Abstract
Background: Conventional medical image retrieval methods treat images and text as independent embeddings, limiting their ability to fully utilize the complementary information from both modalities. This separation often results in suboptimal retrieval performance, as the intricate relationships between images and text remain underexplored. Methods: To address this limitation, we propose a novel retrieval method that integrates medical image and text embeddings using a cross-attention mechanism. Our approach creates a unified representation by directly modeling the interactions between the two modalities, significantly enhancing retrieval accuracy. Results: Built upon the pre-trained BioMedCLIP model, our method outperforms existing techniques across multiple metrics, achieving the highest mean Average Precision (mAP) on the MIMIC-CXR dataset. Conclusions: These results highlight the effectiveness of our method in advancing multimodal medical image retrieval and set the stage for further innovation in the field.
1. Introduction
Medical images such as X-ray images and MRIs have been accumulated in the medical field for many years. Meanwhile, data digitalization has rapidly progressed recently, and high volumes of medical data, including images, can now be efficiently stored and used [1]. Medical images are crucial in diagnosis and treatment decision-making [2], and they are vital data sources for education and research. Furthermore, advances in machine learning and artificial intelligence in recent years have resulted in rapid advances in using medical data [3,4,5,6]. This is expected to reduce physician burden and improve diagnostic accuracy.
Medical data provides various aspects of information for each modality. Image data contains anatomical information and details of lesions, whereas text data includes records of physicians’ judgments and patient conditions [7,8]. Therefore, integrating images and text is crucial for effectively extracting the complementary information possessed by each, and this is expected to improve diagnostic accuracy and make report generation more efficient. Examples in previous research include a proposed method of simultaneously using images and corresponding diagnostic reports to improve the classification accuracy of lesions and a proposed technology for automatically generating diagnostic reports from images [9,10]. Such research has garnered attention as technologies that support decision-making in the medical field by using the interaction between images and text.
In this context, medical image retrieval is a prevailing method for the effective utilization of large volumes of medical data [11,12]. Medical image retrieval is a technology that efficiently retrieves similar cases and related information based on images and text that are given as queries. For example, suppose that a user provides a chest X-ray image related to a specific disease as an input. Then, case images with similar characteristics and related diagnostic reports can be obtained. This technology is crucial for diagnostic support and educational purposes. Diagnostic accuracy is expected to be improved by providing related similar cases. Additionally, providing visually similar cases can improve the quality of education for medical students and residents. Furthermore, in research, an efficient collection of similar images is expected to lead to discoveries and the identification of interesting patterns. Therefore, medical image retrieval is expected to have diverse applications in the medical and research fields, and it is an essential means for the practical and effective utilization of large volumes of medical data.
In recent years, attention has been paid to methods that treat images and text in the same embedding space in medical image retrieval [13,14,15,16]. A representative example is BioMedCLIP [16], which is a method that learns medical image and text embeddings in a unified manner to enable cross-modal search (image-text and text-image). This approach has considerably improved retrieval performance by modeling the interrelationships between image and text modalities. Additionally, Simon et al. [17] conducted a comprehensive review of multimodal AI in radiology, discussing how advanced architectures such as transformers and graph neural networks (GNNs) enable the integration of imaging data with various clinical metadata. They highlight both the potential benefits of these methods and critical challenges, including data scarcity, bias, and the lack of standardized taxonomy in multimodal research.
More recently, Ou et al. [18] proposed a method that integrates medical images and textual reports using a Report Entity Graph and Dual Attention mechanisms. Their approach enhances fine-grained semantic alignment between images and text by structuring radiology reports into entity graphs. This method focuses on cross-modal retrieval tasks, specifically retrieving text from images (image-text) and retrieving images from text (text-image). In addition, Jeong et al. [19] introduced a retrieval-based chest X-ray report generation module that computes an image–text matching score using a multimodal encoder, improving the selection of the most clinically relevant report. While Jeong et al.’s method effectively refines the image-text retrieval process for report generation, our work targets a different scenario: retrieving image–text pairs from a combined image and text query (image & text-image & text). Hence, rather than focusing on text generation or single-modality retrieval, we aim to jointly model the interaction between images and textual descriptions to facilitate richer multimodal queries. However, these methods treat images and text as separate embeddings and compare them in the same space. Therefore, these methods do not consider learning or integrated representations that combine images and text. This is a major limitation in maximizing the information obtained from both images and text. Therefore, existing research provides a foundation for cross-modal approaches in medical image retrieval. Nonetheless, the above findings suggest a need for new methodologies that integrate information between modalities. Unlike previous studies, which primarily focus on cross-modal retrieval tasks (image-text, text-image), our study tackles a distinct challenge: retrieving image-text pairs using a multimodal query (image & text → image & text). Instead of retrieving a single modality, our method models the interaction between image and text modalities jointly. This approach facilitates more robust multimodal fusion and enhances retrieval performance in clinical applications.
In this study, we tackle the challenge of medical image retrieval by proposing an innovative method that integrates image and text embeddings. Our contributions are as follows:
- 1.
- Introduction of a novel image-text fusion strategy;
Conventional medical image retrieval methods treat images and text separately, which limits retrieval effectiveness. The proposed method introduces a cross-attention-based fusion strategy that facilitates more effective multimodal integration by directly modeling the relevance between images and text.
- 2.
- Empirical demonstration of the effectiveness of integrated embeddings in medical image retrieval;
This study empirically demonstrates how a unified image-text representation enhances retrieval accuracy, underscoring the importance of multimodal fusion in medical image retrieval. This insight paves the way for leveraging cross-modal relationships in clinical applications.
- 3.
- Enhancement of retrieval performance through targeted training of additional layers;
By keeping the original BioMedCLIP model and training only the additional cross-attention layers, we demonstrated that retrieval performance can be significantly improved. This finding highlights that even minimal architectural modifications to pre-trained models can yield substantial improvements, making the approach both computationally efficient and practically valuable.
2. Methods
Figure 1 illustrates the proposed model. This model consists of an embedding layer, which comprises a pre-trained BioMedCLIP, and a cross-attention layer that uses a transformer encoder [20]. First, an image-text pair is input, and two embeddings are obtained by the embedding layer. Subsequently, both embeddings are sent to the cross-attention layer. The cross-attention layer emphasizes the important parts using an attention mechanism and outputs a final embedding that reflects the interaction between modalities. Details of each part of the proposed model are described in the following subsections.
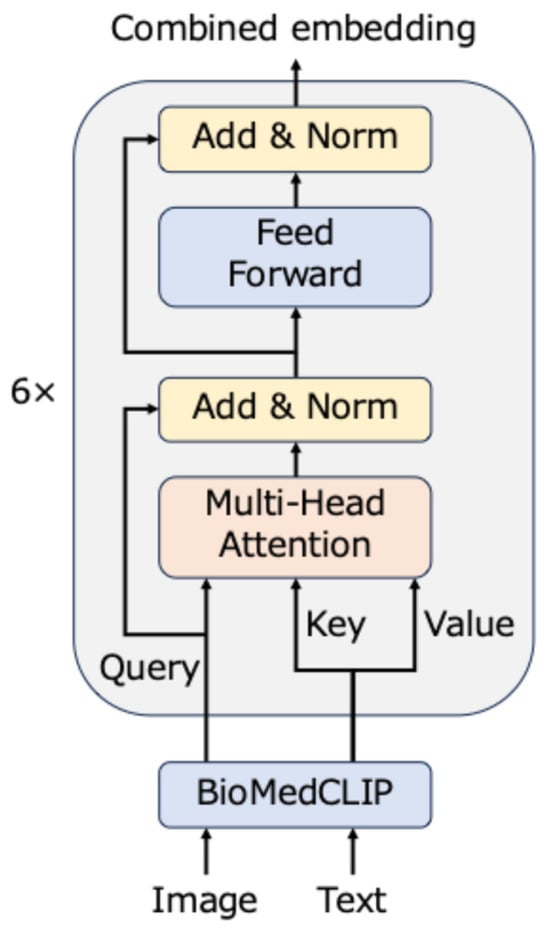
Figure 1.
Framework of the proposed model. The embedding layer converts image-text pairs to embeddings, which are subsequently combined by the cross-attention model.
2.1. Embedding Layer
When an image-text pair is provided, each part is input to the image encoder and text encoder of the pre-trained BioMedCLIP.
2.1.1. Image Encoder
BioMedCLIP uses the Vision Transformer (ViT) [21] as the image encoder. The final ViT layer outputs 768-dimensional embedding. However, in this study, we sought to utilize local information within an image by extracting embeddings for each region from the intermediate ViT layer. Specifically, the input image is divided into 14 × 14 patches (regions), with each patch converted into a 768-dimensional embedding. ViT adds the input patch and a special token for class classification (CLS token) to summarize the overall information. This process results in the entire image being represented as 197 embeddings, including the CLS token (14 × 14 = 196 patches + 1 CLS token), ultimately yielding a (197,768)-dimensional tensor. This embedding in the intermediate layer can be used to model the information of the entire image while preserving its local features.
2.1.2. Text Encoder
The text encoder used by BioMedCLIP is PubMedBERT [22], which is a domain-specific language model that is pre-trained on medical and biomedical text data. PubMedBERT tokenizes the input text and converts each token into a 768-dimensional embedding. Extremely long input texts are truncated, and those that are too short undergo a padding process such that the final text length has a fixed length of 256 tokens. This results in the entire text being represented as a (256,768)-dimensional tensor. In this study, as with the image encoder, the text encoder effectively used local information in the text by extracting per-token embeddings from the intermediate layer of PubMedBERT. Specifically, the encoder tokenized the input text and obtained the 768-dimensional embeddings of each token from the intermediate layer to obtain embeddings that reflect contextual information while preserving the local features of each token. This process provides a foundation for cross-attention integration with image embeddings without losing per-token information.
2.2. Cross-Attention Layer
The cross-attention layer is crucial in integrating image and text embeddings and learning their interrelationship. In this study, we adopted cross-attention, which uses image embeddings as queries and text embeddings as keys and values. This design is based on the characteristics of text and images and the cross-attention mechanism. Specifically, text data has linear and information-dense characteristics, such as physicians’ diagnostic results and symptom descriptions, and attention mechanisms are effective in highlighting important information. Meanwhile, image data often has structures and lesions that are distributed in a multidimensional manner and are ambiguous. When considering this difference, the design with images as queries and text as keys and values is thought to be optimal for effectively referencing and integrating contextual information in text based on local information in images. Furthermore, the cross-attention output is calculated as a weighted sum of values; therefore, the quality of information contained in the values directly affects the results. This characteristic results in a design where using text with high information density as a value contributes to generating optimal embedding representations. Given the above reasons, this study adopts a cross-attention design with images as queries and text as keys and values.
The calculation of cross-attention is conducted as follows.
- 1.
- Create queries, keys, and values;
Convert image embedding into query , and text embedding into key and value :
where are learnable weight matrices.
- 2.
- Calculate attention score;
Calculate the inner product of queries and keys and perform scaling and normalization:
where represents the attention score, indicating which text token each image token should direct its attention towards.
- 3.
- Generate output;
Apply the attention score to the value to obtain an output that integrates image and text embeddings:
where is the combined embedding.
- 4.
- Feedforward network (FFN);
Apply a feedforward network to the output of cross-attention and conduct a nonlinear transformation. This further strengthens the feature representation.
- 5.
- Layer normalization and residual connection;
Layer normalization and residual connections [23] are applied to each layer to ensure learning stability and prevent the gradient vanishing problem.
In this study, we adopted a configuration that does not apply positional embeddings. This decision was made based on the characteristics of medical reports and the relationship between medical images and reports. Medical reports vary in writing style depending on the physician, and different expressions may be used for the same condition. Therefore, word-level information tends to be more important for diagnosis than sentence structure or word order. Additionally, in the medical domain, the presence or absence of specific terms, such as disease names and symptoms, often directly influences the diagnosis more than the overall meaning of the text.
On the other hand, medical images contain localized abnormalities (lesions), but they lack sufficient diagnostic information on their own. To address this, we adopted a design in which local information in images is used to reference medical reports (with images as queries and text as keys and values). To learn which information in the report corresponds to the lesion in an image, word-level relevance is more important than absolute positional information in the text. Consequently, applying positional embeddings may impose constraints on the flexible association of words within the text, potentially making information integration less efficient. A detailed comparative experiment and analysis of this effect are presented in Section 4.
2.3. Loss Function
In this study, we adopted supervised contrastive loss (SupConLoss) [24] to improve the discriminability between classes in the image and text embedding space. This loss function brings embeddings within the same class closer and moves embeddings of different classes further apart, thereby effectively promoting clustering in the embedding space.
SupConLoss brings all positive examples belonging to the same class closer to the anchor and moves negative examples of different classes further apart. The specific form of the loss function is as follows:
where are embedding vectors corresponding to positive examples and all examples (including positive and negative examples), respectively; is the index set of positive examples of the same class as anchor in the batch; is the index set of all examples in the batch except anchor ; and is the temperature scale parameter that controls the scaling of the loss function; is the total number of samples in the batch; is the index set of all samples in the batch. When SupConLoss is adopted, embeddings of the same class are brought closer together, and those of different classes are pushed further apart. This design has the following features compared to the conventional triplet loss [25] and N-pair loss [26]. First, the loss is calculated using all samples in a batch, allowing for more stable learning. In addition, hard mining does not need to be explicitly conducted, which reduces the uncertainty associated with negative example selection. Furthermore, simultaneously considering many positive and negative examples results in the separability between classes in the embedding space being effectively learned. In this study, we adopted a temperature parameter τ = 0.07 and applied SupConLoss to model the embedding space of images and text in an integrated manner. This is expected to improve performance in retrieval and similarity calculation tasks.
2.4. Evaluation Metrics
The performance of the proposed method and the comparison methods in the retrieval task was evaluated using the following metrics. These metrics were selected due to their widespread use in prior research and their ability to comprehensively assess different aspects of retrieval performance.
- P@K (Precision at K)
This is an index that measures the accuracy rate in the top K retrieval results and is crucial for evaluating retrieval performance when the user only checks the top K results:
where “Relevant results” refers to images and text in the same class as the query.
- Mean Average Precision (mAP)
This is an index that calculates the mean of the average precision (AP) for each query and is used to comprehensively evaluate the performance of the entire retrieval results. Here, mAP evaluates the quality of the ranking of retrieval results by considering where related documents appear in the retrieval results:
where R is the total number of candidates in the retrieval results that belong to the same class as the query, P(k) is the precision of the top k results, rel(k) is 1 if the candidate at position k of the retrieval result belongs to the same class and 0 otherwise, and Q is the total number of all queries.
3. Experiments
3.1. Datasets
The experiments in this study used the MIMIC-CXR [27,28,29,30,31], which is a publicly available medical dataset consisting of chest radiographs in DICOM format with free-text radiology reports. The dataset contains 377,110 images corresponding to 227,835 radiographic studies performed at the Beth Israel Deaconess Medical Center in Boston, MA. The data collection and sharing process were approved by the Institutional Review Board (IRB) at the Beth Israel Deaconess Medical Center, and a waiver of informed consent was granted. All data in the dataset are fully de-identified to comply with the Health Insurance Portability and Accountability Act (HIPAA) Safe Harbor requirements. Protected Health Information (PHI) has been removed using both automated and manual de-identification techniques.
Following the methods of MedCLIP [15] and ConVIRT [13], we extracted exclusively positive data for the five classes: atelectasis, cardiomegaly, consolidation, edema, and pleural effusion. Here, “exclusively positive” implies that each datum is positive only for a specific class and negative for the rest. We extracted and used the FINDINGS and IMPRESSION sections in the report for text data. Suppose that these sections did not exist; in that case, we used the last paragraph of the report. In the dataset, 20,713 samples contain both FINDINGS and IMPRESSION sections, while 1285 samples lack both sections. Additionally, 5141 samples contain only the FINDINGS section, whereas 11,102 samples contain only the IMPRESSION section.
The dataset was constructed as follows.
- 1.
- Test data (mimic-5x200)
Here, mimic-5x200 includes 10 images of each class as query data and 200 images of each class as retrieval candidate data for each of the five classes (atelectasis, cardiomegaly, consolidation, edema, and pleural effusion). The dataset consists of 1050 images, and it was used to evaluate the retrieval task.
- 2.
- Training and validation data
The remaining data not included in mimic-5x200 were split at 9:1 and used as training and validation data.
Image and text embeddings were pre-extracted using a pre-trained encoder. Image embeddings were represented as (197,768) dimensions (196 image patches + CLS token), and text embeddings were represented as (256,768) dimensions (256 tokens).
Figure 2 presents examples from the dataset used in this study. As shown in the figure, each report may contain either or both of the FINDINGS and IMPRESSION sections. The FINDINGS section outlines detailed radiological observations, while the IMPRESSION section offers a diagnostic summary based on these observations. If both sections are absent, the final paragraph of the report is used instead, as it may still contain diagnostic information. Additionally, the corresponding chest X-ray images are shown with the reports to illustrate their alignment.
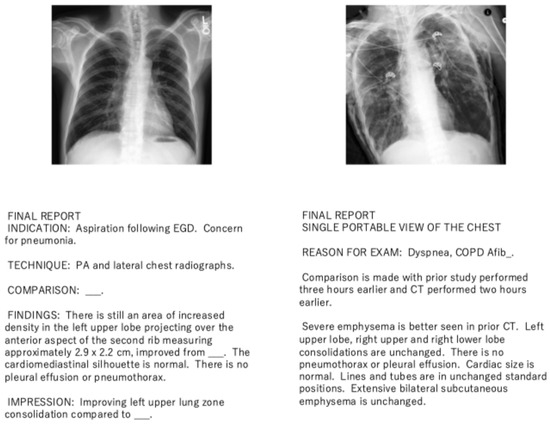
Figure 2.
Example of chest X-ray images and corresponding radiology reports used in this study. The reports may contain FINDINGS and IMPRESSION sections, and if both are absent, the final paragraph is used instead.
3.2. Baselines
The following comparison methods were used to evaluate the performance of the proposed method.
- BioMedCLIP: This is a vision-text integrated model that is specialized for the medical field, where 15 million image-text pairs— PMC-15M—were used for pre-training. The joint embedding of medical images and text achieved high performance in various medical tasks. Reproductions were based on the official code and provided a pre-trained model. A fair comparison was made with the proposed method in this study by adding the self-attention layer to the output embedding of BioMedCLIP and performing additional training by combining images and text. This allowed for a more equitable and more appropriate evaluation of the performance comparison with the proposed method. In addition, we designate BioMedCLIP as our primary baseline for state-of-the-art (SOTA) comparison for the following reasons:
- Many existing studies on multimodal retrieval in the medical domain do not release their code or pre-trained models, making direct replication under the same experimental conditions difficult.
- BioMedCLIP provides an officially released pre-trained model and code, ensuring reproducibility.
- It has demonstrated high performance in various public medical image-text tasks and is currently recognized as one of the most effective open-source models.
- Therefore, we treat BioMedCLIP as a practical SOTA benchmark to highlight the effectiveness of our proposed method, given the challenges in directly comparing with other methods under identical settings.
- Simple combination method: In this method, image and text embeddings were integrated using straightforward techniques such as Concat (concatenation), Sum(α) (weighted sum), and Mul (element-wise multiplication). These combination methods were applied to embeddings extracted from the final layer of BioMedCLIP + Self-Attention to enable fair comparison under additional training conditions. The Concat method performs concatenation of the image and text embeddings along the feature dimension:where and represent the image and text embeddings, respectively. This method allows the model to learn a fused representation from both modalities. In the Sum(α) method, the integrated embedding is computed as follows:where is a weight parameter that controls the balance between the two modalities. In the Mul method, the element-wise multiplication of image and text embeddings is performed:where represents element-wise multiplication. This operation captures the interactions between image and text features and emphasizes overlapping information. By applying these methods, we created three different baseline models to compare with our proposed method. These methods facilitate the analysis of the impact of different fusion strategies on retrieval performance.
3.3. Experiment Details
In this study, BioMedCLIP remained unchanged, and only the newly added Self-Attention and Cross-Attention layers were fine-tuned. The rationale behind this design choice is as follows: BioMedCLIP is a large-scale pre-trained model designed for medical image and text understanding. Since its feature extraction capability is already optimized for a wide range of medical datasets, re-training or fine-tuning the entire model from scratch would be computationally expensive and unnecessary for our specific retrieval task. Instead, we focused on optimizing the newly introduced layers to improve the alignment between image and text embeddings.
For training, we used the mimic-5x200 dataset described in Section 3.1. Specifically, we used 33,471 image-text pairs for training and 3,720 pairs for validation. The training process was conducted using a learning rate of 1 × 10−4, a batch size of 32, and a total of 30 epochs. The Adam optimizer was used with a dropout rate of 0.1. The Cross-Attention layer consisted of 8 attention heads.
We applied Supervised Contrastive Loss (SupConLoss) as described in Section 2.3. During training, image-text pairs from the same disease category were encouraged to have similar embeddings, while pairs from different categories were pushed apart. This contrastive learning approach ensured that the model effectively captured the semantic relationships between medical images and their corresponding reports.
The training convergence was monitored using the loss function, and we observed that the model stabilized around epoch 10 to 12, indicating that additional training beyond this point provided negligible improvements. All experiments were run on an NVIDIA Tesla V100 GPU.
To ensure fair comparisons, we also trained a model where a Self-Attention layer was added instead of a Cross-Attention layer. By doing so, we aimed to verify whether the improvement in performance was specifically due to the interaction modeling capability of Cross-Attention. As discussed in Section 4, our results confirm that fine-tuning the added layers alone is sufficient to achieve superior retrieval accuracy without requiring the full re-training of BioMedCLIP.
3.4. Results
The retrieval performance was evaluated using mAP and Precision@K. As described in Section 2.4, these metrics measure the relevance of retrieved cases.
Table 1 presents the results of the retrieval tasks (Image-Image, Text-Text, Image-Text, and Text-Image) using the proposed and comparison (BioMedCLIP) methods. The proposed method outperformed the comparison method in all tasks. Notably, the proposed method achieved an mAP of 0.999, outperforming BioMedCLIP-based methods, which achieved a maximum mAP of 0.977 in the Text-Text setting. This indicates that integrating image and text embeddings with cross-attention enables more accurate retrieval results. Furthermore, we compared the proposed method both with and without positional embeddings, as shown in Table 1. This comparison is analyzed in Section 4.

Table 1.
Retrieval task results for mimic-5x200 dataset (the best values are boldened).
Table 2 lists the results of comparing the performance of the proposed method with that of a simple combination method. The results indicate that the proposed cross-attention method outperforms simple fusion techniques such as Concat, Sum(α), and Mul. Specifically, even the best-performing weighted addition (Sum(0.1)) achieved an mAP of 0.980, which is lower than the proposed method’s 0.999. These results suggest that straightforward embedding fusion does not effectively capture the interactions between image and text modalities, whereas cross-attention enables a deeper and more meaningful integration.

Table 2.
Comparison with the simple combination method using the mimic-5x200 dataset (the best values are boldened).
We also evaluated the performance of the proposed method when the query and key/value settings were switched. Specifically, when the query was set to text embedding and the key/value was set to image embedding, the mAP dropped to 0.787, indicating a decreased performance. This suggests that using images as queries and text as keys/values represents a more effective configuration for retrieval, which is likely attributable to the higher information density of text data.
Figure 3 presents retrieval results for three different methods: Image&Text-Image&Text (Proposed), Image-Image, and Image-Text. The retrieved results are ranked, with green-bordered elements representing successful retrievals and red-bordered elements indicating errors. Additionally, the dotted-line elements represent information not used in the retrieval process but included for reference. The proposed method utilizes both image and text inputs for retrieval, while the comparison methods rely on either image or text alone. These retrieval results highlight the advantages of the proposed method in multimodal retrieval tasks, demonstrating its ability to leverage both visual and textual information effectively. A detailed analysis of these qualitative results is provided in Section 4.
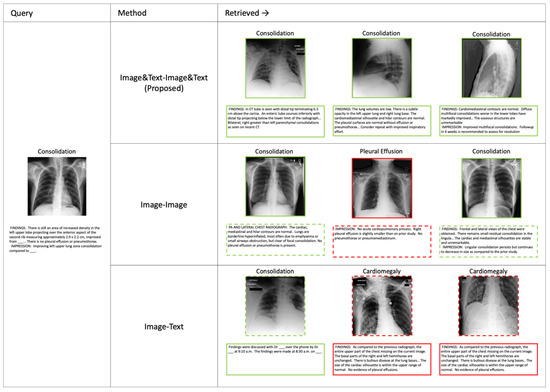
Figure 3.
Retrieval results using different methods. The Image&Text-Image&Text (Proposed), Image-Image, and Image-Text retrieval results are shown. Green-bordered elements indicate correct retrievals, while red-bordered elements indicate incorrect ones.
4. Discussion
In this study, we proposed an integrated representation of images and text and evaluated it in a retrieval task to demonstrate its effectiveness. The proposed method achieved higher performance than conventional methods, with an overwhelming accuracy of 1.00 for P@1, signifying that the appropriate retrieval results could be presented as the first result for all queries. This result suggests that the proposed method could effectively utilize the complementary information of both modalities by treating image and text information in an integrated manner. Conventional methods treated images and text as independent embeddings and retrieved them separately. However, cross-attention enabled the direct modeling of the relevance between images and text in our study. This characteristic allows for retrieval that considers complex information interactions, which produce highly accurate results.
In addition, we investigated the effect of using positional embeddings in the proposed method. Table 1 shows that the proposed method without positional embeddings consistently outperformed the version with positional embeddings. This suggests that incorporating positional embeddings may introduce unnecessary constraints, limiting the flexibility of the attention mechanism. In medical text, keywords such as disease names and symptoms are more critical than sentence structure, making positional information less relevant. Furthermore, since our method aligns image and text embeddings based on semantic relationships rather than strict positional order, removing positional embeddings enhances the model’s ability to extract key information from medical reports effectively. These findings indicate that positional embeddings are not necessarily beneficial when integrating medical images and text in a retrieval system.
Additionally, the proposed method performed exceedingly well compared to the simple combination method, supporting the effectiveness of advanced information fusion using cross-attention. The simple combination method only mechanically combines image and text embeddings and does not sufficiently consider the relevance and interaction between them. We speculated that this limits the expressive power of the integrated embedding and decreases retrieval accuracy.
We also evaluated changes to the query and key/value settings in the proposed method and uncovered that it performed highest when images were used as queries and text as keys/values. Meanwhile, using text as the query and images as the keys/values resulted in an mAP of 0.787, confirming a decrease in performance. This result is possibly due to the fact that text has a high information density and can obtain highly accurate embedding representations when used as values. Specifically, in cross-attention, values are the basis for the final output; therefore, the information density of the data used as values directly influences the results. Using text as a value effectively utilizes its characteristics; however, performance improvement is assumed to be limited for images due to their ambiguity.
Furthermore, Figure 3 presents qualitative retrieval results that complement the quantitative findings. The proposed Image&Text-Image&Text method consistently retrieves correct results, highlighting the advantage of leveraging multimodal information. In contrast, the Image-Image method frequently retrieves visually similar but incorrect images, as it relies solely on image features without textual context. This suggests that the lack of text-based information limits its ability to retrieve relevant cases accurately. Meanwhile, the Image-Text method struggles to align image and text embeddings effectively, leading to incorrect retrievals. These qualitative results support the superiority of the proposed cross-attention-based method in medical image retrieval, demonstrating its ability to integrate both modalities to improve search accuracy.
The retrieval method proposed in this study, which handles images and text in an integrated manner, is useful in education for residents and case collection due to its characteristics. The proposed method requires images and text as queries and has limitations in diagnostic support; however, the following application scenarios reveal its high practicality.
- 1.
- Application in medical education
Using similar previous cases is effective when medical students and residents conduct case studies [11]. The proposed method provides an opportunity to specifically learn about the diagnostic process and characteristics of cases by inputting images and corresponding diagnostic reports as queries and retrieving similar cases with high accuracy.
- 2.
- Application in case collection
Regarding medical research, a retrieval method that considers the interaction between images and text can increase the selection of cases as research and survey subjects. The proposed method can efficiently and accurately collect highly relevant cases using images and text. This is expected to improve the quality of research by enabling the identification of cases that existing methods may overlook.
- 3.
- Limitations in diagnostic support
Only images are assumed to be used as queries for diagnostic support; therefore, the direct application of the proposed method is limited. Particularly, text information is often unavailable before the physician writes the diagnosis, and using the characteristics of the proposed method would be challenging in this case. However, the applicable scenario can be expanded using this method in a complementary manner to existing diagnostic support systems.
Thus, we proposed a new retrieval method that handles images and text in an integrated manner and demonstrated its effectiveness; nevertheless, limitations and challenges remain.
- 1.
- Applicability and limitations of the proposed method
The proposed method requires images and text as queries; therefore, its application may be limited as diagnostic support. Only images are assumed to be prepared as queries when a physician makes a diagnosis, making the direct use of the proposed method challenging. This aspect needs to be resolved by complementary use with a diagnostic support system or the development of technology to complement text information in the future.
- 2.
- Bias due to characteristics of the experimental dataset
In this study, we conducted experiments using the mimic-5x200 dataset that was extracted from the MIMIC-CXR dataset. However, only five specific classes (atelectasis, cardiomegaly, consolidation, edema, and pleural effusion) were targeted; thus, its applicability to other diseases and more diverse classes needs to be evaluated. In addition, the MIMIC-CXR dataset is collected from a single hospital system in the United States and may reflect specific demographic characteristics and clinical practices. This can lead to biases in disease prevalence, imaging protocols, and patient demographics, making the model less generalizable to other regions or healthcare settings. Furthermore, MIMIC-CXR contains English text data, so its versatility is limited regarding non-English-language data. Consequently, models trained on this dataset may exhibit performance degradation when applied to medical data in other languages or when the clinical documentation style differs significantly. These forms of dataset bias potentially impact real-world applications. For instance, if the model is deployed in a region with different patient populations or languages, diagnostic performance and retrieval accuracy could decrease. Similarly, focusing on five classes in the present experiments may not cover complexities such as comorbidities or rarer diseases that appear in real-world clinical settings. Simon et al. [17] also highlighted such issues in their review of multimodal AI, noting that data scarcity and potential biases in publicly available datasets (e.g., MIMIC) remain critical challenges. In agreement with their observations, our study underscores how limited disease categories and single-institution data can constrain the generalizability of retrieval performance. Although this study focused on a limited number of conditions, our approach is not fundamentally limited to these specific classes. Since the method learns the relationships between image and text representations, it has the potential to generalize to other diseases by incorporating additional and diverse training data. Future work will include experiments on a broader set of conditions to validate its generalizability and address the need for multi-language, multi-regional datasets to reduce biases.
The following research is necessary for overcoming these limitations:
- Design of a flexible model that can handle cases where images and text are partially missing.
- Additional experiments using medical datasets in other languages.
- Extension and evaluation of the model to handle more diverse diseases and classes.
Resolving these challenges would enable the proposed method to evolve into a technology with wider applicability and promote its use in medical settings and research fields.
5. Conclusions
In this study, we proposed a new retrieval method that handles images and text in an integrated manner and verified its effectiveness in a retrieval task. The proposed method demonstrated overwhelmingly higher performance than the conventional method, suggesting its high practicality in applications such as education and case collection. Meanwhile, limitations to the method’s direct application as a diagnostic support indicate the need for additional research to expand its applicability. Future research would evaluate the method using more diverse datasets and develop a model that can handle cases where images and text are missing.
Author Contributions
Conceptualization, I.S.; Methodology, I.S. and M.K.; Software, I.S. and M.K.; Validation, I.S., M.A. and M.K.; Formal Analysis, I.S., M.A. and M.K.; Investigation, I.S.; Resources, I.S. and M.K.; Data Curation, I.S., M.A. and M.K.; Writing—Original Draft Preparation, I.S.; Writing—Review & Editing, I.S., M.A. and M.K.; Visualization, I.S.; Supervision, M.A.; Project Administration, M.A. and M.K.; Funding Acquisition, M.A. All authors have read and agreed to the published version of the manuscript.
Funding
METI R&D Support Program for Growth-oriented Technology SMEs Grant Number JPJ005698.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The MIMIC-CXR dataset used in this study is available on PhysioNet at https://physionet.org/content/mimic-cxr/2.0.0/ (accessed on 16 February 2025). Access to the dataset is restricted and requires the following: (1) credentialed user status, (2) completion of the CITI Data or Specimens Only Research training, and (3) signing of the Data Use Agreement (DUA). Further details on accessing the dataset can be found at the provided link. The code used in this study is available at the GitHub repository “MultimodalRetrieval” (https://github.com/193sata/MultimodalRetrieval (accessed on 16 February 2025)) under the MIT License. This release complies with the requirements of the Data Use Agreement (DUA) for the MIMIC-CXR dataset. Further inquiries can be directed to the corresponding author.
Acknowledgments
We would like to thank Yoshiharu Nakayama and Manabu Sugawara from Y’s READING Co., Ltd. for their valuable advice on the interpretation of medical data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Çallı, E.; Sogancioglu, E.; van Ginneken, B.; van Leeuwen, K.G.; Murphy, K. Deep learning for chest X-ray analysis: A survey. Med. Image Anal. 2021, 72, 102125. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. “Medical Imaging”. U.S. Food and Drug Administration. Available online: https://www.fda.gov/radiation-emitting-products/radiation-emitting-products-and-procedures/medical-imaging (accessed on 2 December 2024).
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Webster, D.R. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Ronneberger, O. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Lungren, M.P. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef] [PubMed]
- Moor, M.; Banerjee, O.; Abad, Z.S.H.; Krumholz, H.M.; Leskovec, J.; Topol, E.J.; Rajpurkar, P. Foundation models for generalist medical artificial intelligence. Nature 2023, 616, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Azizi, S.; Driess, D.; Schaekermann, M.; Amin, M.; Chang, P.C.; Natarajan, V. Towards generalist biomedical AI. NEJM AI 2024, 1, AIoa2300138. [Google Scholar] [CrossRef]
- Schlegl, T.; Waldstein, S.M.; Vogl, W.D.; Schmidt-Erfurth, U.; Langs, G. Predicting semantic descriptions from medical images with convolutional neural networks. In Proceedings of the International Conference on Information Processing in Medical Imaging, Isle of Skye, UK, 28 June–3 July 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 437–448. [Google Scholar]
- Shin, H.C.; Lu, L.; Kim, L.; Seff, A.; Yao, J.; Summers, R.M. Interleaved text/image deep mining on a large-scale radiology database for automated image interpretation. J. Mach. Learn. Res. 2016, 17, 1–31. [Google Scholar]
- Müller, H.; Michoux, N.; Bandon, D.; Geissbuhler, A. A review of content-based image retrieval systems in medical applications—Clinical benefits and future directions. Int. J. Med. Inform. 2004, 73, 1–23. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, H.; Miura, Y.; Manning, C.D.; Langlotz, C.P. Contrastive learning of medical visual representations from paired images and text. In Proceedings of the Machine Learning for Healthcare Conference, Durham, NC, USA, 5–6 August 2022; PMLR: New York, NY, USA, 2022; Volume 182, pp. 2–25. [Google Scholar]
- Huang, S.C.; Shen, L.; Lungren, M.P.; Yeung, S. Gloria: A multimodal global-local representation learning framework for label-efficient medical image recognition. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 3942–3951. [Google Scholar]
- Wang, Z.; Wu, Z.; Agarwal, D.; Sun, J. Medclip: Contrastive learning from unpaired medical images and text. arXiv 2022, arXiv:2210.10163. [Google Scholar]
- Zhang, S.; Xu, Y.; Usuyama, N.; Xu, H.; Bagga, J.; Tinn, R.; Poon, H. BiomedCLIP: A multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs. arXiv 2023, arXiv:2303.00915. [Google Scholar]
- Simon, B.D.; Ozyoruk, K.B.; Gelikman, D.G.; Harmon, S.A.; Türkbey, B. The future of multimodal artificial intelligence models for integrating imaging and clinical metadata: A narrative review. Diagn. Interv. Radiol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Ou, W.; Chen, Y.; Liang, L.; Gou, J.; Xiong, J.; Zhang, J.; Lai, L.; Zhang, L. Cross-modal retrieval of chest X-ray images and diagnostic reports based on report entity graph and dual attention. Multimed. Syst. 2025, 31, 58. [Google Scholar] [CrossRef]
- Jeong, J.; Tian, K.; Li, A.; Hartung, S.; Adithan, S.; Behzadi, F.; Calle, J.; Osayande, D.; Pohlen, M.; Rajpurkar, P. Multimodal Image-Text Matching Improves Retrieval-based Chest X-ray Report Generation. In Medical Imaging with Deep Learning; PMLR: Nashville, TN, USA, 2023; Volume 227, pp. 978–990. [Google Scholar]
- Vaswani, A. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5999–6009. [Google Scholar]
- Dosovitskiy, A. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Poon, H. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans. Comput. Healthc. 2021, 3, 1–23. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Khosla, P.; Teterwak, P.; Wang, C.; Sarna, A.; Tian, Y.; Isola, P.; Krishnan, D. Supervised contrastive learning. Adv. Neural Inf. Process. Syst. 2020, 33, 18661–18673. [Google Scholar]
- Weinberger, K.Q.; Saul, L.K. Distance metric learning for large margin nearest neighbor classification. J. Mach. Learn. Res. 2009, 10, 207–244. [Google Scholar]
- Sohn, K. Improved deep metric learning with multi-class n-pair loss objective. Adv. Neural Inf. Process. Syst. 2016, 29, 1857–1865. [Google Scholar]
- Johnson, A.; Lungren, M.; Peng, Y.; Lu, Z.; Mark, R.; Berkowitz, S.; Horng, S. MIMIC-CXR-JPG—Chest Radiographs with Structured Labels. PhysioNet 2019, Version 2.0.0. Available online. Available online: https://physionet.org/content/mimic-cxr-jpg/2.0.0/ (accessed on 2 December 2024).
- Johnson, A.E.; Pollard, T.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.Y.; Peng, Y.; Horng, S. MIMIC-CXR-JPG: A large publicly available database of labeled chest radiographs. arXiv 2019, arXiv:1901.07042. [Google Scholar]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.C.; Mark, R.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Pollard, T.; Mark, R.; Berkowitz, S.; Horng, S. MIMIC-CXR Database. PhysioNet 2019, Version 2.1.0. Available online: https://physionet.org/content/mimic-cxr/2.1.0/ (accessed on 2 December 2024).
- Johnson, A.E.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.Y.; Horng, S. MIMIC-CXR: A de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).