Effective Machine Learning Techniques for Non-English Radiology Report Classification: A Danish Case Study
Abstract
1. Introduction
- (1)
- Develop a string-matching algorithm for the automatic identification of positive and negative mentions of abnormalities in Danish chest X-ray reports.
- (2)
- Compare the performance of the above method against various BERT-based machine learning models, including models trained on multi-lingual datasets versus models fine-tuned on the target language.
- (3)
- Present an overview of the experimental results on the annotation effort needed to achieve the desired performance.
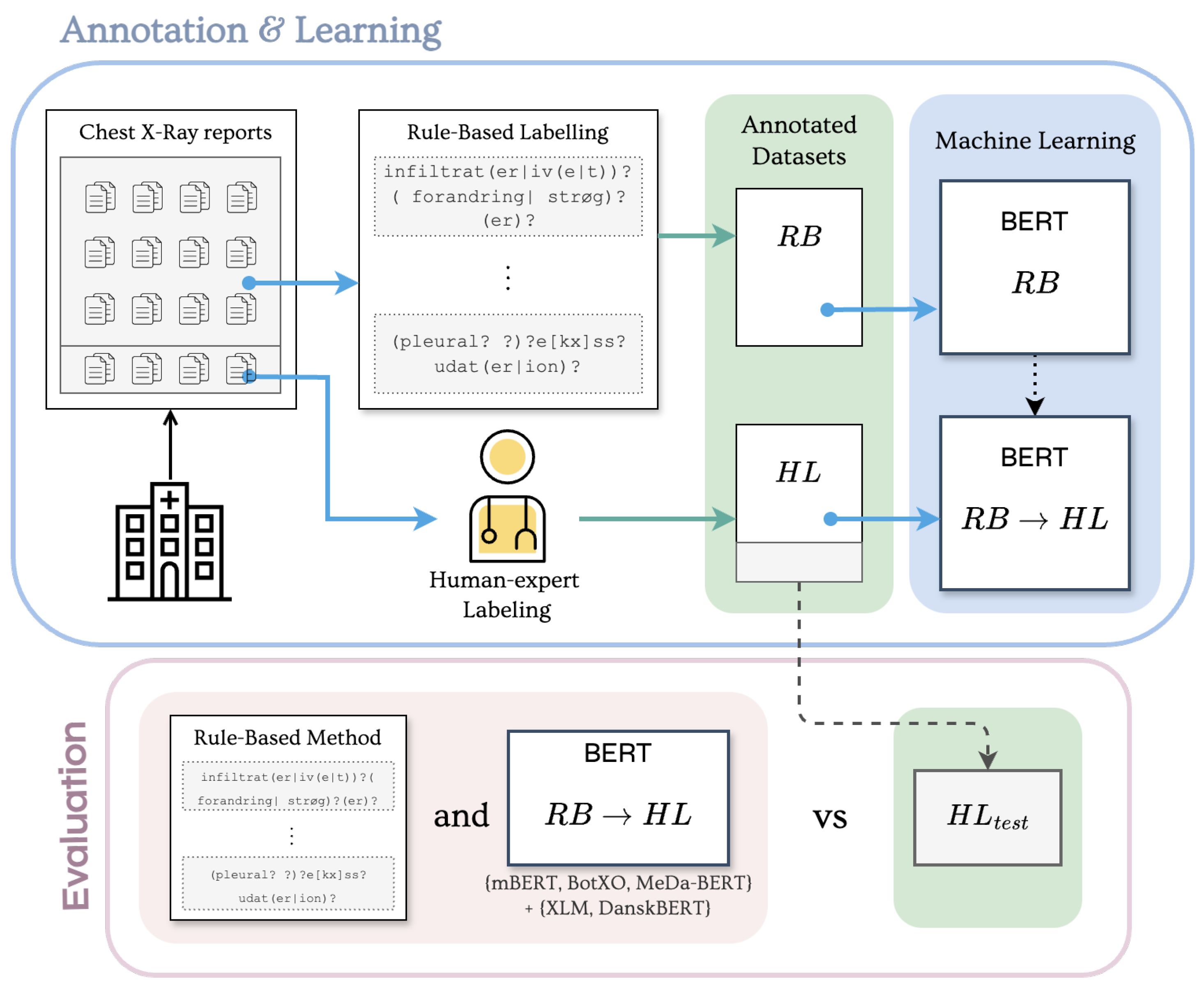
2. Materials and Methods
2.1. Materials: Data Collection
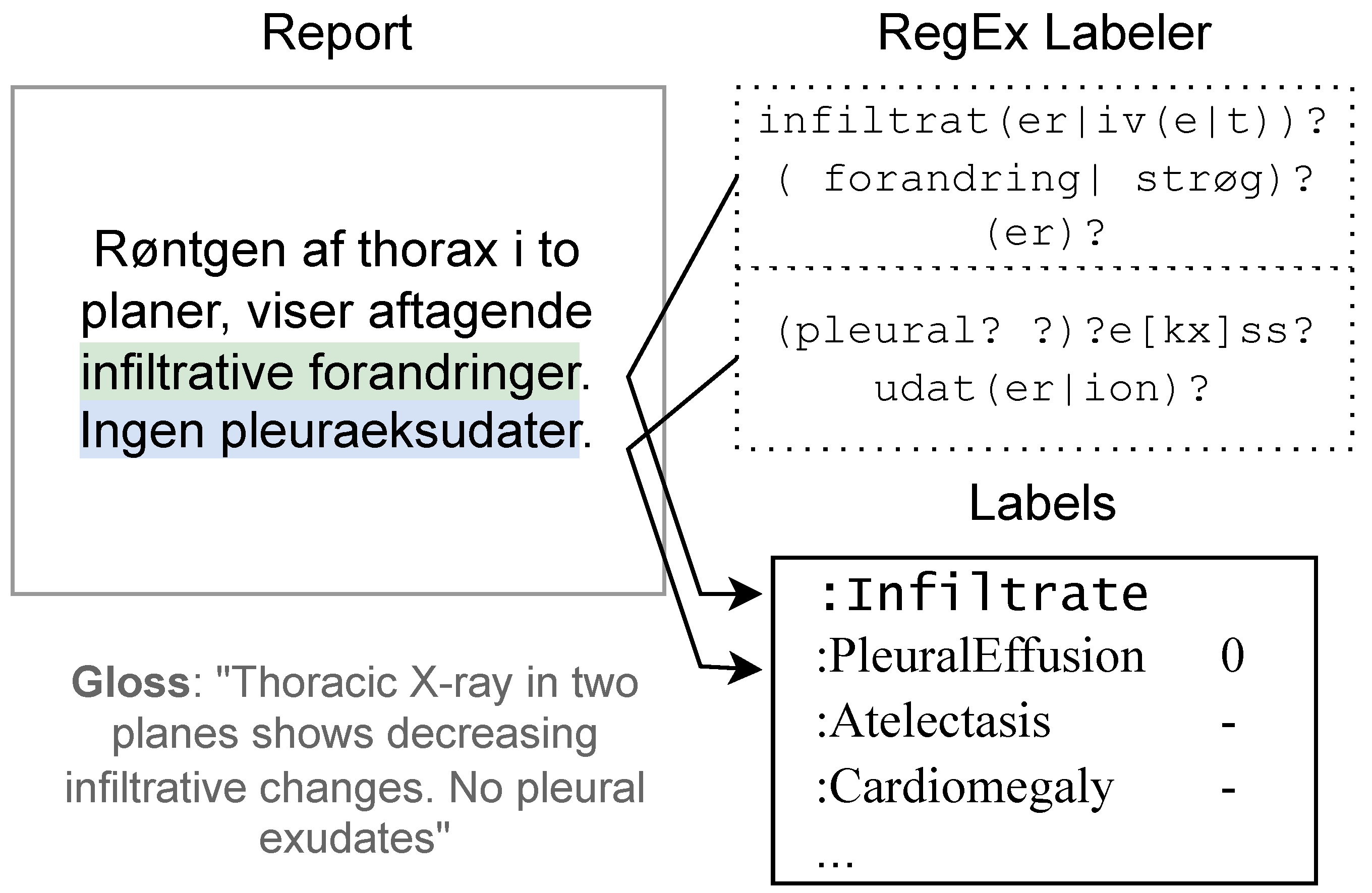
2.2. Methods: Regular Expressions ()
2.3. Methods: Pre-Trained Language Models
| Pre-training | Models are trained with unstructured, unlabeled data by predicting missing parts of the input as a next word prediction task. |
| Fine-tuning | A pre-trained LLM is further trained to specialize it for a specific task, e.g., radiology report classification. |
| Token | Fundamental unit of text in LLMs, representing words or subwords. |
| Tokenizer | A function that converts the input text into tokens. |
| Context | Amount of text, in tokens, that the model can process at any one time. |
2.4. Experimental Protocol
2.4.1. Rule-Based Labels ()
2.4.2. Human Expert Labels ()
2.4.3. Transfer Learning ()
2.5. Resource-Driven Experiments
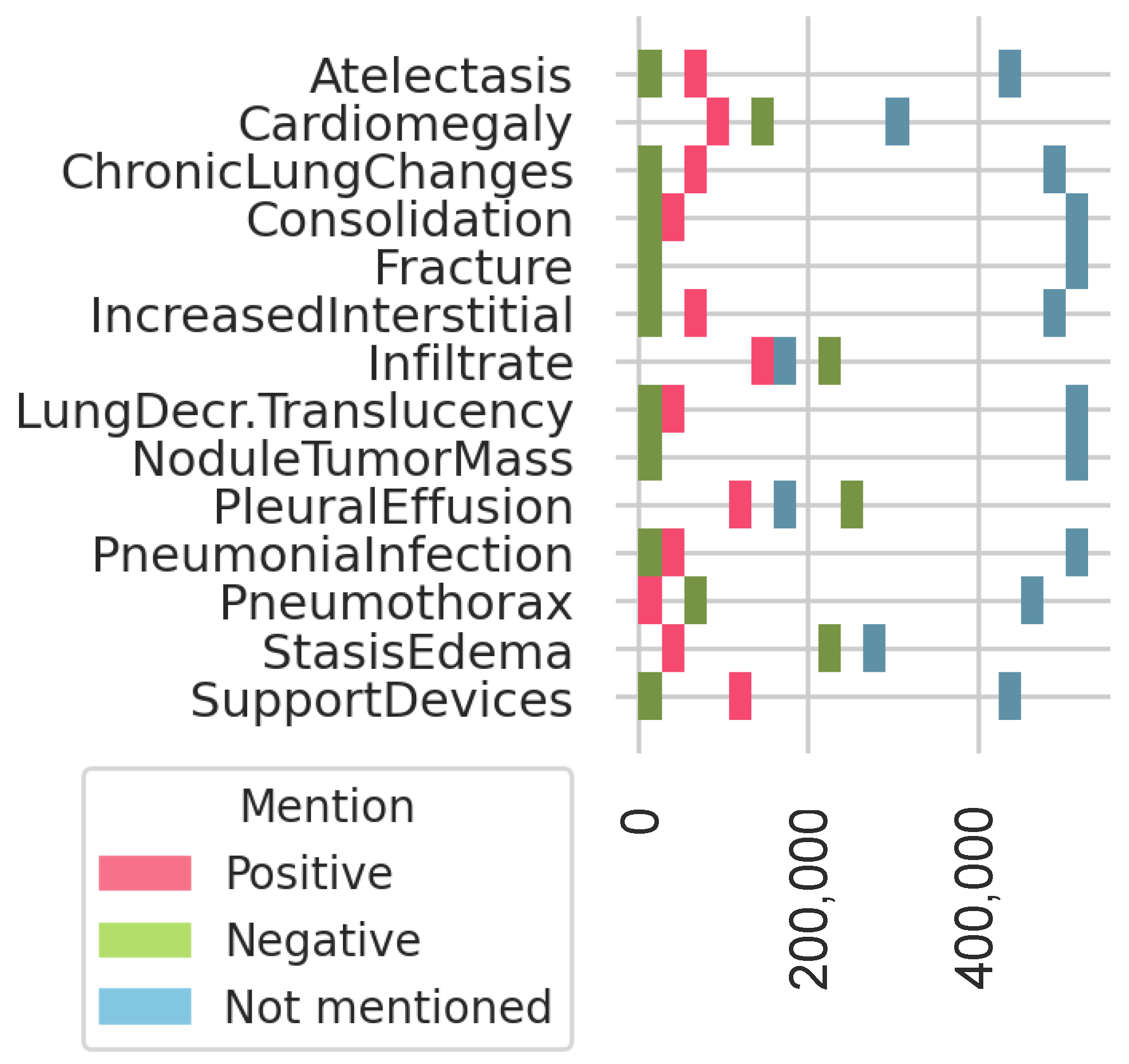
2.5.1. Definition of Most Frequent Findings
2.5.2. Definition of Model Ensemble
2.5.3. Definition of Data Ablation
3. Results
3.1. Evaluation Metrics
3.2. RegEx and Transfer Learning
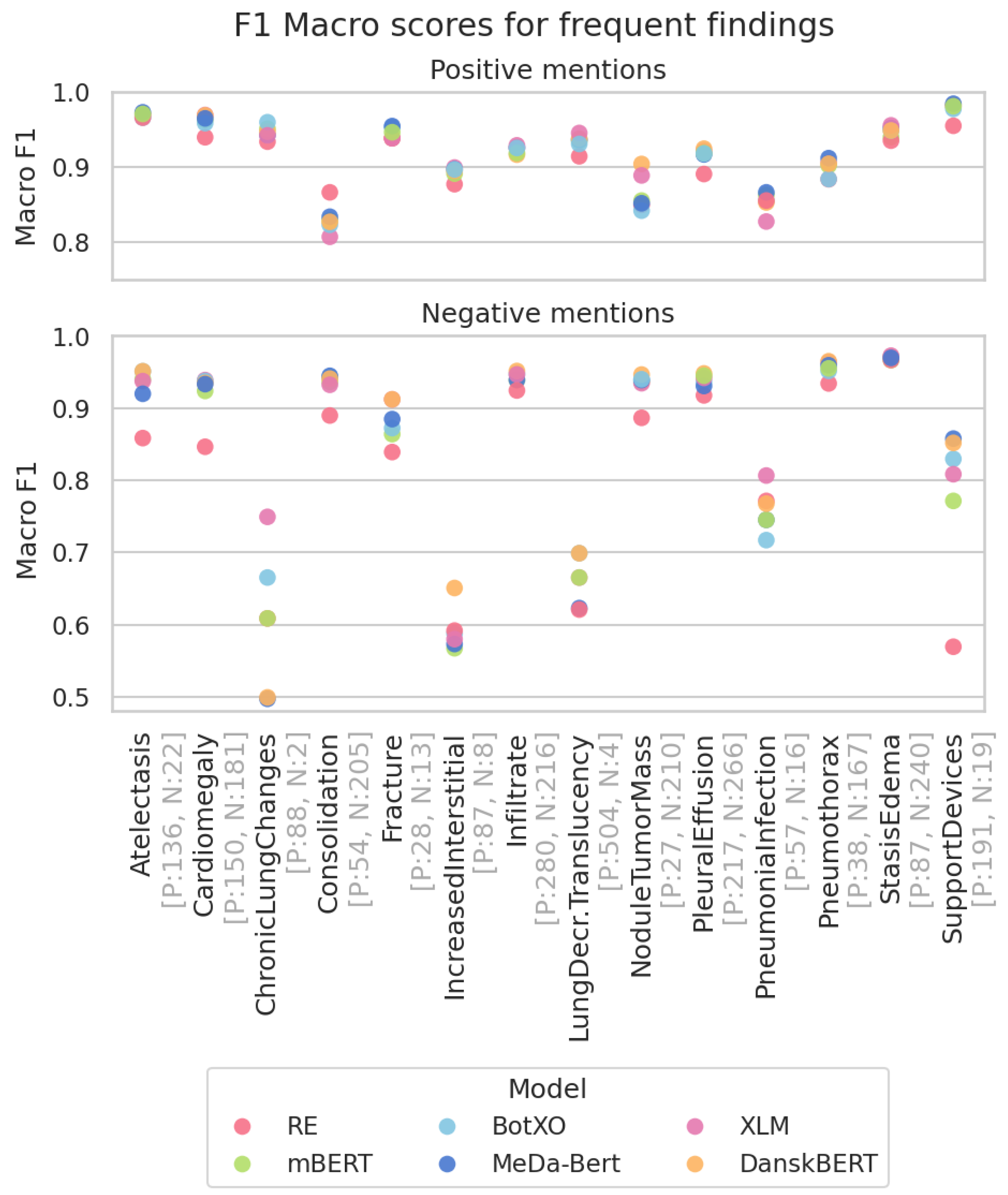
3.3. Most Frequent Findings Subanalysis
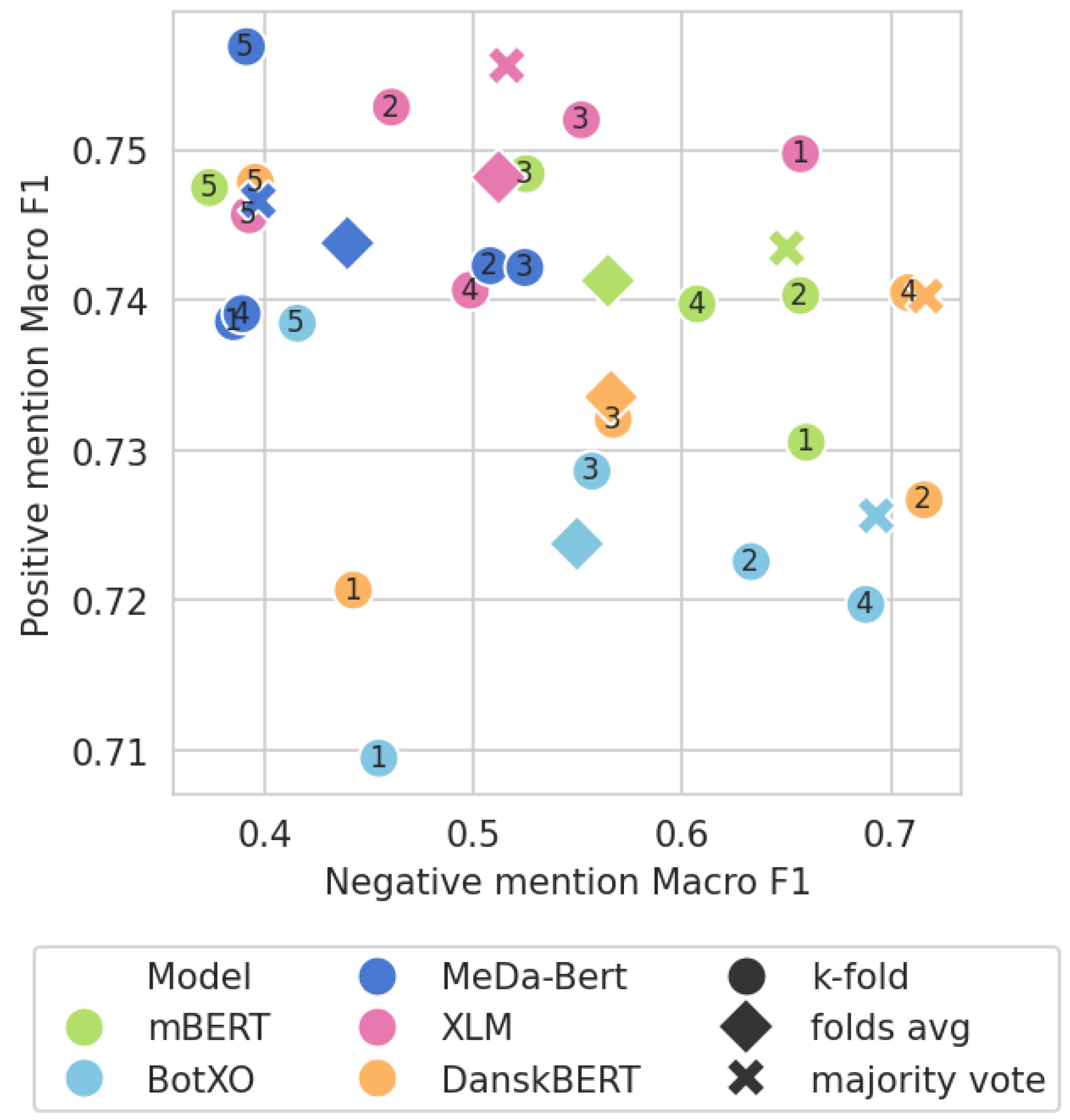
3.4. Model Ensemble Subanalysis
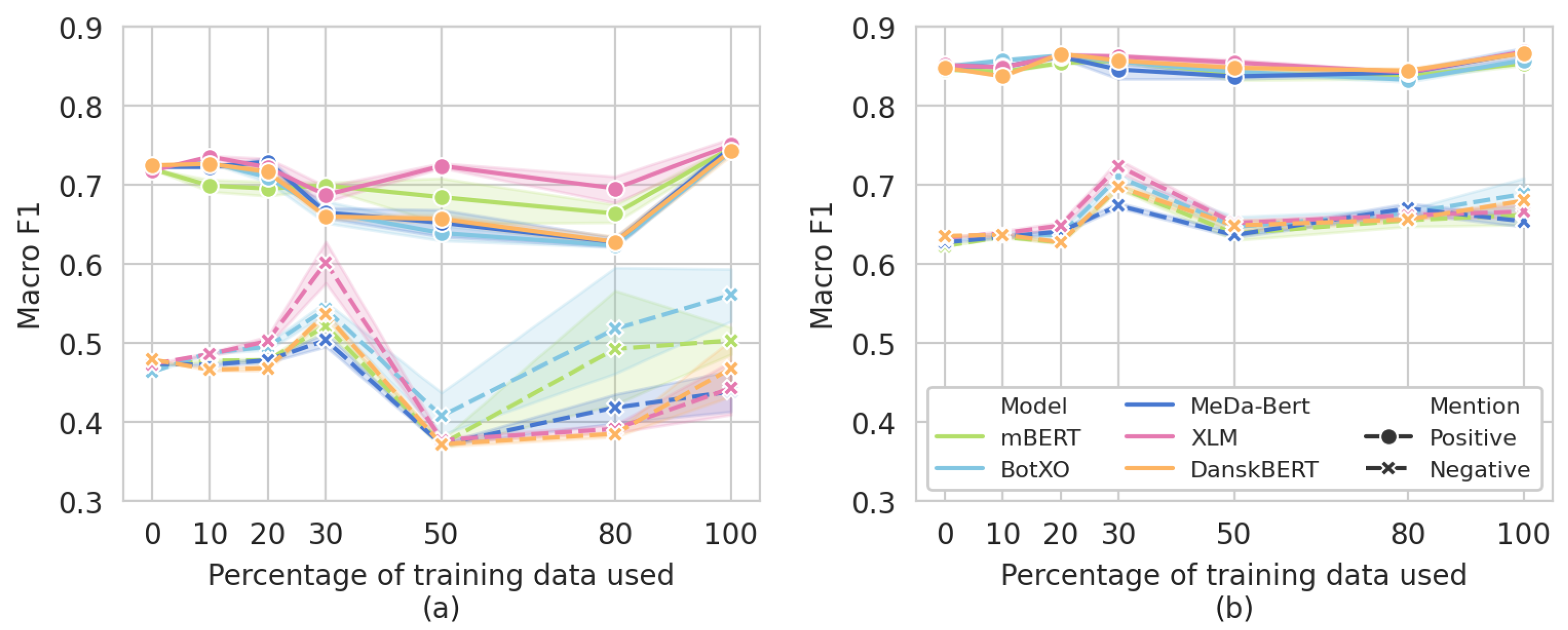
3.5. Data Ablation Subanalysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Data Description

| Listing A1. Hierarchical tree structure defined for the annotation of chest X-ray reports, translated from Danish into English for readability. |
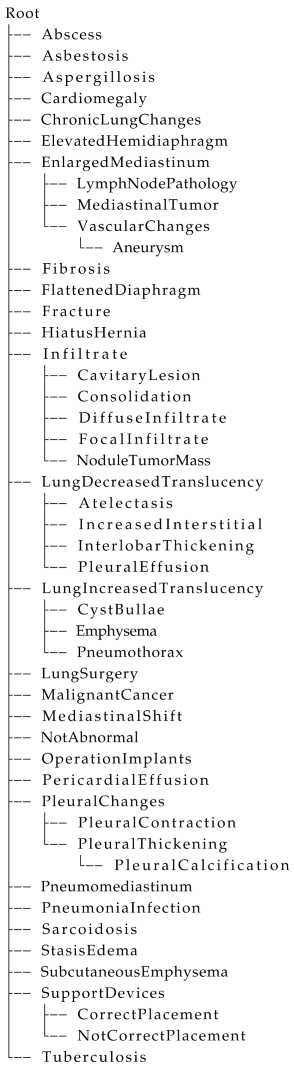 |
Appendix B. Implementation Details
Appendix B.1. Regular Expressions
Appendix B.2. Large Language Models
Appendix C. Detailed Results
| Positive F1 | Negative F1 | Weighted F1 | |
|---|---|---|---|
| RE | 0.721 (0.65–0.79) | 0.478 (0.40–0.55) | 0.667 (0.61–0.73) |
 mBERT mBERT | 0.491 (0.39–0.59) | 0.308 (0.20–0.41) | 0.566 (0.48–0.65) |
 BotXO BotXO | 0.455 (0.35–0.56) | 0.279 (0.18–0.38) | 0.511 (0.42–0.60) |
 MeDa-BERT MeDa-BERT | 0.447 (0.34–0.55) | 0.326 (0.22–0.43) | 0.540 (0.45–0.63) |
 XLM XLM | 0.471 (0.37–0.57) | 0.299 (0.19–0.40) | 0.532 (0.44–0.62) |
 DanskBERT DanskBERT | 0.466 (0.36–0.57) | 0.309 (0.21–0.41) | 0.535 (0.44–0.63) |
| Positive F1 | Negative F1 | Weighted F1 | |
|---|---|---|---|
| RE | 0.721 (0.65–0.79) | 0.478 (0.40–0.55) | 0.667 (0.61–0.73) |
 mBERT mBERT | 0.720 (0.65–0.79) | 0.471 (0.40–0.55) | 0.702 (0.64–0.76) |
 BotXO BotXO | 0.722 (0.65–0.79) | 0.463 (0.39–0.54) | 0.700 (0.64–0.76) |
 MeDa-BERT MeDa-BERT | 0.723 (0.65–0.79) | 0.474 (0.40–0.55) | 0.704 (0.64–0.77) |
 XLM XLM | 0.718 (0.65–0.79) | 0.473 (0.40–0.55) | 0.701 (0.64–0.76) |
 DanskBERT DanskBERT | 0.725 (0.65–0.80) | 0.479 (0.40–0.55) | 0.707 (0.64–0.77) |
| Positive F1 | Negative F1 | Weighted F1 | |
|---|---|---|---|
| RE | 0.848 (0.80–0.89) | 0.635 (0.46–0.81) | 0.844 (0.80–0.89) |
 mBERT mBERT | 0.857 (0.80–0.91) | 0.682 (0.50–0.87) | 0.874 (0.83–0.92) |
 BotXO BotXO | 0.860 (0.80–0.92) | 0.721 (0.57–0.87) | 0.881 (0.84–0.93) |
 MeDa-BERT MeDa-BERT | 0.877 (0.83–0.92) | 0.663 (0.46–0.87) | 0.883 (0.84–0.93) |
 XLM XLM | 0.868 (0.81–0.92) | 0.653 (0.42–0.88) | 0.879 (0.82–0.94) |
 DanskBERT DanskBERT | 0.867 (0.82–0.92) | 0.679 (0.47–0.88) | 0.883 (0.84–0.93) |
| Positive F1 | Negative F1 | Weighted F1 | |
|---|---|---|---|
| RE | 0.721 (0.65–0.79) | 0.478 (0.40–0.55) | 0.667 (0.61–0.73) |
 mBERT mBERT | 0.743 (0.67–0.81) | 0.650 (0.57–0.73) | 0.766 (0.71–0.82) |
 BotXO BotXO | 0.726 (0.65–0.80) | 0.693 (0.62–0.77) | 0.763 (0.71–0.82) |
 MeDa-BERT MeDa-BERT | 0.747 (0.67–0.82) | 0.397 (0.28–0.51) | 0.727 (0.66–0.79) |
 XLM XLM | 0.756 (0.68–0.83) | 0.516 (0.41–0.62) | 0.748 (0.69–0.81) |
 DanskBERT DanskBERT | 0.740 (0.67–0.81) | 0.717 (0.63–0.80) | 0.778 (0.72–0.83) |
| 10% | 20% | 30% | 50% | 80% | ||
|---|---|---|---|---|---|---|
 mBERT mBERT | Negative F1 | 0.478 | 0.478 | 0.521 | 0.372 | 0.493 |
| Positive F1 | 0.699 | 0.695 | 0.699 | 0.684 | 0.664 | |
| Weighted F1 | 0.690 | 0.692 | 0.710 | 0.685 | 0.696 | |
 BotXO BotXO | Negative F1 | 0.488 | 0.495 | 0.543 | 0.408 | 0.518 |
| Positive F1 | 0.728 | 0.709 | 0.666 | 0.639 | 0.623 | |
| Weighted F1 | 0.711 | 0.708 | 0.695 | 0.666 | 0.677 | |
 MeDa-BERT MeDa-BERT | Negative F1 | 0.472 | 0.478 | 0.504 | 0.372 | 0.418 |
| Positive F1 | 0.723 | 0.729 | 0.665 | 0.652 | 0.628 | |
| Weighted F1 | 0.712 | 0.715 | 0.702 | 0.676 | 0.673 | |
 XLM XLM | Negative F1 | 0.486 | 0.503 | 0.602 | 0.377 | 0.391 |
| Positive F1 | 0.736 | 0.722 | 0.687 | 0.724 | 0.695 | |
| Weighted F1 | 0.722 | 0.720 | 0.722 | 0.711 | 0.700 | |
 DanskBERT DanskBERT | Negative F1 | 0.466 | 0.468 | 0.536 | 0.372 | 0.385 |
| Positive F1 | 0.726 | 0.716 | 0.660 | 0.657 | 0.628 | |
| Weighted F1 | 0.703 | 0.705 | 0.699 | 0.675 | 0.665 |
Appendix D. Negative Results
| Positive F1 | Negative F1 | Weighted F1 | |
|---|---|---|---|
| DanskBERT() | 0.72 | 0.65 | 0.76 |
| DanskBERT() | 0.04 | 0.30 | 0.28 |
| DanskBERT() | 0.003 | 0.94 | 0.87 |
References
- Ait Nasser, A.; Akhloufi, M.A. A Review of Recent Advances in Deep Learning Models for Chest Disease Detection Using Radiography. Diagnostics 2023, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Chou, K.; Yeung, S.; Naik, N.; Madani, A.; Mottaghi, A.; Liu, Y.; Topol, E.; Dean, J.; Socher, R. Deep learning-enabled medical computer vision. NPJ Digit. Med. 2021, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Rädsch, T.; Reinke, A.; Weru, V.; Tizabi, M.D.; Schreck, N.; Kavur, A.E.; Pekdemir, B.; Roß, T.; Kopp-Schneider, A.; Maier-Hein, L. Labelling instructions matter in biomedical image analysis. Nat. Mach. Intell. 2023, 5, 273–283. [Google Scholar] [CrossRef]
- Wang, H.; Jin, Q.; Li, S.; Liu, S.; Wang, M.; Song, Z. A comprehensive survey on deep active learning in medical image analysis. Med. Image Anal. 2024, 95, 103201. [Google Scholar] [CrossRef] [PubMed]
- Chng, S.Y.; Tern, P.J.W.; Kan, M.R.X.; Cheng, L.T.E. Automated labelling of radiology reports using natural language processing: Comparison of traditional and newer methods. Health Care Sci. 2023, 2, 120–128. [Google Scholar] [CrossRef]
- Pereira, S.C.; Mendonça, A.M.; Campilho, A.; Sousa, P.; Teixeira Lopes, C. Automated image label extraction from radiology reports—A review. Artif. Intell. Med. 2024, 149, 102814. [Google Scholar] [CrossRef] [PubMed]
- Zingmond, D.; Lenert, L.A. Monitoring free-text data using medical language processing. Comput. Biomed. Res. 1993, 26, 467–481. [Google Scholar] [CrossRef] [PubMed]
- George Hripcsak, M.; Carol Friedman, P.; Philip, O.; Alderson, M.; William DuMouchel, P.; Stephen, B.; Johnson, P.; Paul, D.; Clayton, P. Unlocking Clinical Data from Narrative Reports: A Study of Natural Language Processing. Ann. Intern. Med. 1995, 122, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.L.; Knirsch, C.A.; Friedman, C.; Hripcsak, G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. In Proceedings of the AMIA Annual Fall Symposium, Washington, DC, USA, 26–30 October 1996; p. 542. [Google Scholar]
- Peng, Y.; Wang, X.; Lu, L.; Bagheri, M.; Summers, R.; Lu, Z. NegBio: A high-performance tool for negation and uncertainty detection in radiology reports. arXiv 2017, arXiv:cs.CL/1712.05898. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv 2019. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar] [CrossRef]
- Reis, E.P.; de Paiva, J.P.Q.; da Silva, M.C.B.; Ribeiro, G.A.S.; Paiva, V.F.; Bulgarelli, L.; Lee, H.M.H.; Santos, P.V.; Brito, V.M.; Amaral, L.T.W.; et al. BRAX, Brazilian labeled chest x-ray dataset. Sci. Data 2022, 9, 487. [Google Scholar] [CrossRef]
- Nguyen, T.; Vo, T.M.; Nguyen, T.V.; Pham, H.H.; Nguyen, H.Q. Learning to diagnose common thorax diseases on chest radiographs from radiology reports in Vietnamese. PLoS ONE 2022, 17, e0276545. [Google Scholar] [CrossRef] [PubMed]
- Wollek, A.; Hyska, S.; Sedlmeyr, T.; Haitzer, P.; Rueckel, J.; Sabel, B.O.; Ingrisch, M.; Lasser, T. German CheXpert Chest X-ray Radiology Report Labeler. arXiv 2023, arXiv:cs.CL/2306.02777. [Google Scholar] [CrossRef]
- Smit, A.; Jain, S.; Rajpurkar, P.; Pareek, A.; Ng, A.Y.; Lungren, M.P. CheXbert: Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT. arXiv 2020. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Rehm, G.; Berger, M.; Elsholz, E.; Hegele, S.; Kintzel, F.; Marheinecke, K.; Piperidis, S.; Deligiannis, M.; Galanis, D.; Gkirtzou, K.; et al. European Language Grid: An Overview. In Proceedings of the Twelfth Language Resources and Evaluation Conference, Marseille, France, 11–16 May 2020; pp. 3366–3380. [Google Scholar]
- Li, D.; Pehrson, L.M.; Lauridsen, C.A.; Tøttrup, L.; Fraccaro, M.; Elliott, D.; Zając, H.D.; Darkner, S.; Carlsen, J.F.; Nielsen, M.B. The Added Effect of Artificial Intelligence on Physicians’ Performance in Detecting Thoracic Pathologies on CT and Chest X-ray: A Systematic Review. Diagnostics 2021, 11, 2206. [Google Scholar] [CrossRef]
- Li, D.; Pehrson, L.M.; Tøttrup, L.; Fraccaro, M.; Bonnevie, R.; Thrane, J.; Sørensen, P.J.; Rykkje, A.; Andersen, T.T.; Steglich-Arnholm, H.; et al. Inter- and Intra-Observer Agreement When Using a Diagnostic Labeling Scheme for Annotating Findings on Chest X-rays—An Early Step in the Development of a Deep Learning-Based Decision Support System. Diagnostics 2022, 12, 3112. [Google Scholar] [CrossRef]
- Li, D.; Pehrson, L.M.; Bonnevie, R.; Fraccaro, M.; Thrane, J.; Tøttrup, L.; Lauridsen, C.A.; Butt Balaganeshan, S.; Jankovic, J.; Andersen, T.T.; et al. Performance and Agreement When Annotating Chest X-ray Text Reports—A Preliminary Step in the Development of a Deep Learning-Based Prioritization and Detection System. Diagnostics 2023, 13, 1070. [Google Scholar] [CrossRef]
- Bustos, A.; Pertusa, A.; Salinas, J.M.; De La Iglesia-Vaya, M. Padchest: A large chest x-ray image dataset with multi-label annotated reports. Med. Image Anal. 2020, 66, 101797. [Google Scholar] [CrossRef] [PubMed]
- Chapman, W.W.; Bridewell, W.; Hanbury, P.; Cooper, G.F.; Buchanan, B.G. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. J. Biomed. Inform. 2001, 34, 301–310. [Google Scholar] [CrossRef] [PubMed]
- von der Mosel, J.; Trautsch, A.; Herbold, S. On the Validity of Pre-Trained Transformers for Natural Language Processing in the Software Engineering Domain. IEEE Trans. Softw. Eng. 2023, 49, 1487–1507. [Google Scholar] [CrossRef]
- Sechidis, K.; Tsoumakas, G.; Vlahavas, I. On the stratification of multi-label data. In Proceedings of the Machine Learning and Knowledge Discovery in Databases: European Conference, ECML PKDD 2011, Athens, Greece, 5–9 September 2011; Proceedings, Part III 22. Springer: Berlin/Heidelberg, Germany, 2011; pp. 145–158. [Google Scholar]
- Dietterich, T.G. Ensemble methods in machine learning. In Proceedings of the International Workshop on Multiple Classifier Systems, Cagliari, Italy, 21–23 June 2000; pp. 1–15. [Google Scholar]
- Mohammed, A.; Kora, R. A comprehensive review on ensemble deep learning: Opportunities and challenges. J. King Saud-Univ.-Comput. Inf. Sci. 2023, 35, 757–774. [Google Scholar] [CrossRef]
- Kær Jørgensen, R.; Hartmann, M.; Dai, X.; Elliott, D. mDAPT: Multilingual Domain Adaptive Pretraining in a Single Model. In Proceedings of the Findings of the Association for Computational Linguistics: EMNLP 2021, Punta Cana, Dominican Republic, 16–20 November 2021; pp. 3404–3418. [Google Scholar] [CrossRef]
- Chen, H.; Miao, S.; Xu, D.; Hager, G.D.; Harrison, A.P. Deep Hierarchical Multi-label Classification of Chest X-ray Images. In Proceedings of the 2nd International Conference on Medical Imaging with Deep Learning, PMLR, London, UK, 8–10 July 2019; Volume 102, pp. 109–120. [Google Scholar]
- Del Moral, P.; Nowaczyk, S.; Pashami, S. Why is multiclass classification hard? IEEE Access 2022, 10, 80448–80462. [Google Scholar] [CrossRef]
- Nielsen, D. ScandEval: A Benchmark for Scandinavian Natural Language Processing. In Proceedings of the 24th Nordic Conference on Computational Linguistics (NoDaLiDa), Tórshavn, Faroe Islands, 22–24 May 2023; pp. 185–201. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Dai, X.; Chalkidis, I.; Darkner, S.; Elliott, D. Revisiting Transformer-based Models for Long Document Classification. In Proceedings of the Findings of the Association for Computational Linguistics: EMNLP 2022, Abu Dhabi, United Arab Emirates, 7–11 December 2022; pp. 7212–7230. [Google Scholar] [CrossRef]
- BotXO, CertainlyIO. Danish BERT. 2020. Available online: https://github.com/botxo/nordic_bert (accessed on 4 April 2024).
- Pedersen, J.; Laursen, M.; Vinholt, P.; Savarimuthu, T.R. MeDa-BERT: A medical Danish pretrained transformer model. In Proceedings of the 24th Nordic Conference on Computational Linguistics (NoDaLiDa), Tórshavn, Faroe Islands, 22–24 May 2023; pp. 301–307. [Google Scholar]
- Conneau, A.; Khandelwal, K.; Goyal, N.; Chaudhary, V.; Wenzek, G.; Guzmán, F.; Grave, E.; Ott, M.; Zettlemoyer, L.; Stoyanov, V. Unsupervised Cross-lingual Representation Learning at Scale. arXiv 2019, arXiv:abs/1911.02116. [Google Scholar]
- Snæbjarnarson, V.; Simonsen, A.; Glavaš, G.; Vulić, I. Transfer to a Low-Resource Language via Close Relatives: The Case Study on Faroese. In Proceedings of the 24th Nordic Conference on Computational Linguistics (NoDaLiDa), Tórshavn, Faroe Islands, 22–24 May 2023. [Google Scholar]


 indicates multi-lingual data,
indicates multi-lingual data,  stands for Danish, and
stands for Danish, and  for Danish medical data. The models are presented from the least amount of Danish data used in their pre-training (mBERT) to the most (DanskBERT).
for Danish medical data. The models are presented from the least amount of Danish data used in their pre-training (mBERT) to the most (DanskBERT).
 indicates multi-lingual data,
indicates multi-lingual data,  stands for Danish, and
stands for Danish, and  for Danish medical data. The models are presented from the least amount of Danish data used in their pre-training (mBERT) to the most (DanskBERT).
for Danish medical data. The models are presented from the least amount of Danish data used in their pre-training (mBERT) to the most (DanskBERT).| Pre-Training Data | Model Name | Parameters | Tokens/Words |
|---|---|---|---|
 | mBERT | 110 M | ≈200 M words 1 |
 | BotXO | 110 M | 1.6 B words |
 | MeDa-BERT | 110 M | +123 M tokens |
 | XML-RoBERTa | 125 M | 7.8 B tokens |
 | DanskBERT | 125 M | +1 B words |
| Positive F1 | Negative F1 | Weighted F1 | ||
|---|---|---|---|---|
| All Findings | Most Frequent | |||
| RE | 0.721 | 0.478 | 0.667 | 0.846 |
 mBERT mBERT | 0.742 ± 0.003 | 0.477 ± 0.008 | 0.732 ± 0.003 | 0.869 ± 0.001 |
 BotXO BotXO | 0.745 ± 0.007 | 0.509 ± 0.012 | 0.737 ± 0.004 | 0.876 ± 0.002 |
 MeDa-BERT MeDa-BERT | 0.739 ± 0.005 | 0.480 ± 0.006 | 0.742 ± 0.003 | 0.873 ± 0.002 |
 XLM XLM | 0.738 ± 0.007 | 0.498 ± 0.004 | 0.736 ± 0.004 | 0.884 ± 0.001 |
 DanskBERT DanskBERT | 0.738 ± 0.011 | 0.524 ± 0.008 | 0.744 ± 0.007 | 0.882 ± 0.003 |
| Positive F1 | Negative F1 | Weighted F1 | |
|---|---|---|---|
| RE | 0.721 | 0.478 | 0.667 |
 mBERT mBERT | 0.743 | 0.650 | 0.766 |
 BotXO BotXO | 0.726 | 0.693 | 0.763 |
 MeDa-BERT MeDa-BERT | 0.747 | 0.397 | 0.727 |
 XLM XLM | 0.756 | 0.516 | 0.748 |
 DanskBERT DanskBERT | 0.740 | 0.717 | 0.778 |
| Ensemble | ||||
|---|---|---|---|---|
| RE | 0.667 | - | - | - |
 mBERT mBERT | 0.702 | 0.566 | 0.732 | 0.766 |
 BotXO BotXO | 0.700 | 0.511 | 0.737 | 0.763 |
 MeDa-BERT MeDa-BERT | 0.704 | 0.540 | 0.742 | 0.727 |
 XLM XLM | 0.701 | 0.532 | 0.736 | 0.748 |
 DanskBERT DanskBERT | 0.707 | 0.535 | 0.744 | 0.778 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schiavone, A.; Pehrson, L.M.; Ingala, S.; Bonnevie, R.; Fraccaro, M.; Li, D.; Nielsen, M.B.; Elliott, D. Effective Machine Learning Techniques for Non-English Radiology Report Classification: A Danish Case Study. AI 2025, 6, 37. https://doi.org/10.3390/ai6020037
Schiavone A, Pehrson LM, Ingala S, Bonnevie R, Fraccaro M, Li D, Nielsen MB, Elliott D. Effective Machine Learning Techniques for Non-English Radiology Report Classification: A Danish Case Study. AI. 2025; 6(2):37. https://doi.org/10.3390/ai6020037
Chicago/Turabian StyleSchiavone, Alice, Lea Marie Pehrson, Silvia Ingala, Rasmus Bonnevie, Marco Fraccaro, Dana Li, Michael Bachmann Nielsen, and Desmond Elliott. 2025. "Effective Machine Learning Techniques for Non-English Radiology Report Classification: A Danish Case Study" AI 6, no. 2: 37. https://doi.org/10.3390/ai6020037
APA StyleSchiavone, A., Pehrson, L. M., Ingala, S., Bonnevie, R., Fraccaro, M., Li, D., Nielsen, M. B., & Elliott, D. (2025). Effective Machine Learning Techniques for Non-English Radiology Report Classification: A Danish Case Study. AI, 6(2), 37. https://doi.org/10.3390/ai6020037







