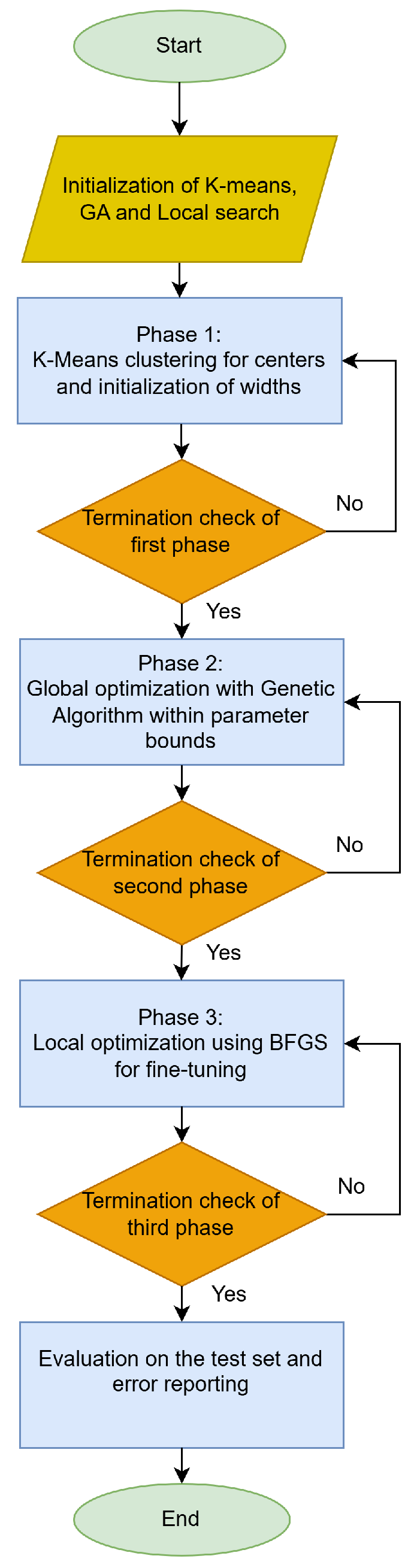
4.1. Experimental Results
The experiments were conducted on a Debian Linux system with 128 GB of RAM, and all the necessary code was implemented in the C++ programming language. Also, the OPTIMUS computing environment [
101], available from
https://github.com/itsoulos/GlobalOptimus.git (accessed on 9 October 2025), was used for the optimization methods. Ten-fold cross-validation was employed to validate the experimental results. The average classification error is calculated as follows:
The set
T denotes the associated test set, where
. Similarly, the average regression error has the following definition:
Table 3 contains the values for each parameter of this method.
In the results tables that follow, the columns and rows have the following meanings:
The column DATASET is used to represent the name of the used dataset.
The results from the incorporation of the BFGS procedure [
102] to train an artificial neural network [
103,
104] with 10 weights are presented in the column under the title BFGS.
The ADAM column presents the results obtained by training a 10-weight neural network using the ADAM local optimization technique [
105,
106].
The column RBF-KMEANS is used here to denote the usage of the initial training method of RBF networks to train an RBF network with 10 nodes.
The column NEAT (NeuroEvolution of Augmenting Topologies) [
107] stands for the method NEAT incorporated in the training of neural networks.
The column DNN stands for the incorporation of a deep neural network, as implemented in the Tiny Dnn library, which can be downloaded freely from
https://github.com/tiny-dnn/tiny-dnn (accessed on 9 October 2025). The optimization method AdaGrad [
108] was incorporated for the training of the neural network.
The BAYES column presents results obtained using the Bayesian optimizer from the open-source BayesOpt library [
109], applied to train a neural network with 10 processing nodes.
The column GENRBF stands for the method introduced in [
110] for RBF training.
The column PROPOSED is used to represent the results obtained by the current work.
The row denoted as AVERAGE summarizes the mean regression or classification error calculated over all datasets.
In the experimental results, boldface highlighting was used to make it clear which of the machine learning techniques has the lowest error on each dataset.
Table 4 compares the performance of eight methods on thirty-three classification datasets. The mean percentage error clearly shows that the proposed method is best overall at 19.45%, followed by DNN at 25.52% and then BAYES at 27.21%, RBF-KMEANS at 28.54%, NEAT at 32.77%, BFGS at 33.50%, ADAM at 33.73%, and GENRBF at 34.89%. Relative to the strongest competitor, DNN, the proposed method lowers the average error by 6.07 points, about 24%. The reduction versus the classical BFGS and ADAM is about 14 points, roughly 42%, and versus RBF-KMEANS about 9.1 points, roughly 32%.
At the level of individual datasets, the proposed method delivers strikingly low errors in several cases. On Spiral, it drops to 13.26% while others are around 45–50%; on Wine it reaches 9.47% versus 21–60%; and on Wdbc it achieves 5.54% versus 7–35%. On Z_F_S, ZO_NF_S, ZONF_S, and Cleveland it attains the best or tied-best results. On Heart, HeartAttack, Statheart, Regions2, Saheart, Pima, Australian, Alcohol, and HouseVotes, the results are also highly competitive, usually best or within the top two. There are, however, datasets where other methods prevail: DNN clearly leads on Segment and HouseVotes and is very strong on Dermatology; RBF-KMEANS is best on Appendicitis; and ADAM wins narrowly on Student and Balance. In cases like Balance, Popfailures, Dermatology, and Segment, the proposed method is not the top performer, though it remains competitive.
In summary, the proposed method not only attains the lowest average error but also consistently outperforms a broad range of classical and contemporary baseline methods. Despite local exceptions where DNN, ADAM, or RBF-KMEANS come out ahead, the approach appears more generalizable and stable, achieving systematically low errors and large improvements on challenging datasets, which supports its practical use as a default choice for classification.
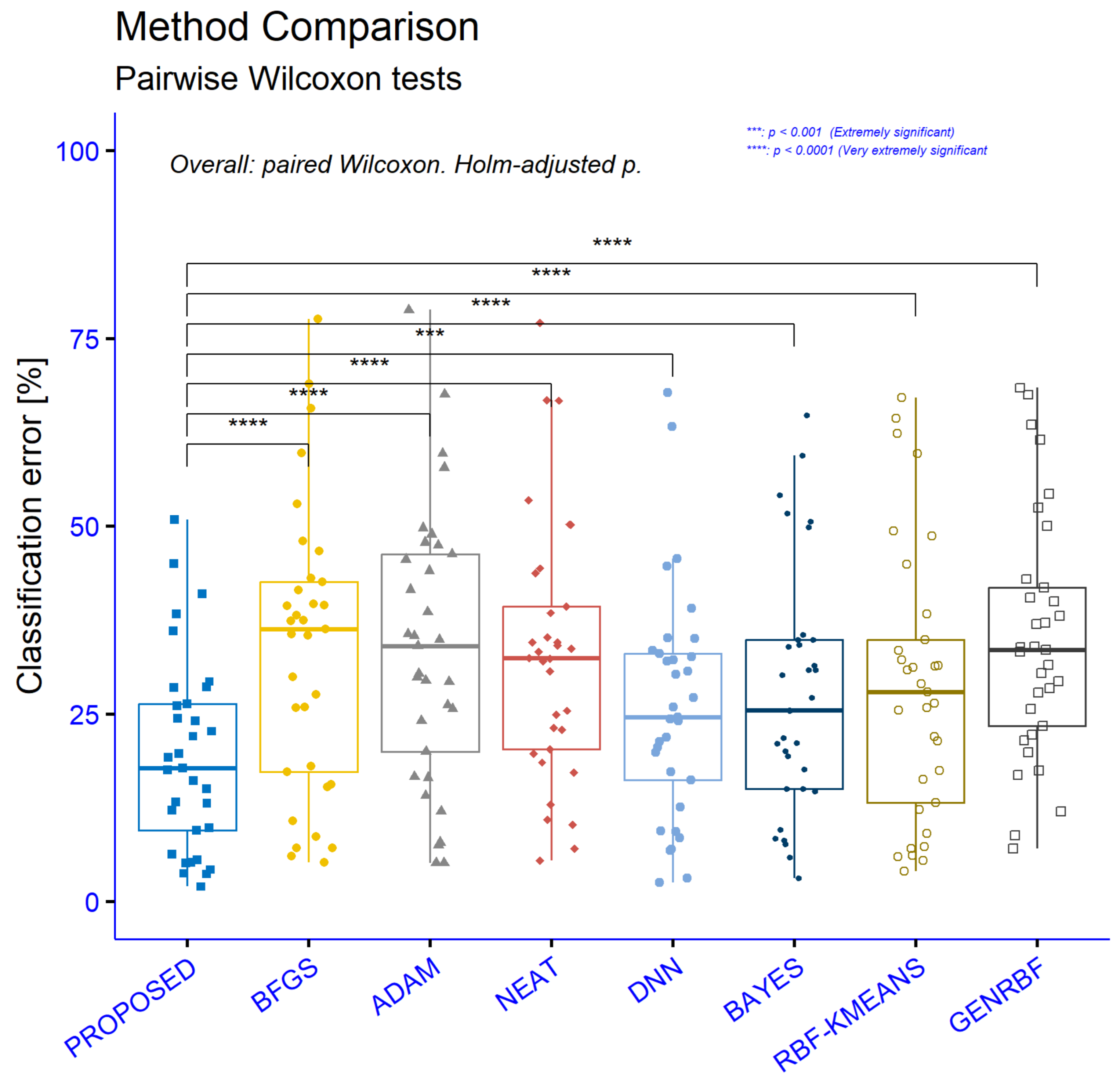
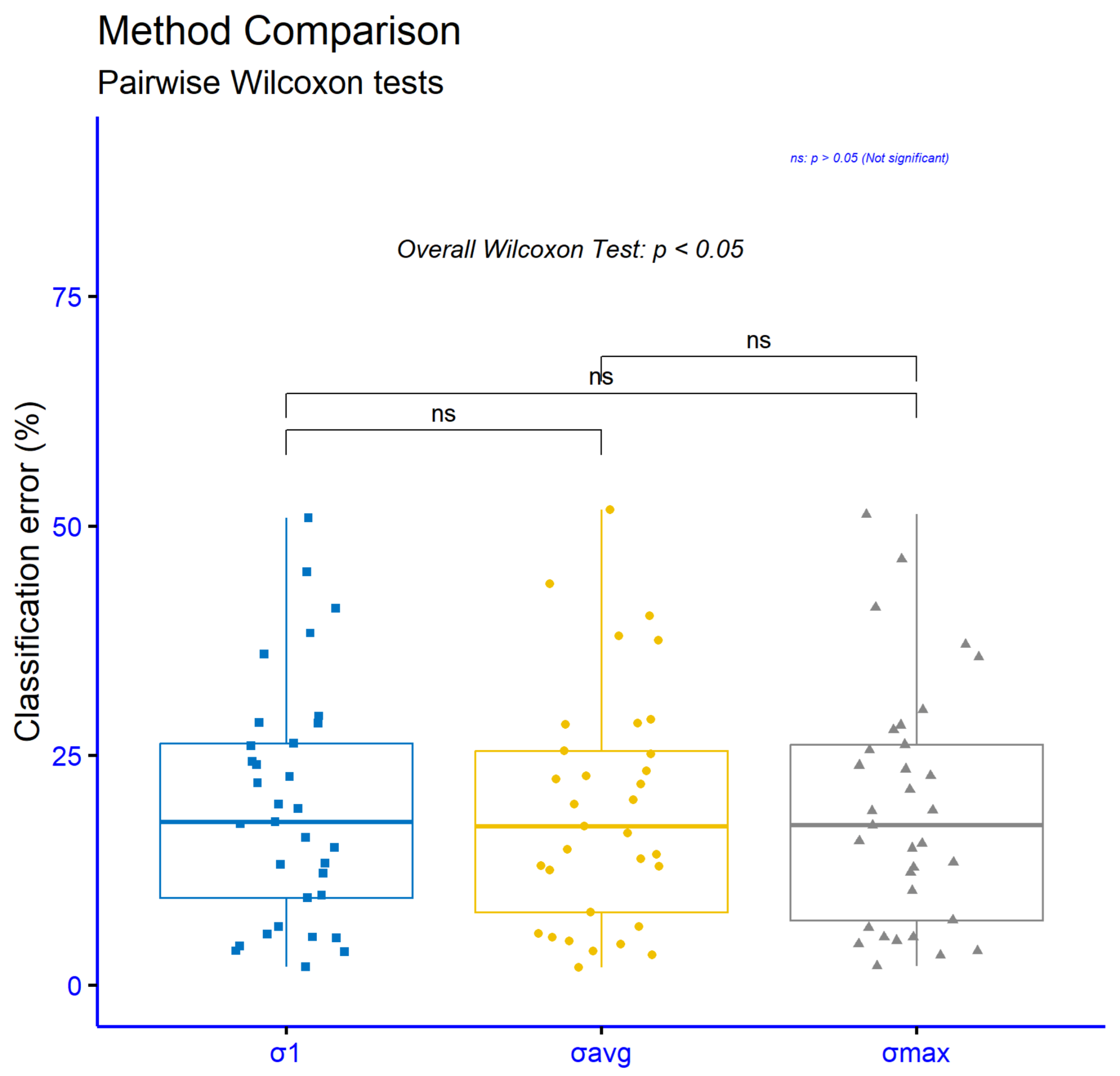
Figure 6 and
Table 5 summarize paired Wilcoxon signed-rank tests comparing the PROPOSED method against each competitor on the same 33 classification datasets. In all statistical figures The asterisks correspond to
p-value thresholds as follows:
ns: p > 0.05 (not statistically significant)
*: (significant)
**: (highly significant)
***: (extremely significant)
****: (very extremely significant)
The column n is the number of paired datasets, V is the Wilcoxon signed-rank statistic, is the rank-biserial effect size (range −1 to 1, with more a negative value meaning PROPOSED has a lower error), and give the 95% Hodges–Lehmann confidence interval for the median paired difference (PROPOSED–competitor) in percentage point error, p is the raw p-value, is the Holm-adjusted p-value, and is the significance code. Because all confidence intervals are entirely negative, the PROPOSED method consistently shows a lower error than each baseline, not just statistical significance but a stable direction of effect across datasets. Adjusted p-values remain very small in every comparison, from (vs. GENRBF) up to (vs. DNN), yielding **** everywhere except the DNN comparison, which is ***. Effect sizes are uniformly large in magnitude. The strongest difference is against GENRBF, with r ≈ −0.998 and a 95% CI for the median error reduction of roughly −19.12 to −12.01 percentage points. Very large effects also appear versus NEAT (r ≈ −0.980, CI ≈ [−18.40, −7.15]) and RBF-KMEANS (r ≈ —0.977, CI ≈ [−12.11, −4.90]). Comparisons with BFGS (r ≈ −0.954, CI ≈ [−18.51, −7.96]) and ADAM (r ≈ −0.943, CI ≈ [−19.42, −8.03]) remain strongly favorable. The smallest, yet still large, effect is against DNN (r ≈ −0.882), with a clearly negative CI ≈ [−8.93, −3.09]. Taken together, the results show consistent, substantial reductions in classification error for the PROPOSED method across all baselines, with very large effect sizes, tight negative confidence intervals, and significance that survives multiple-comparison correction.
Table 6 further illustrates the comparison of precision and recall on the classification datasets between the conventional RBF training method and the proposed technique.
Table 7 evaluates the performance of eight regression methods on twenty-one datasets using absolute errors. The average error shows a clear overall advantage for the proposed method at 5.87, followed by BAYES at 9.18, RBF-KMEANS at 9.56, DNN at 11.82, NEAT at 13.99, GENRBF at 13.38, ADAM at 21.39, and BFGS at 28.82. Relative to the best-competing average, BAYES, the proposed method reduces the error by about 3.31 points (≈36%). The reduction versus RBF-KMEANS is about 3.69 points (≈39%), versus DNN about 5.95 points (≈50%), and relative to NEAT and GENRBF the drops are roughly 58% and 56%, respectively. The advantage is even larger against ADAM and BFGS, where the mean error is nearly halved or more.
Across individual datasets, the proposed method attains the best value in roughly two thirds of the cases. It is clearly first on Auto, BL, Concrete, Dee, Housing, FA, HO, Mortgage, PL, Plastic, Quake, Stock, and Treasury, with particularly large margins on Housing and Stock where errors fall to 15.36 and 1.44 while other methods range from tens to hundreds. On Airfoil it is essentially tied with the best at 0.004, while BFGS is slightly lower at 0.003. There are datasets where other methods lead, such as Abalone where ADAM and BAYES are ahead; Friedman and Laser where BFGS gives the best value; BK where DNN and RBF-KMEANS lead; and PY where RBF-KMEANS is lower. Despite these isolated exceptions, the proposed method remains consistently among the top performers and most often the best.
Overall, the proposed approach combines a very low average error with broad superiority across diverse problem types and error scales, from thousandths to very large magnitudes. The consistency of the gains and the size of the margins over all baselines indicate that it is the most efficient and generalizable choice among the regression methods considered.
Figure 7 and
Table 8 summarize paired Wilcoxon signed-rank tests between PROPOSED and each method on the same regression datasets. In every comparison the 95% confidence interval is entirely negative, so PROPOSED consistently attains a lower error than each baseline. Holm-adjusted
p-values range from about
to 0.0063, yielding ** or *** across all pairings, indicating strong though not extreme significance. Effect sizes are very large in absolute value, implying a consistent sign of the differences across datasets. The strongest dominance is against NEAT with r ≈ −1 and V = 0, meaning that in every non-tied pair PROPOSED was better, with a confidence interval of roughly [−9.93, −0.16]. Similarly large effects appear against GENRBF (r ≈ −0.976, CI [−10.25, −0.098]) and RBF-KMEANS (r ≈ −0.945, CI [−4.52, −0.034]); the upper bound near zero indicates that the typical improvement can range from very small to several points depending on the dataset. Against BFGS and ADAM the effects remain very large (r ≈ −0.889 and r ≈ −0.857, respectively) with wider intervals [−20.51, −0.316] and [−27.34, −0.0416], showing substantial heterogeneity in the magnitude of error reduction while the direction remains in favor of PROPOSED. The most challenging comparison is with DNN: although |r| is still very large (≈0.900), the CI is narrow and close to zero [−11.09, −0.012], implying that while superiority is consistent, the typical error reduction may be small in many cases.
Overall, the results demonstrate that PROPOSED systematically outperforms all alternatives on regression, with very large rank-based effect sizes, negative and robust confidence intervals, and significance that survives multiple-comparison correction. The strength of the improvement varies by problem and is more modest against DNN, but the direction of the effect is consistently in favor of PROPOSED across all comparisons.
4.2. Experiments with Different Values of Scale Factor F
In order to evaluate the stability and reliability of the current work when its critical parameters are altered, a series of additional experiments were executed. In one of them, the stability of the technique was studied with the change in the scale factor F. This factor regulates the range of network parameter values and is scaled as a multiple of the initial estimates obtained from the first-phase K-means method. In this series of experiments, the value of F was altered in the range .
The effect of the scale factor
F on the performance of the proposed machine learning model is presented in
Table 9. The parameter
F takes four different values, 1, 2, 4, and 8, and for each dataset the classification error rate is reported. Analyzing the mean values, it is observed that
and
achieve the best overall performance, with average errors of 19.45% and 18.53%, respectively, compared to 20.99% for
and 18.60% for
. This indicates that selecting an intermediate value of the initialization factor improves performance, reducing the error by about two percentage points relative to the baseline case of
. At the individual dataset level, interesting patterns emerge. For example, on Sonar the error drops significantly from 32.90% at
to 18.75% at
, suggesting that the parameter
F strongly influences performance in certain problems. In contrast, on Spiral increasing
F worsens the results, as the error rises from 12.03% at
to 23.56% at
. Similarly, on the Australian dataset a gradual increase in
F from 1 to 8 systematically improves performance, reducing the error from 24.04% to 20.59%. Overall, the data show that the effect of the scale factor is not uniform across all problems, but the general trend indicates improvement when
F increases from 1 to 2 or 4. Choosing
appears to yield the best mean result, although the difference compared with
is very small. Therefore, it can be concluded that tuning this parameter plays an important role in the stability and accuracy of the model and that intermediate values such as 4 constitute a good general choice.
Table 10 shows the effect of the scale factor
F on the performance of the proposed regression model. Based on the mean errors, the best overall performance occurs at
with an average error of 5.68, while the values for
and
are 5.94, 5.87, and 5.78, respectively. The differences across the four settings are not large, but they indicate that intermediate values and especially
tend to offer the best accuracy–stability trade-off. At the level of individual datasets, substantial variations are observed. For Friedman the reduction is dramatic, with the error dropping from 6.74 at
to 1.41 at
, highlighting that proper tuning of
F can have a strong impact on performance. Laser shows a similarly large improvement, from 0.027 at
to just 0.0024 at
. Mortgage also improves markedly, from
at
to 0.015 at
. By contrast, on some datasets the value of
F has little practical effect, such as Quake and HO, where errors remain nearly constant regardless of F. There are also cases like Housing where increasing
F degrades performance, with the error rising from 14.64 at
to 18.48 at
. Overall, the results indicate that the scale factor
F has a significant but nonuniform influence on model performance. On some datasets it sharply reduces error, while on others its impact is negligible or even negative. Nevertheless, the aggregate picture based on the mean errors suggests that
and
yield the most reliable results, with
being the preferred choice for a general-purpose setting.
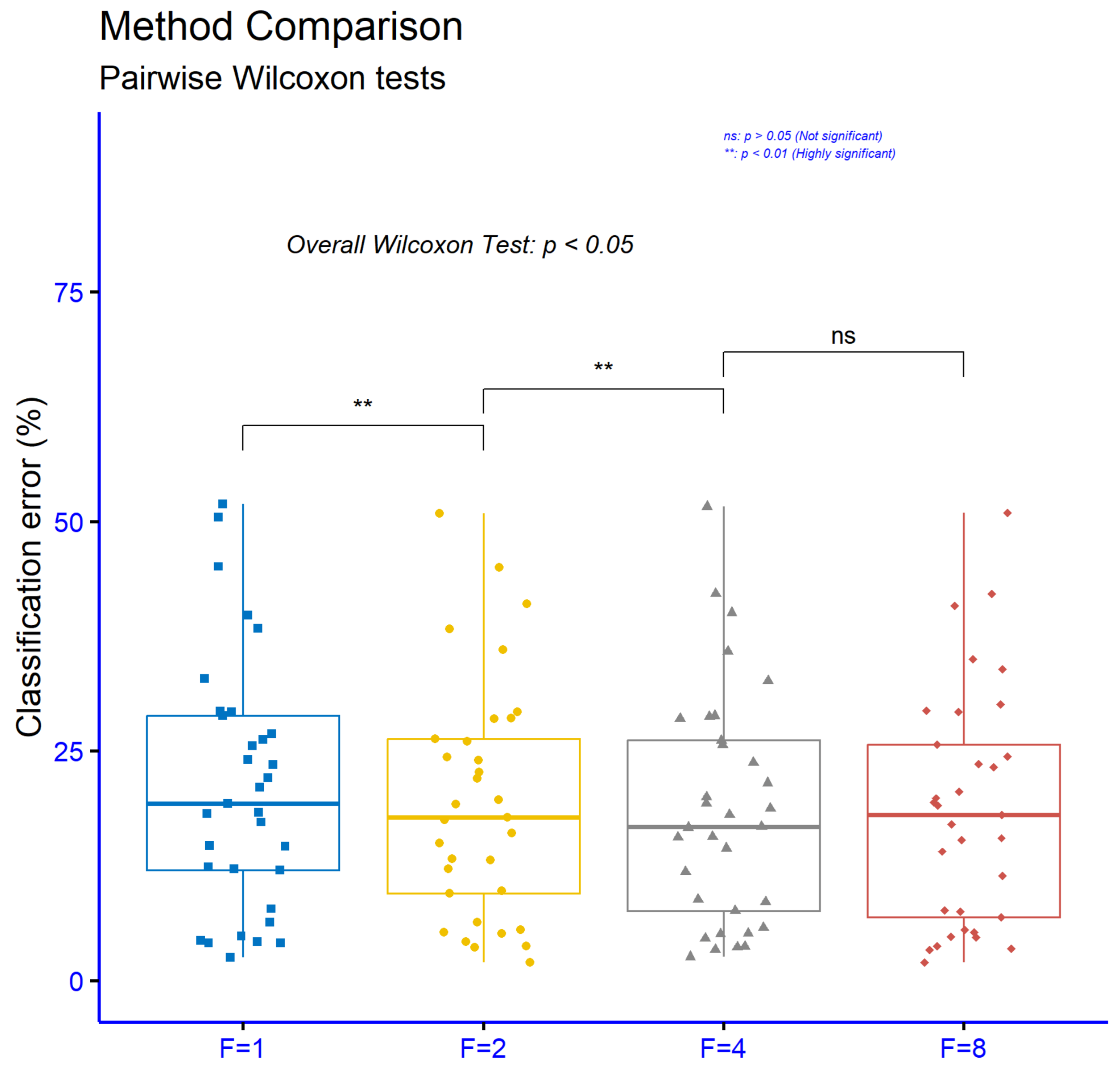
The significance levels for comparisons among different values of the parameter
F in the proposed machine learning method, using the classification datasets, are shown in
Figure 8. The analysis shows that the comparison between
and
results in high statistical significance with
, indicating that the transition from the initial value to
has a substantial impact on performance. Similarly, the comparison between
and
also shows high statistical significance with
, suggesting that further increasing the parameter continues to positively affect the results. However, the comparison between
and
is characterized as not statistically significant, since
, which means that increasing the parameter beyond
does not bring a meaningful difference in performance. Overall, the findings indicate that smaller values of
F play a critical role in improving the model, while increases beyond 4 do not lead to further statistically significant improvements.
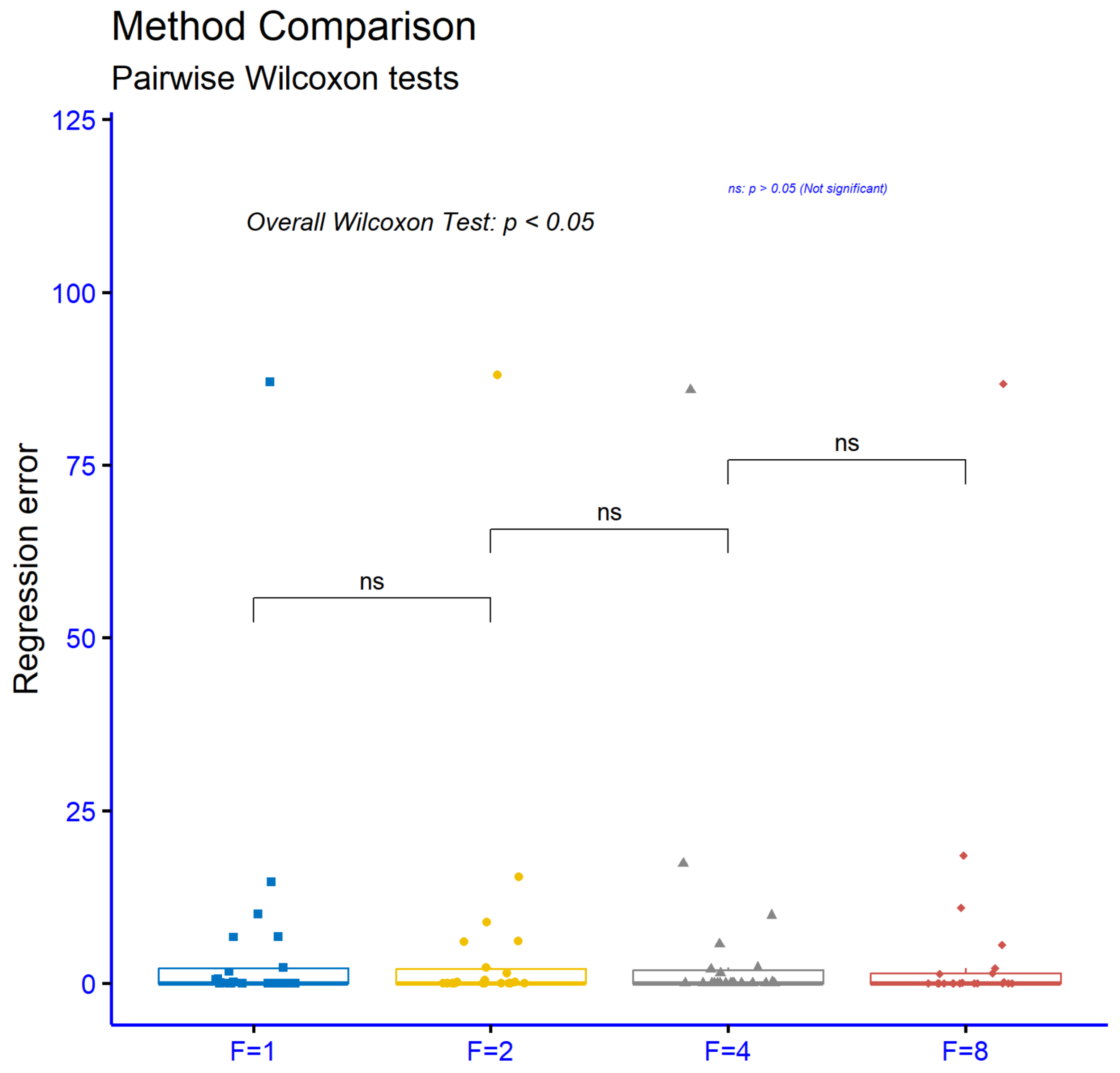
The significance levels for comparisons among different values of the parameter
F in the proposed method, using the regression datasets, are presented in
Figure 9. The results show that none of the comparisons
vs.
,
vs.
, and
vs.
exhibit statistically significant differences, since in all cases
. These results suggest that variations in the parameter
F do not significantly influence the performance of the model on regression tasks. Therefore, it can be concluded that the choice of the
F value is not of critical importance for these datasets and that the model remains stable regardless of the specific setting of this parameter.
4.3. Experiments with Differential Initialization Methods for Variances
The stability of the proposed method was also evaluated by employing a different procedure to determine the range of
parameters for the radial functions. Here, the
parameters were initially estimated using the variance calculated by the K-means algorithm. This calculation scheme is denoted as
in the following experimental tables. In this additional set of experiments, two more techniques were used, which will be denoted as
and
in the following tables. In
the following calculation is performed:
Subsequently
is used to determine the range of values of the
parameters of the radial functions of the network. In
the following quantity is calculated:
This quantity is then employed to define the range of
parameter values for the radial functions.
Table 11 presents the effect of three different calculation techniques for the
parameters used in the radial basis functions of the RBF model. The techniques are a fixed value
, the mean distance-based initialization
, and the maximum distance-based initialization
. Based on the mean errors, the maximum distance technique yields the lowest overall error at 19.18%. Very close is the mean distance technique at 19.27%, while the simple
initialization has a slightly higher error of 19.45%. Although the differences among the three approaches are small, the two adaptive methods
tend to produce marginally better overall performance. At the individual dataset level, behaviors vary. For example, on Wine the
choice reduces error to 7.06%, far below the 9.47% obtained with
. On Dermatology,
performs better than the other two, whereas on Segment the mean-based option is preferable. In some cases the differences are minor, e.g., Circular, Pima, and Popfailures, where all techniques are comparable; in others the choice of technique materially affects performance, as in Transfusion, where error drops from 26.04% with
to about 22.78% with the other two methods. Overall, the statistical picture indicates that no single technique dominates across all datasets. Nevertheless, methods that adapt
to the geometry of the data
tend to yield more reliable and stable results, while the fixed value lags slightly. The average differences are modest, but for certain problems the choice can significantly impact final performance.
Table 12 presents the effect of three different calculation techniques for the
parameters used in the radial basis functions of the RBF model. Based on the mean errors, the average distance method yields the lowest overall error at 5.81. Very close is the fixed value
with a mean error of 5.87, while the maximum distance method shows a slightly higher mean error of 5.96. The difference among the three methods is small, indicating that all can deliver comparable performance at a general level, with a slight advantage for the average distance approach. At the level of individual datasets, however, significant variations are observed. For example, on Mortgage the
method reduces the error dramatically from 0.23 with
to 0.021, while
also provides a much better result with 0.041. On Treasury the improvement is again substantial, as the error decreases from 0.47 with
to just 0.08 using
. On Stock the reduction is clear, from 1.44 to 1.23, while on Plastic both
and
yield lower errors than
. On the other hand, on datasets such as Housing, the use of
worsens performance, increasing the error from 15.36 with
to 19.45. Similarly, on Auto and Baseball the lowest errors are obtained with
, whereas the alternative techniques result in slightly worse performance. Overall, the results show that the choice of calculation technique for
can significantly affect performance in certain problems, while in others the difference is negligible. Although no method consistently outperforms the others across all datasets, the average distance method appears slightly more reliable overall, while the maximum distance method can in some cases produce very large improvements but in others lead to a degradation in performance.
The significance levels for comparisons of various computation methods for the
parameters in the radial basis functions of the proposed model, using the classification datasets, are presented in
Figure 10. The comparisons performed,
vs.
,
vs.
, and
vs.
, did not show any statistically significant differences, since in all cases
. These results indicate that variations in the computation method for the
parameters do not significantly influence the performance of the model on classification tasks. Therefore, it can be concluded that the model maintains stable performance regardless of which of the three computation techniques is used.
The significance levels for comparisons of various methods for computing the
parameters in the radial basis functions of the proposed model, using the regression datasets, are presented in
Figure 11. The comparisons examined,
vs.
,
vs.
, and
vs.
, did not show any statistically significant differences, since in all cases
. This means that the choice of computation method for the
parameters does not have a substantial impact on the performance of the model in regression problems. Therefore, it can be concluded that the model demonstrates stable and consistent behavior regardless of which initialization technique is applied.
4.4. Experiments with the Number of Generations
An additional experiment was executed, where the number of generations was altered from
to
.
Table 13 presents the effect of the number of generations
on the performance of the proposed model. The overall trend is downward: the mean error decreases from 20.56% at 50 generations to 19.46% at 100, essentially stabilizes at 200 with 19.45%, and improves slightly further at 400 to 19.11%. Thus, the largest gain arrives early, from 50 to 100 generations (about 1.1 points), after which returns diminish, with small but tangible additional gains. At the dataset level the behavior varies. There are cases with clear improvements as
increases, such as Alcohol (34.11%→27.02%), Australian (25.23%→21.39%), Ionosphere (13.94%→11.17%), Spiral (16.66%→12.45%), and Z_O_N_F_S (45.14%→38.26%), where more generations yield substantial benefits. In other problems the best value occurs around 100–200 generations and then plateaus or slightly worsens, as on Wdbc (best 4.84% at 100), Student (4.85% at 100), Lymography (21.64% at 100), ZOO (8.70% at 100), ZONF_S (1.98% at 200), and Z_F_S (3.73% at 200). A few datasets show mild degradation with higher
, such as Wine (7.59%→10.24%), Parkinsons (17.32%→17.63%), and to a lesser extent Saheart, indicating that beyond a point further search is not beneficial for all problems. Overall, 100 generations deliver the major error reduction and represent an efficient “sweet spot,” while 200–400 generations extract modest additional gains and, on some datasets, meaningful improvements, at the cost of more computation and occasional local regressions.
Table 14 examines the effect of the number of generations
on the performance of the proposed regression model. At the level of average error, the best value appears at 100 generations with 5.61, marginally better than 50 generations at 5.65, while at 200 and 400 generations the mean error increases slightly to 5.87 and 5.86, respectively. This suggests that most of the benefit is achieved early and that further increasing the number of generations does not yield systematic improvement and may even introduce a small deterioration in overall performance.
Across individual datasets the picture is heterogeneous. Clear improvements with more generations are observed on Abalone, where the error steadily drops to 5.88 at 400 generations; on Friedman, with a continuous decline to 5.66; on Stock, improving to 1.33; and on Treasury, where performance stabilizes at 0.47 from 200 generations onward. In other problems the “sweet spot” is around 200 generations: for example, on Mortgage the error falls from 0.66 to 0.23 at 200 before rising again, on Housing it improves to 15.36 at 200 but worsens at 400, on BL the minimum 0.0004 occurs at 200, and on Concrete and HO there is a small but real improvement near 200. There are also cases where more generations seem to burden performance, such as Baseball and BK, where the error rises as increases. On several datasets the number of generations has little practical effect, with near-constant values on Airfoil, Quake, SN, and PL and only minor fluctuations on Dee, FA, Laser, and FY.
Overall, 100 generations provide an efficient and safe choice with the lowest mean error, while 200 generations can deliver the best results on specific datasets at the risk of small regressions elsewhere. Further increasing to 400 generations does not offer a general gain and may lead to slight degradation in some problems, pointing to diminishing returns and possible overfitting or instability depending on the dataset.
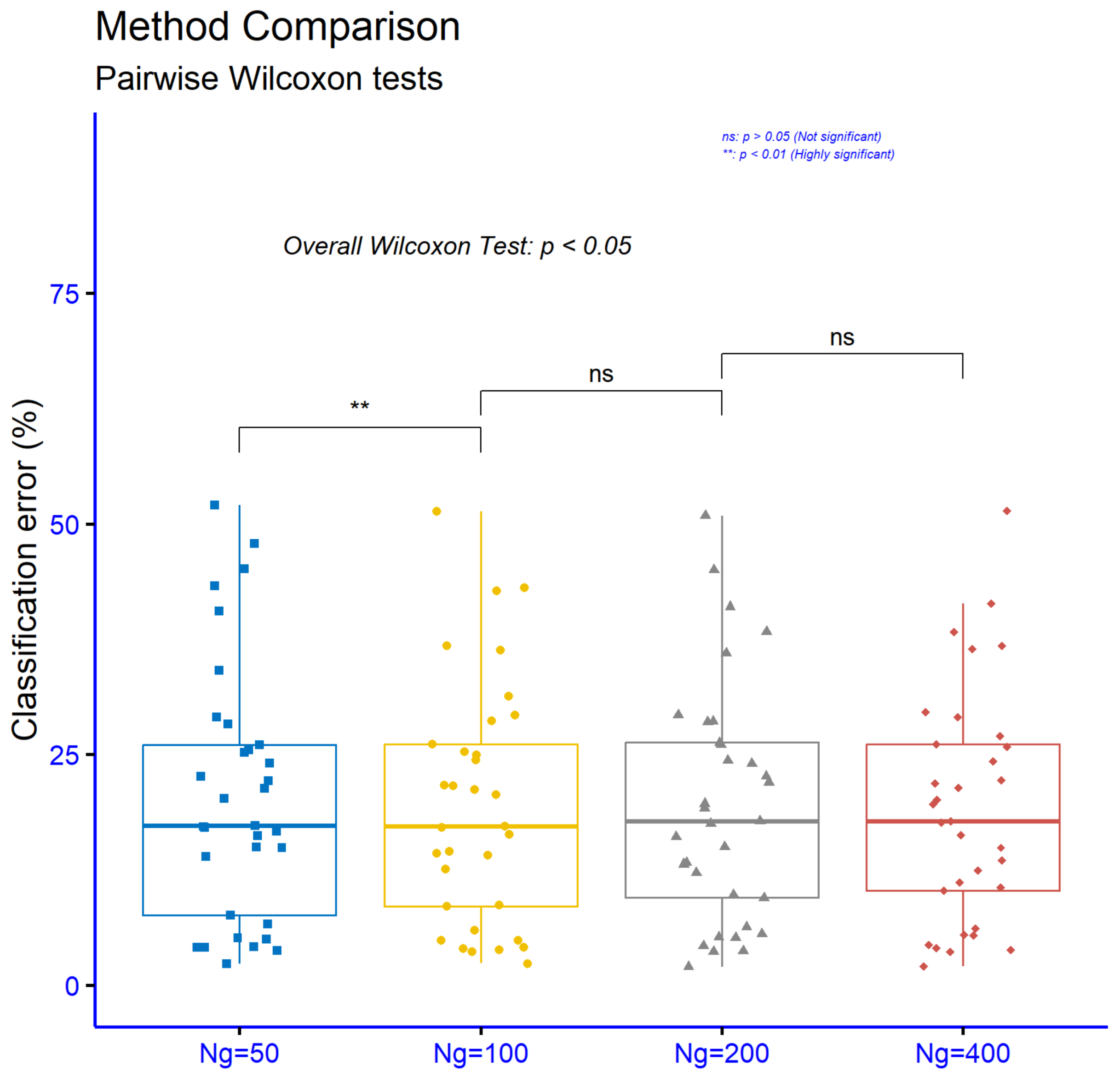
Figure 12 shows that increasing the number of generations from
to
yields a statistically significant improvement (
, **), indicating a meaningful reduction in error in this range. By contrast, the comparisons
vs.
and
vs.
are not statistically significant (
, ns), which means that further increasing generations beyond 100 does not produce a consistent additional gain in performance. Overall, the results suggest that the main benefit is achieved early up to about 100 generations, after which returns diminish and the differences are not statistically meaningful.
In
Figure 13, the
p-value analysis on the regression datasets shows that the comparison between
and
is not statistically significant (
, ns), so increasing generations in this range does not yield a consistent improvement. By contrast, moving from
to
is statistically significant (
, *), indicating a measurable reduction in error around 200 generations. Finally, the comparison between
and
is not statistically significant (
, ns), suggesting diminishing returns beyond 200 generations. Overall, the findings indicate that for regression problems the benefit concentrates around 200 generations, while further increases in
do not guarantee additional consistent gains.