What We Know About the Role of Large Language Models for Medical Synthetic Dataset Generation
Abstract
1. Introduction
2. Methodology
2.1. Literature Search Strategy
| (“ChatGPT*” OR “Chat-GPT*” OR “genAI*” OR “llms*” OR “large language model*” OR “generative artificial intelligence*” OR “GAI” OR “GPT-based model*” OR “transformer model*”) AND (“synthetic data*” OR “synthetic text*” OR “text data*” OR “medical conversation*” OR “text dataset augmentation*” OR “clinical text*” OR “medical text*”) AND (“gener*” OR “creat*”) AND (“AI” OR “NLP” OR “natural language processing*”) AND (“biomed*” OR “medic*” OR “health*” OR “clinic*” OR “hospital*” OR “clinical documentation*” OR “clinical annotation frameworks*” OR “SOAP*” OR “Calgary-Cambridge”) AND (“ethics*” OR “bias*” OR “privacy*” OR “HIPAA*” OR “GDPR” OR “data compliance*” OR “AI hallucination*”) |
| (“ChatGPT*” OR “Chat-GPT*” OR “genAI*” OR “llms*” OR “large language model*” OR “GPT-based model*” OR “generative artificial intelligence*” OR “GAI”) AND (“text data*” OR “text synthetic*” OR “text gener*” OR “synthetic data*” OR “clinical text*” OR “medical text*”) AND (“AI” OR “NLP” OR “natural language processing*”) AND (“biomed*” OR “medic*” OR “health*” OR “clinic*” OR “hospital*”) |
| (“ChatGPT*” OR “genAI*” OR “llms*” OR “large language model*” OR “generative artificial intelligence*” OR “GPT-based model*”) AND (“synthetic data*” OR “synthetic text*” OR “text data*” OR “synthetic datasets for medic*” OR “biomedical text generation*”) AND (“ethic*” OR “bias*” OR “privacy*” OR “HIPAA*” OR “GDPR” OR “data compliance*” OR “AI hallucination*” OR “responsible AI*”) AND (“AI” OR “NLP” OR “biomedical NLP*” OR “healthcare NLP*”) |
| (“ChatGPT*” OR “Chat-GPT*” OR “genAI*” OR “llms*” OR “large language model*” OR “generative artificial intelligence*” OR “GAI” OR “GPT-based model*” OR “Claude*” OR “Gemini*” OR “GatorTron*” OR “BioGPT*” OR “transformer model*”) AND (“synthetic data*” OR “synthetic text*” OR “text data*” OR “structured annotation datasets*” OR “clinical text*”) AND (“gener*” OR “creat*” OR “augment*”) AND (“AI” OR “NLP” OR “biomedical NLP*” OR “healthcare NLP*”) AND (“comparison*” OR “evaluation*” OR “benchmarks*” OR “framework*”) |
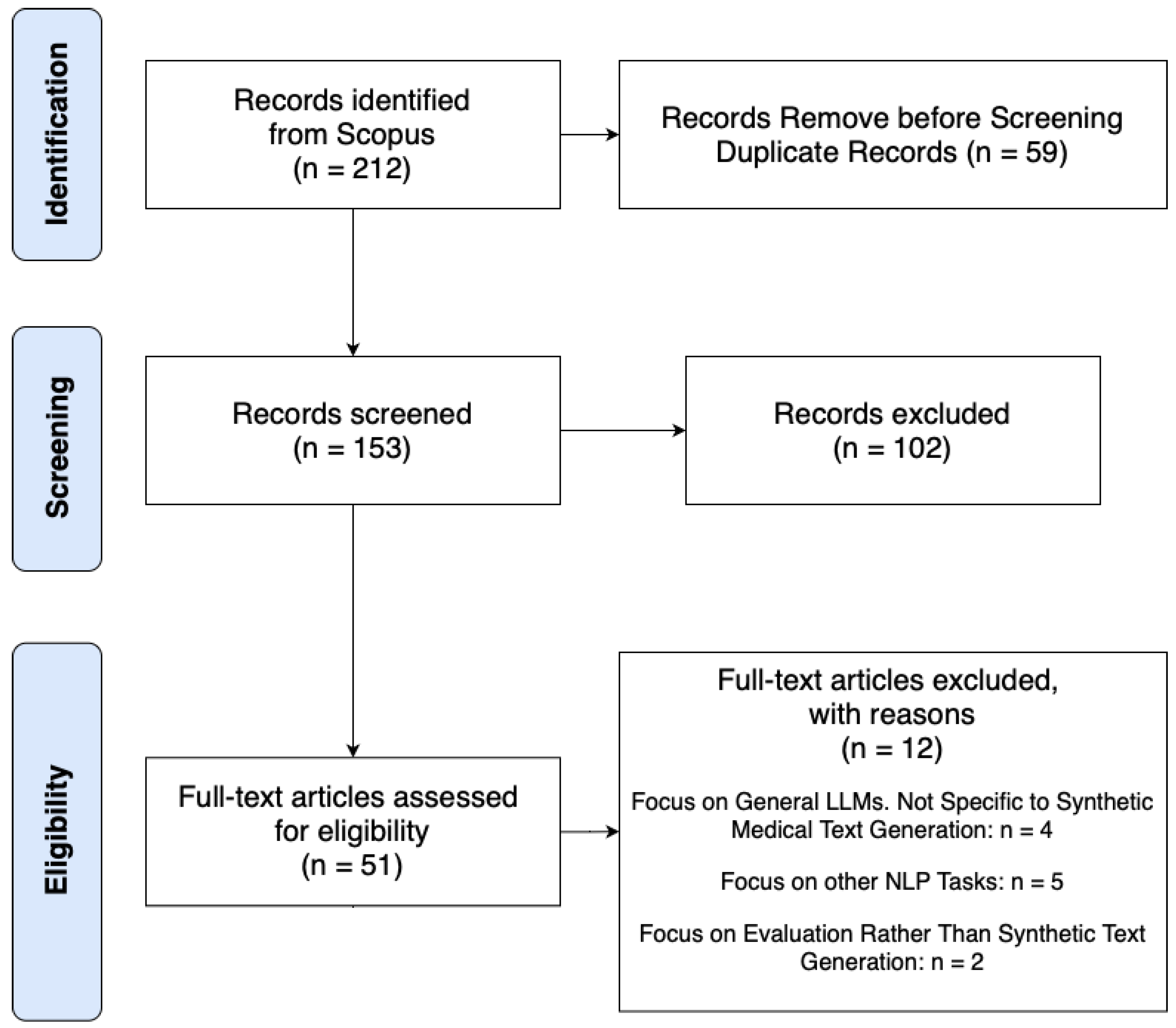
2.2. Screening Process and Data Extraction
3. Developments in Synthetic Medical Text Generation
3.1. Approaches to Synthetic Medical Text Generation
3.2. Benchmarking Synthetic Medical Text Generation
4. Discussion
4.1. Applications of LLMs in Clinical NLP
4.2. Limitations in Medical LLMs
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chlap, P.; Bagheri, A.P.; Owens, S.P.; Dagan, D.M.; Nguyen, Q.; Drummond, T. A survey of synthetic data generation methods for medical imaging. Comput. Med. Imaging Graph. 2021, 94, 101997. [Google Scholar] [CrossRef]
- Goncalves, A.; Ray, P.; Soper, B.; Stevens, J.; Coyle, L.; Sales, A.P. Generation and evaluation of synthetic patient data. BMC Med. Res. Methodol. 2020, 20, 108. [Google Scholar] [CrossRef]
- Ratner, A.; Bach, S.H.; Ehrenberg, H.; Fries, J.; Wu, S.; Ré, C. Snorkel: Rapid Training Data Creation with Weak Supervision. Proc. Vldb Endow. 2020, 13, 1971–1983. [Google Scholar] [CrossRef]
- Gilardi, F.; Kühner, M.; Grass, L. Large Language Models for Synthetic Dataset Generation: A Comparative Analysis. J. Artif. Intell. Res. 2023, 67, 145–162. [Google Scholar]
- Sun, Y.; Yang, L.; Tang, J. Evaluating LLM-Generated Synthetic Datasets for NLP: Challenges and Opportunities. Trans. Assoc. Comput. Linguist. 2023, 11, 202–219. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P. Language Models are Few-Shot Learners: Applications in Synthetic Dataset Generation. Adv. Neural Inf. Process. Syst. (Neurips) 2023, 34, 1877–1890. [Google Scholar] [CrossRef]
- Liu, Y.; Ju, S.; Wang, J. Exploring the Potential of ChatGPT in Medical Dialogue Summarization: A Study on Consistency with Human Preferences. BMC Med. Inform. Decis. Mak. 2024, 24, 75. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, H.; Liu, H.; Jiang, Y.; Ran, T.; Jin, X.; Xiao, X.; Lin, Z.; Chen, H.; Niu, Z. Benchmarking ChatGPT for Biomedical Text Generation. Bioinformatics 2023, 39, btad557. [Google Scholar] [CrossRef]
- Tian, Y.; Gan, R.; Song, Y.; Zhang, J.; Zhang, Y. CHIMED-GPT: A Chinese Medical Large Language Model with Full Training Regime and Better Alignment to Human Preferences. arXiv 2024, arXiv:2311.06025. [Google Scholar]
- Zhang, X.; Zhao, G.; Ren, Y.; Wang, W.; Cai, W.; Zhao, Y.; Liu, J. Data-Augmented Large Language Models for Medical Record Generation. Appl. Intell. 2025, 55, 88. [Google Scholar] [CrossRef]
- Zhu, Z.; Liu, S.; Zhang, R. Examining the Persuasive Effects of Health Communication in Short Videos: Systematic Review. J. Med. Internet Res. 2023, 25, e48508. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Madden, S.; Cafarella, M. DataSynth: Generating Synthetic Data Using Declarative Constraints. Proc. Vldb Endow. 2022, 13, 2071–2083. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Tang, J. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. J. Biomed. Inform. 2022, 127, 103994. [Google Scholar] [CrossRef]
- Weed, L. Medical Records, Medical Education, and Patient Care: The Problem-Oriented Record as a Basic Tool. J. Am. Med. Assoc. 2004, 292, 1066–1070. [Google Scholar]
- Silverman, J.; Kurtz, S.; Draper, J. The Calgary-Cambridge Guide to the Medical Interview: Communication for Clinical Practice. Med. Educ. 2008, 42, 673–679. [Google Scholar] [CrossRef]
- Montenegro, L.; Gomes, L.M.; Machado, J.M. AI-Based Medical Scribe to Support Clinical Consultations: A Proposed System Architecture. In EPIA Conference on Artificial Intelligence; Springer: Cham, Switzerland, 2023; pp. 1–12. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, T.P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Falagas, M.E.; Pitsouni, E.I.; Malietzis, G.A.; Pappas, G. Comparison of PubMed, Scopus, Web of Science, and Google Scholar: Strengths and weaknesses. FASEB J. 2008, 22, 338–342. [Google Scholar] [CrossRef]
- Goyal, M.; Mahmoud, Q. Synthetic Data Generation: Trends, Challenges, and Future Directions. Expert Syst. Appl. 2024, 229, 120462. [Google Scholar] [CrossRef]
- Ghebrehiwet, I.; Zaki, N.; Damseh, R.; Mohamad, M.S. Revolutionizing personalized medicine with generative AI: A systematic review. Artif. Intell. Rev. 2024, 57, 128. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, W.; Leroy, G.; Wang, S.; Cohen, T. Retrieval augmentation of large language models for lay language generation. J. Biomed. Inform. 2024, 149, 104580. [Google Scholar] [CrossRef]
- Xie, C.; Lin, Z.; Backurs, A.; Gopi, S.; Yu, D.; Inan, H.; Nori, H.; Jiang, H.; Zhang, H.; Lee, Y.T.; et al. Differentially Private Synthetic Data via Foundation Model APIs. In Proceedings of the 41st International Conference on Machine Learning (ICML), Vienna, Austria, 21–27 July 2024. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems (NeurIPS); Curran Associates, Inc.: San Francisco, CA, USA, 2017; Volume 30, pp. 5998–6008. [Google Scholar]
- Moser, D.; Bender, M.; Sariyar, M. Generating synthetic healthcare dialogues in emergency medicine using large language models. Stud. Health Technol. Inform. 2024, 321, 235–239. [Google Scholar] [PubMed]
- Schlegel, V.; Li, H.; Wu, Y.; Subramanian, A.; Nguyen, T.T.; Kashyap, A.R.; Beck, D.; Zeng, X.; Batista-Navarro, R.T.; Winkler, S.; et al. PULSAR at MEDIQA-Sum 2023: Large Language Models Augmented by Synthetic Dialogue Convert Patient Dialogues to Medical Records. arXiv 2023, arXiv:2307.02006. [Google Scholar]
- Almutairi, M.; Alghamdi, A.; Alkadi, S. Multi-Agent Large Language Models for Arabic Medical Dialogue Generation: A Culturally Adapted Approach. J. Artif. Intell. Healthc. 2024, 18, 112–134. [Google Scholar]
- Lund, J.A.; Burman, J.; Woldaregay, A.Z.; Jenssen, R.; Mikalsen, K. Instruction-Guided Deidentification with Synthetic Test Cases for Norwegian Clinical Text. In Proceedings of the 5th Northern Lights Deep Learning Conference (NLDL), Tromsø, Norway, 9–11 January 2024. [Google Scholar]
- Zecevic, A.; Haug, C.; Schenk, L.; Zaveri, K.; Heuss, L.T.; Raemy, E.; Tzovara, A.; Oezcan, I.M. Privacy-Preserving Synthetic Text Generation Using Differentially Private Fine-Tuning of BioGPT. Artif. Intell. Med. 2024, 150, 102649. [Google Scholar]
- Abdel-Khalek, S.; Algarni, A.D.; Amoudi, G.; Alkhalaf, S.; Alhomayani, F.M.; Kathiresan, S. Leveraging AI-Generated Content for Synthetic Electronic Health Record Generation with Deep Learning-Based Diagnosis Model. IEEE Trans. Consum. Electron. 2024. [CrossRef]
- Latif, S.; Kim, Y.B. Augmenting Clinical Text with Large Language Models: A Comparative Study on Synthetic Data Generation for Medical NLP. J. Biomed. Inform. 2024, 145, 104567. [Google Scholar]
- Frei, J.; Kramer, F. Annotated Dataset Creation through General Purpose Language Models for Non-English Medical NLP. arXiv 2023, arXiv:2308.14493. [Google Scholar]
- Li, R.; Zhang, H.; Zhang, J.; Wu, Y. LlamaCare: Instruction-Tuned Large Language Models for Clinical Text Generation and Prediction. arXiv 2024, arXiv:2401.12345. [Google Scholar]
- Tian, S.; Jin, Q.; Yeganova, L.; Lai, P.T.; Zhu, Q.; Chen, X.; Yang, Y.; Chen, Q.; Kim, W.; Comeau, D.C.; et al. Opportunities and Challenges for ChatGPT and Large Language Models in Biomedicine and Health. Briefings Bioinform. 2024, 25, bbad493. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Mao, K.; Zhang, Y.; Chen, J. CALLM: Enhancing Clinical Interview Analysis Through Data Augmentation with Large Language Models. IEEE J. Biomed. Health Inform. 2024, 28, 7531–7542. [Google Scholar] [CrossRef]
- Wang, N.; Lee, H.; Patel, R.; Johnson, M. Taxonomy-Based Prompt Engineering for Synthetic Patient Portal Messages. J. Biomed. Inform. 2024, 160, 104752. [Google Scholar] [CrossRef] [PubMed]
- Ghanadian, H.; Nejadgholi, I.; Al Osman, H. Socially Aware Synthetic Data Generation for Suicidal Ideation Detection Using Large Language Models. IEEE Access 2024, 12, 3358206. [Google Scholar] [CrossRef]
- Serbetçi, G.; Leser, U. Challenges and Solutions in Multilingual Clinical Data Analysis. J. Glob. Health Inform. 2023, 15, 210–222. [Google Scholar]
- Xu, R.; Cui, H.; Yu, Y.; Kan, X.; Shi, W.; Zhuang, Y.; Wang, M.D.; Jin, W.; Ho, J.C.; Yang, C. Knowledge-Infused Prompting: Assessing and Advancing Clinical Text Data Generation with Large Language Models. In Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing (EMNLP), Miami, FL, USA, 12–16 November 2024; pp. 321–339. [Google Scholar]
- Yu, Y.; Zhuang, Y.; Zhang, J.; Meng, Y.; Ratner, A.; Krishna, R.; Shen, J.; Zhang, C. Large Language Model as Attributed Training Data Generator: A Tale of Diversity and Bias. Adv. Neural Inf. Process. Syst. 2023, 36, 55734–55784. [Google Scholar]
- Zafar, A.; Sahoo, S.K.; Bhardawaj, H.; Das, A.; Ekbal, A. KI-MAG: A Knowledge-Infused Abstractive Question Answering System in the Medical Domain. Neurocomputing 2024, 571, 127141. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, K.; Huang, Y.; Wang, L. Performance Evaluation of LLMs in Biomedical Text Generation. J. Med. Inform. 2024, 48, 102–118. [Google Scholar]
| Study | Model Used | Application | Optimization | Evaluation Metrics |
|---|---|---|---|---|
| Moser et al. (2024) [24] | Zephyr-7b-beta, GPT-4 Turbo | Emergency medicine dialogues | Multi-stage pipeline | Accuracy (94% → 87%) |
| Latif et al. (2024) [30] | ChatGPT, BART, T5 | Clinical text augmentation | Zero-shot prompting, LLM-based rephrasing | ROUGE-1, ROUGE-2, ROUGE-L |
| Frei et al.(2023) [31] | GPT-NeoX (20B) | Synthetic NER- annotated text | Few-shot prompting, XML-based entity tagging | F1-score |
| Abdel-Khalek et al. (2024) [29] | ChatGPT (SEHRG-DLD) | Synthetic EHRs | Deep Belief Network (DBN), HHO, GJO | Classification accuracy (97%) |
| Li et al. (2024) [32] | LlamaCare (Llama 2-7B) | Discharge summary generation, clinical text classification | Instruction tuning, self-instruction, LoRA | ROUGE-L, BLEU-4, AUROC |
| Tian et al. (2024) [33] | ChiMed-GPT | Structured dialogue, NER, QA | Pre-training, RLHF, rejection sampling | Accuracy, BLEU-1, ROUGE-L |
| Schlegel et al. (2023) [25] | GPT-3.5 (PULSAR) | Medical summarization | Domain-specific fine-tuning | ROUGE, BERTScore |
| Chen et al. (2023) [8] | DialoGPT (DoPaCos) | Doctor–patient conversations | Pre-training on synthetic dialogues | ROUGE, BERTScore |
| Zhang et al. (2025) [10] | Qwen-7B-Chat | Data-to-text, summarization | Faithfulness Score for fine-tuning | D2T (+19.72%), MTS (+19.33%) |
| Wu et al. (2024) [34] | CALLM | PTSD transcript generation | T-A-A partitioning, Response–Reason prompting | bACC (77%), F1 (0.70), AUC (0.78) |
| Almutairi et al. (2024) [26] | Multi-Agent System | Arabic medical dialogues | Multi-agent refinement | BERT Score (0.834), human acceptability (90%) |
| Lund et al. (2024) [27] | GPT-4 | PHI de-identification | Automated deidentification | F1-score (0.983) |
| Zecevic et al. (2024) [28] | BioGPT | Privacy-preserving text generation | Differential Privacy (DP) | Textual similarity, privacy protection |
| Study | Benchmarking Method | Model(s) Evaluated | Evaluation Metric | Key Findings |
|---|---|---|---|---|
| Serbetçi et al. (2023) [37] | Generalization | GPTNERMED | Maximum Mean Discrepancy (MMD), Domain Adversarial Training Loss (DAT), Entity Consistency Score | Limited generalization to real-world datasets |
| Li et al. (2024) [32] | Instruction-Tuned LLM Evaluation | LlamaCare (Llama 2-7B) | ROUGE-L, BLEU-4, AUROC | Improved domain adaptation and coherence |
| Tian et al. (2024) [33] | Domain-Specific Benchmarking | ChiMed-GPT | Multi-choice QA Accuracy, NER F1-score, BLEU | Domain adaptation improved factual consistency |
| Guo et al. (2024) [21] | Retrieval-Augmented Evaluation | RALL (Wikipedia, UMLS retrieval) | Precision at Rank 1, Recall in Top 5, Mean Reciprocal Rank | Higher factual accuracy, reduced fluency |
| Latif et al. (2024) [30] | Augmentation Performance | ChatGPT, BART, T5 | ROUGE-1, ROUGE-2, ROUGE-L | BART-augmented datasets outperformed back-translation |
| Frei et al. (2023) [31] | NER Model Evaluation | GPT-NeoX (20B) | F1-score (GBERT, GottBERT, German-MedBERT) | Synthetic annotations improved NER performance |
| Zafar et al. (2024) [40] | Knowledge-Infused Benchmarking | KI-MAG | BLEU-1, BLEU-2, BLEU-3, BLEU-4 | Improved factual consistency in synthetic QA datasets |
| Zhang et al. (2025) [10] | Fine-Tuning and Faithfulness | Qwen-7B-Chat | Faithfulness Score, Contrastive Learning, Gradient-Based Adversarial Fine-Tuning | Reduced hallucination, improved factual consistency |
| Xu et al. (2024) [38] | Domain-Specific Faithfulness | CLINGEN | Faithfulness Score, Domain-Specific Accuracy | Hallucination reduction in clinical NLP |
| Chen et al. (2023) [8] | Realism Evaluation | GatorTronGPT | Physicians’ Turing Test | Synthetic text indistinguishable from real data |
| Lund et al. (2024) [27] | PHI De-Identification | GPT-4 | PHI Removal F1-score | High F1, limitations in complex PHI detection |
| Xie et al. (2024) [22] | Privacy-Preserving Evaluation | AUG-PE (API-based DP) | Privacy–Utility Trade-off | Improved privacy–utility balance |
| Challenge | Issue in LLM-Generated Synthetic Text | Solution via Structured Medical Frameworks |
|---|---|---|
| Faithfulness and Clinical Relevance | LLM-generated dialogues often lack structured clinical progression and deviate from real-world medical interactions. | Training LLMs with structured models (e.g., Calgary–Cambridge, SOAP) ensures logical, medical questioning and progression, improving clinical training and research dataset usability. |
| Hallucinations in Clinical Conversations | LLMs frequently introduce fabricated symptoms, test results, or diagnoses that were not present in the input data. Lack of structured constraints leads to unpredictable factual inconsistencies. | Embedding structured consultation formats constrains LLM outputs to follow expected medical interactions, reducing the risk of hallucinated symptoms and fabricated patient histories. |
| Dataset Generalization | Synthetic medical text lacks adaptability across clinical settings, specialties, and languages. Models trained on domain-specific, unstructured synthetic data struggle with real-world clinical tasks. | Structured LLM fine-tuning with consultation frameworks (SOAP, SBAR (Situation, Background, Assessment, and Recommendation), and Calgary–Cambridge) improves dataset standardization and enhances cross-domain generalization for multiple medical specialties. |
| Category | Applications of LLM-Generated Synthetic Text | Challenges |
|---|---|---|
| Synthetic Medical Dialogues | Generates structured doctor–patient conversations, enhancing fluency in clinical interactions. | Often introduces fabricated symptoms and medical details, reducing factual consistency. |
| Fine-tuned to support clinical summarization models. | Lacks structured medical reasoning, failing to follow consultation models like SOAP or Calgary–Cambridge. | |
| Synthetic EHR and Medical Reports | Generates structured EHRs incorporating clinical attributes (e.g., BMI, blood pressure) for training disease prediction models. | Lacks built-in privacy mechanisms, raising concerns about patient data protection. |
| Used in clinical decision support systems for data augmentation. | Alternative privacy-preserving models (e.g., AUG-PE) outperform LLMs in privacy–utility balance. | |
| Medical Summarization and Abstraction | Produces fluent medical summaries, improving documentation efficiency. | Performs worse than specialized models (e.g., BART) in factual accuracy (ROUGE-1: −14.94%, ROUGE-2: −53.48%). |
| Automates discharge summary and progress note generation. | Requires retrieval-augmented generation (RAG) and knowledge-infused prompting to improve factual consistency. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montenegro, L.; Gomes, L.M.; Machado, J.M. What We Know About the Role of Large Language Models for Medical Synthetic Dataset Generation. AI 2025, 6, 109. https://doi.org/10.3390/ai6060109
Montenegro L, Gomes LM, Machado JM. What We Know About the Role of Large Language Models for Medical Synthetic Dataset Generation. AI. 2025; 6(6):109. https://doi.org/10.3390/ai6060109
Chicago/Turabian StyleMontenegro, Larissa, Luis M. Gomes, and José M. Machado. 2025. "What We Know About the Role of Large Language Models for Medical Synthetic Dataset Generation" AI 6, no. 6: 109. https://doi.org/10.3390/ai6060109
APA StyleMontenegro, L., Gomes, L. M., & Machado, J. M. (2025). What We Know About the Role of Large Language Models for Medical Synthetic Dataset Generation. AI, 6(6), 109. https://doi.org/10.3390/ai6060109








