adjustment for potential confounding variables. As a result, some associations identified may be influenced by underlying covariates that were not accounted for, potentially limiting the robustness and generalizability of the findings. While this approach aligns with the platform’s initial focus on demonstrating core functionality and query interpretability, future versions of AI-HOPE-TGFbeta will incorporate multivariate regression models to support confounder-adjusted analyses and enhance the statistical rigor of clinical–genomic interpretations.
While this study focused primarily on validating the technical and analytical capabilities of AI-HOPE-TGFbeta, it is important to recognize that key dimensions of clinical communication—such as patient understanding, emotional tone, and ethical risk—were not formally evaluated. These factors are critical for ensuring that AI-generated outputs support effective, compassionate, and responsible communication, particularly in health contexts involving sensitive information or vulnerable populations. AI-HOPE-TGFbeta is intended strictly for research use only and is not designed to provide clinical guidance or patient-facing communication. As AI systems increasingly interface with researchers, patients, and clinicians, future work should incorporate structured assessments of how these tools impact comprehension, emotional response, and ethical acceptability.
As this study represents an initial demonstration of the AI-HOPE-TGFbeta platform, the number of showcased queries is intentionally limited and exploratory in nature. While the system was tested with a broader set of inputs during development, we selected a focused subset of seven representative queries that best illustrate the platform’s core functionalities—particularly in stratified survival analysis, mutation enrichment, and population-specific genomic insights related to the TGF-β pathway in colorectal cancer. These examples were chosen based on their alignment with previously published findings by our team and their ability to validate known clinical–genomic relationships, especially within early-onset colorectal cancer. We also acknowledge that the dataset used is relatively small, in part due to our focus on populations who are significantly underrepresented in current genomic databases. This limitation has been noted to ensure transparency and contextualize the scope of the current evaluation. Future benchmarking efforts will incorporate larger and more systematically designed query sets to further assess the scalability, robustness, and generalizability of the AI-HOPE-TGFbeta platform.
While AI-HOPE-TGFbeta successfully replicated known clinical–genomic associations involving SMAD4, TGFBR2, and BMPR1A—validated through prior peer-reviewed studies from our group—the current demonstration is based on a relatively small and exploratory cohort. This limitation is partially attributable to the underrepresentation of minority populations, such as Hispanic/Latino patients, in public genomic datasets. We acknowledge that broader clinical validation using larger, more diverse, and independent cohorts is essential to further establish the clinical significance and generalizability of our findings. Future work will focus on extending the platform’s reach by integrating additional datasets and validating outputs across external cohorts to enhance reproducibility, equity, and translational impact in precision oncology.
The observed enrichment of BMPR1A mutations in Hispanic/Latino patients with EOCRC underscores the potential of AI-HOPE-TGFbeta to reveal population-specific genomic patterns. However, we recognize that these conclusions are based on a relatively small sample size, which limits the statistical power and generalizability of the findings. This reflects a broader challenge in precision oncology: the persistent underrepresentation of racial and ethnic minority groups in genomic databases. To strengthen the validity of these insights, future research will prioritize the inclusion of larger, ancestrally diverse cohorts and multi-institutional datasets. Such efforts are critical for ensuring that AI-driven platforms like AI-HOPE-TGFbeta contribute meaningfully to equitable and generalizable precision medicine.
As AI-HOPE-TGFbeta enables population-specific genomic exploration, particularly in underserved groups such as Hispanic/Latino patients, it is critical to approach these analyses with ethical responsibility. The interpretation of genetic data from underrepresented populations must be conducted with caution to avoid reinforcing stereotypes, misrepresentation, or stigmatization. Moreover, while our platform utilizes de-identified, publicly available data (e.g., from cBioPortal), we acknowledge the ongoing responsibility to protect patient privacy and promote transparency in how subgroup findings are generated and communicated. Future versions of AI-HOPE-TGFbeta will incorporate ethical oversight frameworks and stakeholder input, especially from communities affected by disparities, to guide responsible data use and foster trust in AI-driven precision oncology.
While AI-HOPE-TGFbeta provides a suite of informative outputs—including survival curves, odds ratio plots, and mutation heatmaps—this platform is explicitly intended for research use only and not as a clinical decision support system. Its current functionality is designed to assist investigators in exploring pathway-specific alterations and generating population-aware hypotheses using harmonized genomic and clinical data. The translation of these outputs into actionable insights for clinical decision-making remains outside the current scope of the platform. However, future iterations of AI-HOPE-TGFbeta will incorporate modules aimed at evaluating clinical applicability more systematically, including integration with clinical guidelines, interpretability for care teams, and potential alignment with decision-support frameworks. We acknowledge this current boundary and have added it as a key consideration for future development.
The clinical implications of the prognostic biomarkers and subgroup patterns identified by AI-HOPE-TGFbeta—such as SMAD4 mutations, TGFBR2 alterations, and MSI status—are supported by prior peer-reviewed publications from our group, which investigated these associations in early-onset colorectal cancer using traditional statistical approaches. Notably, two studies previously published serve as empirical references that both informed and validated key query outputs presented in this manuscript. These prior studies enhance the interpretability and translational relevance of our AI-driven analyses. Nevertheless, we acknowledge that broader clinical validation, especially across diverse ethnic groups and larger cohorts, remains necessary. Future research will focus on systematically evaluating the predictive and therapeutic value of these findings to inform personalized treatment strategies and support equitable precision oncology regarding the TGF-β pathway in CRC.
It is also important to note that this study was conducted entirely within a research setting and does not reflect the operational complexities of real-world clinical environments. As such, the findings and applications of AI-HOPE-TGFbeta are not currently generalizable to routine clinical practice. The platform was developed as a research tool to support clinical–genomic exploration, translational oncology, and hypothesis generation. It has not been validated for direct clinical use or decision-making. Future work will be necessary to assess its performance, usability, and safety in clinical contexts, including integration with electronic health records, alignment with clinical workflows, and evaluation under regulatory standards.
Nevertheless, there are important areas for continued development. Expanding the platform to incorporate additional omics layers—including transcriptomics, proteomics, and spatial data—would enable deeper mechanistic insight into TGF-β-driven tumor biology. Integration with federated learning frameworks and secure, privacy-preserving data environments will be essential for real-world clinical deployment, especially in multi-institutional settings where patient-level data must remain decentralized. Further comparative evaluations against other AI-powered bioinformatics agents are needed to assess the generalizability of AI-HOPE-TGFbeta beyond CRC and the TGF-β pathway, paving the way for a modular suite of disease- and pathway-specific AI agents.
Finally, while AI-HOPE-TGFbeta was designed to lower the barriers to entry for researchers working at the intersection of genomics and health disparities, broader efforts to support training, community engagement, and interdisciplinary collaboration will be key to maximizing its impact. The platform is not a substitute for diverse data generation but rather a catalyst for extracting meaningful insights from the data that already exist—highlighting the need for ongoing investment in both algorithmic innovation and inclusive data science.