1. Introduction
Clinical decision support systems are motivated not only by the potential to improve care quality, but also by the increasing workload pressures on healthcare providers [
1]. For example, a national survey found that 62.8% of U.S. physicians reported experiencing burnout in 2021, up from 38.2% in 2020, with documentation burden and excessive clerical tasks cited as key contributors [
2]. Similarly, physicians spend nearly 50% of their workday interacting with the electronic health record (EHR), including both direct use and clerical work, compared with only 27% of their time spent on direct patient care [
3]. This imbalance contributes to inefficiency, dissatisfaction, and medical errors. Other studies have estimated that U.S. physicians spend an average of 1.84 h per day outside clinic hours completing EHR “pajama time” tasks, further compounding fatigue [
4].
While non-AI-based approaches (e.g., rule-based decision trees, keyword retrieval, or static knowledge bases) can address certain narrow clinical support tasks, they are brittle and difficult to scale for the highly variable, context-dependent nature of clinical dialogue. Clinical conversations involve unstructured language, diverse terminology, and context-dependent reasoning, which traditional systems often fail to capture [
5,
6]. Prior reviews have shown that such approaches are prone to errors when applied to complex medical narratives and are insufficient for robust decision support [
7,
8]. Moreover, the complexity and heterogeneity of real-world clinical data—including free-text notes, structured fields, and event logs—further limit the utility of static or rule-based solutions [
9]. Given this context, AI-driven tools that can reduce the burden of information retrieval, documentation, and knowledge synthesis hold promise for alleviating workload pressures and supporting more efficient, patient-centered care.
AI-driven approaches enable systems to dynamically retrieve relevant biomedical evidence and generate contextually appropriate responses. This approach has been shown to improve factual grounding and performance on knowledge-intensive tasks compared to generative-only models [
10]. Such adaptability is particularly important in healthcare, where the knowledge base is vast and continuously evolving—for example, during the COVID-19 pandemic, the volume of biomedical publications grew at an unprecedented pace, underscoring the need for retrieval-based systems that can keep up with rapidly changing evidence [
11]. Together, these factors suggest that AI is not only beneficial but necessary for building flexible, scalable, and clinically meaningful dialogue assistants, since non-AI methods remain too rigid for the complexity of real-world medical consultation.
Natural language processing (NLP) has a long history in the clinical domain, with early work focusing on information extraction from EHR text and its use in clinical decision support [
7,
8,
12]. More recently, transformer-based models have been adapted to healthcare, including ClinicalBERT and BioBERT variants trained on EHR notes and biomedical corpora, enabling contextual embeddings tailored to medical applications [
13,
14]. Large-scale resources such as MIMIC-III [
9] have played a key role in enabling such advances. At the same time, the adoption of AI in healthcare has raised significant concerns about privacy and security, leading to work on frameworks such as k-anonymity [
15], regulatory perspectives on medical big data [
16], and technical solutions such as federated learning for privacy-preserving clinical AI [
17].
The emergence of large language models (LLMs) has transformed NLP, particularly in domains requiring nuanced understanding, such as healthcare. However, despite their power, LLMs suffer from hallucinations and lack of grounding in up-to-date domain-specific information. Furthermore, healthcare professionals are increasingly confronted with complex clinical and administrative demands that require timely access to accurate, contextually relevant information. The proliferation of unstructured medical data, largely driven by the widespread adoption of Electronic Health Records (EHRs), has intensified the cognitive load on clinicians. As a result, physicians often face challenges in efficiently retrieving actionable insights, leading to increased documentation workload, diminished patient interaction, and elevated rates of burnout.
To alleviate these challenges, artificial intelligence (AI) has emerged as a transformative force in healthcare, offering innovative solutions to streamline clinical workflows and enhance decision making. Among various AI paradigms, Retrieval-Augmented Generation (RAG) stands out as a particularly promising approach. RAG addresses this by integrating an external knowledge retriever with a generative model, enabling factually accurate and context-sensitive responses, an especially valuable capability in clinical environments. For instance, early applications of RAG in healthcare have focused on clinical question answering and decision support, demonstrating how RAG-based models outperform traditional neural QA systems on biomedical benchmarks like BioASQ, particularly when dealing with rare diseases and complex treatments.
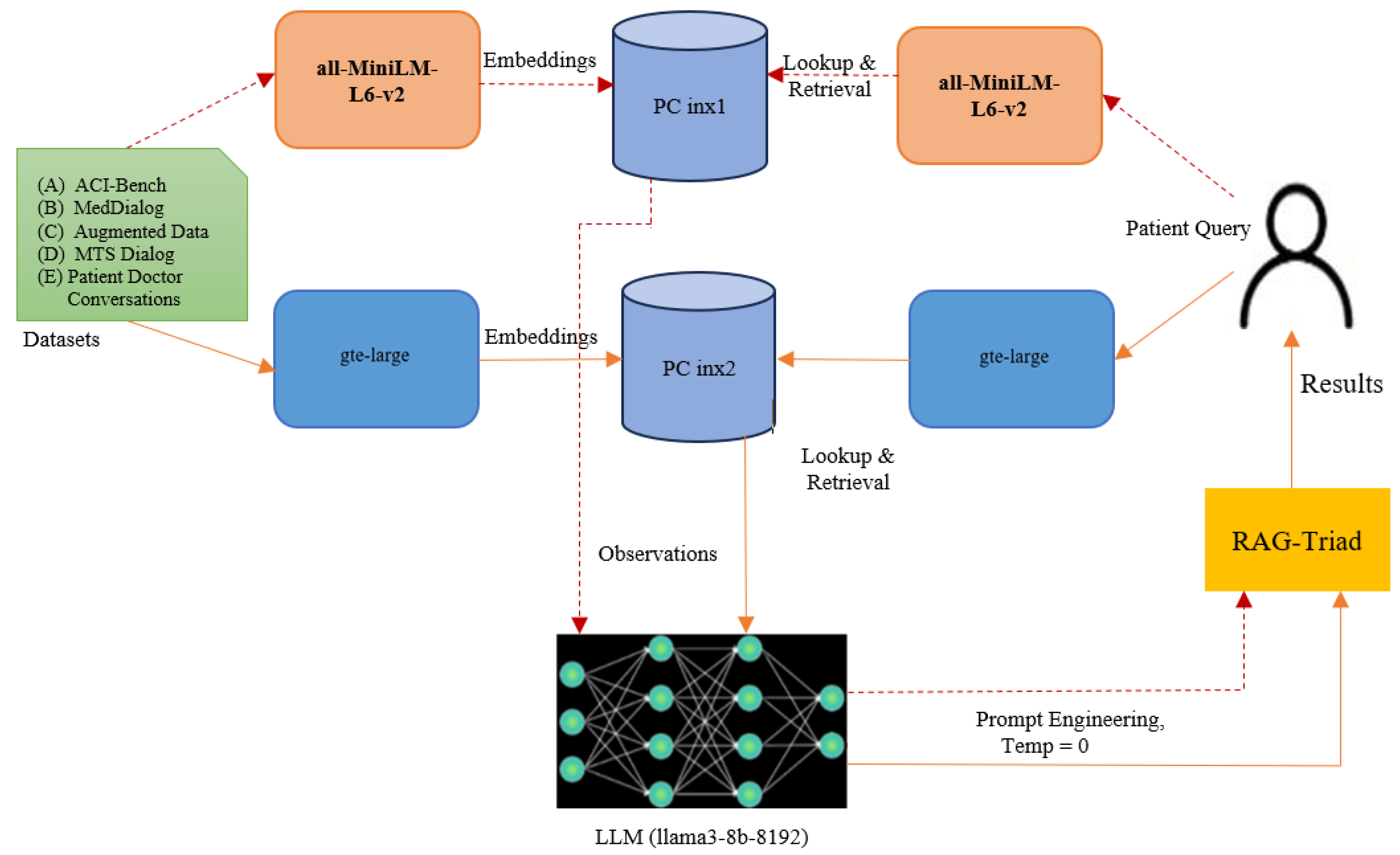
RAG systems combine two core components: a retrieval mechanism that identifies semantically relevant information from a database, and a large language model (LLM) that generates context-aware responses based on the retrieved data. This architecture enables the generation of fluent, factually grounded answers that are both informative and contextually appropriate. In this work, we present a RAG-based medical AI assistant called ‘RAGMed’, designed to support healthcare workflows by automating routine yet critical administrative tasks, including
Responding to frequently asked patient medical questions;
Facilitating patient appointment scheduling;
Summarizing clinical notes for healthcare providers.
Through the integration of semantic search with generative reasoning, RAGMed offers a scalable and interpretable solution for reducing the administrative workload in healthcare. This integration supports the development of more efficient and trustworthy AI-driven tools, enhancing provider workflows and patient engagement in digital health ecosystems. In addition to improving the accessibility and clarity of clinical information, the system fosters greater trust in AI-assisted decision making by delivering responsive and interpretable healthcare solutions. The AI-driven tools employed in our proposed system, RAGMed, are discussed in detail below.
1.1. Objective
The primary objective of our study is to examine how the choice of embedding model influences the informativeness, reliability, and clinical value of RAG-generated responses. To address this objective, the RAGMed system leverages the Pinecone vector database to store and retrieve dense medical embeddings and utilizes the LLaMA3-8B-8192 large language model to generate high-quality responses. To investigate the impact of retrieval quality on system performance, we compare two embedding models—gte-large and all-MiniLM-L6-v2—using 18 queries. In pursuing this objective, our work advances the field through several key contributions, outlined in the next subsection.
1.2. Contributions
This work makes the following key contributions:
Application of RAG in healthcare dialogue and workflows: We demonstrate a retrieval-augmented assistant in a safety-critical setting, focusing on factual grounding for patient FAQs, administrative support, and clinical documentation.
Prototype implementation: We present a working system that integrates medical question answering, natural-language appointment scheduling, and clinical note summarization, illustrating potential to reduce documentation and administrative burden.
Clinically meaningful evaluation framework: We adopt the RAG-Triad metrics (answer relevance, context precision, context recall), moving beyond surface-overlap metrics like BLEU/ROUGE to assess grounding quality.
The remainder of this paper is organized as follows.
Section 2 provides background on large language models, embedding models, vector databases, and similarity search.
Section 3 introduces the design of the proposed RAGMed system, including its novel architecture, datasets, and RAG-Triad evaluation framework.
Section 4 outlines the methodology, while
Section 5 presents experimental results and a comparison of embedding models using the RAG-Triad metrics.
Section 6 reviews related work, and
Section 7 discusses the contributions and limitations of our approach. Finally,
Section 8 concludes the paper and suggests directions for future research.
2. Background
2.1. Large Language Model
To support high-quality response generation within our RAG-based system, we selected LLaMA3-8B-8192, a state-of-the-art large language model developed by Meta. This model offers a balanced combination of advanced reasoning capabilities, low inference latency, and a significantly extended context window, up to 8192 tokens. These characteristics make it highly suitable for clinical applications that demand both real-time responsiveness and the ability to process complex, long-form medical text. LLaMA3-8B-8192 is particularly well-suited for tasks that require integrating and synthesizing detailed contextual information, such as interpreting nuanced patient histories or explaining complex treatment options. Its relatively efficient model size, when compared to larger-scale LLMs, also supports deployment in latency-sensitive environments like digital health assistants, where rapid turnaround and cost-efficiency are essential. Consequently, LLaMA3-8B serves as an ideal backbone for generating contextually grounded, medically accurate responses in our RAG architecture.
2.2. Embedding Models
A fundamental component of the RAG pipeline is the generation of high-quality vector embeddings, which translate unstructured textual data into a numerical format amenable to semantic similarity search. This step allows our system to bridge the linguistic gap between user-submitted patient queries and physician-authored clinical observations by encoding both as dense vectors in the same semantic space. We evaluated two pretrained sentence embedding models for this purpose:
all-MiniLM-L6-v2: A lightweight and high-speed model that produces 384-dimensional embeddings. It is optimized for fast inference and performs well on tasks such as semantic search, clustering, and sentence similarity. Its compact size and efficiency make it particularly advantageous for real-time clinical applications and shorter user queries.
GTE-Large: A more expressive and higher-capacity model that generates 1024-dimensional embeddings. It is designed for general-purpose semantic tasks that demand a deeper understanding of linguistic nuance. GTE-Large excels in capturing subtle similarities across longer and more complex input texts, making it well-suited for detailed medical questions that require advanced contextual reasoning.
Both models were implemented using industry-standard libraries: the SentenceTransformers package for MiniLM and HuggingFace Transformers for GTE-Large. A unified preprocessing pipeline was applied to both models to ensure consistency in comparative evaluation. Each patient query and clinical observation was converted into a fixed-length vector, regardless of input size, thus enabling scalable semantic indexing and retrieval.
2.3. Vector Database and Indexing
Following embedding generation, we implemented semantic indexing to efficiently organize and access the vector representations of physician observations. Given the diversity and complexity of the language used in medical documentation, a robust and flexible indexing system is critical for high-performance retrieval. To this end, we integrated Pinecone, a cloud-native vector database optimized for large-scale, real-time similarity search. Pinecone enables the efficient storage and retrieval of high-dimensional embeddings using Hierarchical Navigable Small World (HNSW) indexing, an approximate nearest neighbor (ANN) algorithm known for its high recall and low latency. This architecture supports scalable retrieval without the need to scan the entire dataset, significantly improving search efficiency. Upon receiving a patient query, the system uses the corresponding embedding to retrieve the top-k most semantically relevant physician records in milliseconds. These documents are then supplied to the large language model as contextual input for grounded response generation. Pinecone’s elastic architecture also supports the seamless scaling of our system to accommodate increasing volumes of medical data over time. Each embedding model maintains a dedicated Pinecone index, enabling independent performance evaluation and retrieval pipelines (e.g., PC-index1 for MiniLM and PC-index2 for GTE-Large).
2.4. Similarity Search
Semantic similarity search lies at the core of our system’s retrieval process. This mechanism ensures that the most contextually relevant physician observations are retrieved from a large and heterogeneous dataset, enabling accurate and grounded response generation. To perform similarity matching, we employed cosine similarity, a widely used metric that calculates the angle between two vectors to quantify their semantic alignment. Unlike traditional keyword-based retrieval methods, cosine similarity assesses conceptual proximity rather than literal term overlap, which is an essential advantage in clinical settings where users may phrase the same problem in diverse and often non-standardized ways.
When a patient submits a question, the system encodes it into a dense vector using the selected embedding model. This query vector is then compared with precomputed physician observation embeddings in the vector database. Cosine similarity scores are calculated for each embedding pair, and the system retrieves the top-k results with the highest scores. This method ensures that the retrieved context is semantically aligned with the intent of the query, even when the terminology, phrasing, or syntax differ significantly. By focusing on the meaning behind the words rather than their exact matches, cosine similarity improves the precision and relevance of the input provided to the LLM. This, in turn, results in higher-quality, trustworthy, and informative responses that align with both clinical expectations and user needs.
2.5. Model Evaluation
To evaluate retrieval quality and the correctness of system outputs, we adopted the RAG-Triad framework. Unlike surface-level metrics such as BLEU, ROUGE, or F1, RAG-Triad emphasizes clinically meaningful aspects of performance, including retrieval utility, factual grounding, and answer relevance. This makes it well suited for healthcare dialogue, where reliability and safety are paramount. A detailed description of the framework and its three evaluation dimensions is provided in
Section 3.3.
5. Experiments and Results
For the evaluation, we use a benchmark of 18 real-world medical queries representative of typical patient and clinician interactions. Each query is processed using both embedding models—gte-large and all-MiniLM-L6-v2—to investigate how embedding dimensionality and semantic richness impact system performance across the three dimensions. Comparative results provide insight into how embedding model selection influences the overall quality of in terms of the relevance, accuracy, factual grounding, informativeness, reliability, and clinical value of RAG-generated system responses. We conducted a comprehensive study of the two embedding models, all-MiniLM-L6-v2 and gte-large. Each embedding model is associated with its vector index, which contains approximately 45000 doctor observations encoded by them. Every input patient query is converted into a numerical embedding by both models, and using the cosine similarity metric, we retrieved the top-five relevant observations. Based on the prior research findings, we selected cosine similarity because it performed better than other alternatives like Euclidean distance and dot product, specifically in healthcare-related tasks.
These retrieved top-five observations are then passed as context to LLM (llama3-8b-8192). Using prompt engineering, the LLM is instructed to generate a summarized response with a fixed temperature of 0. Utilizing the RAG-Triad evaluation framework, we evaluated each model’s pipeline by measuring three important metrics: context relevance, answer relevance, and groundedness. This configuration enabled us to examine the impact of retrieval quality on the final responses’ accuracy, clarity, and reliability. To assess the performance and efficiency of the proposed system, we conducted experiments across three representative healthcare tasks—(1) medical question answering, (2) appointment scheduling, and (3) clinical note summarization—as detailed in
Section 5.3,
Section 5.4 and
Section 5.5. To further clarify our experimental design, we next describe the rationale behind our choice of the temperature parameter (
Section 5.1) and outline the evaluation metrics used in our study (
Section 5.2).
5.1. Temperature Parameter Choice
For all the experiments, the temperature parameter of the language model was set to 0, with the goal of minimizing randomness and encouraging more deterministic, reproducible outputs. While temperature = 0 does not guarantee strict determinism (since factors like non-deterministic GPU operations and beam search randomness can still introduce variation), it significantly reduces variability compared to higher values.
We selected this setting to ensure consistency in the comparative evaluation across embedding models, so that differences in output quality could be attributed primarily to retrieval and grounding rather than sampling noise. Although other temperature values (e.g., 0.3, 0.7) were briefly tested during exploratory runs and yielded more diverse but less stable responses, they were not adopted for formal evaluation. Future work may explore how controlled diversity in generation (via non-zero temperatures or nucleus sampling) could influence answer quality, especially in patient-facing scenarios where nuanced expression is beneficial.
5.2. Evaluation Metrics—Methods Section
We evaluated the system using the RAG-Triad framework, which provides three complementary dimensions essential for assessing retrieval-augmented systems in healthcare: Answer Relevance (whether the generated response correctly addresses the clinical query), Context Precision (whether the retrieved passages are genuinely useful for answering the query), and Context Recall (whether the assistant retrieves enough relevant knowledge to support an appropriate answer). Unlike n-gram–based measures such as BLEU, ROUGE, and F1, which emphasize surface text overlap, RAG-Triad captures whether the system is both retrieving the right medical knowledge and grounding its responses appropriately, which is an essential aspect for clinical decision support. As this study was designed as a proof-of-concept study and as our aim was to highlight the ability of the system to retrieve relevant medical knowledge and generate grounded, clinically appropriate responses, we did not compute additional statistical measures (e.g., confidence intervals, significance testing) and instead focused on retrieval and grounding quality as clinically meaningful indicators of system performance.
5.3. Medical Question Answering
We tested both embedding models across 18 real-world medical queries. Below, we present a few of those examples to demonstrate the key differences.
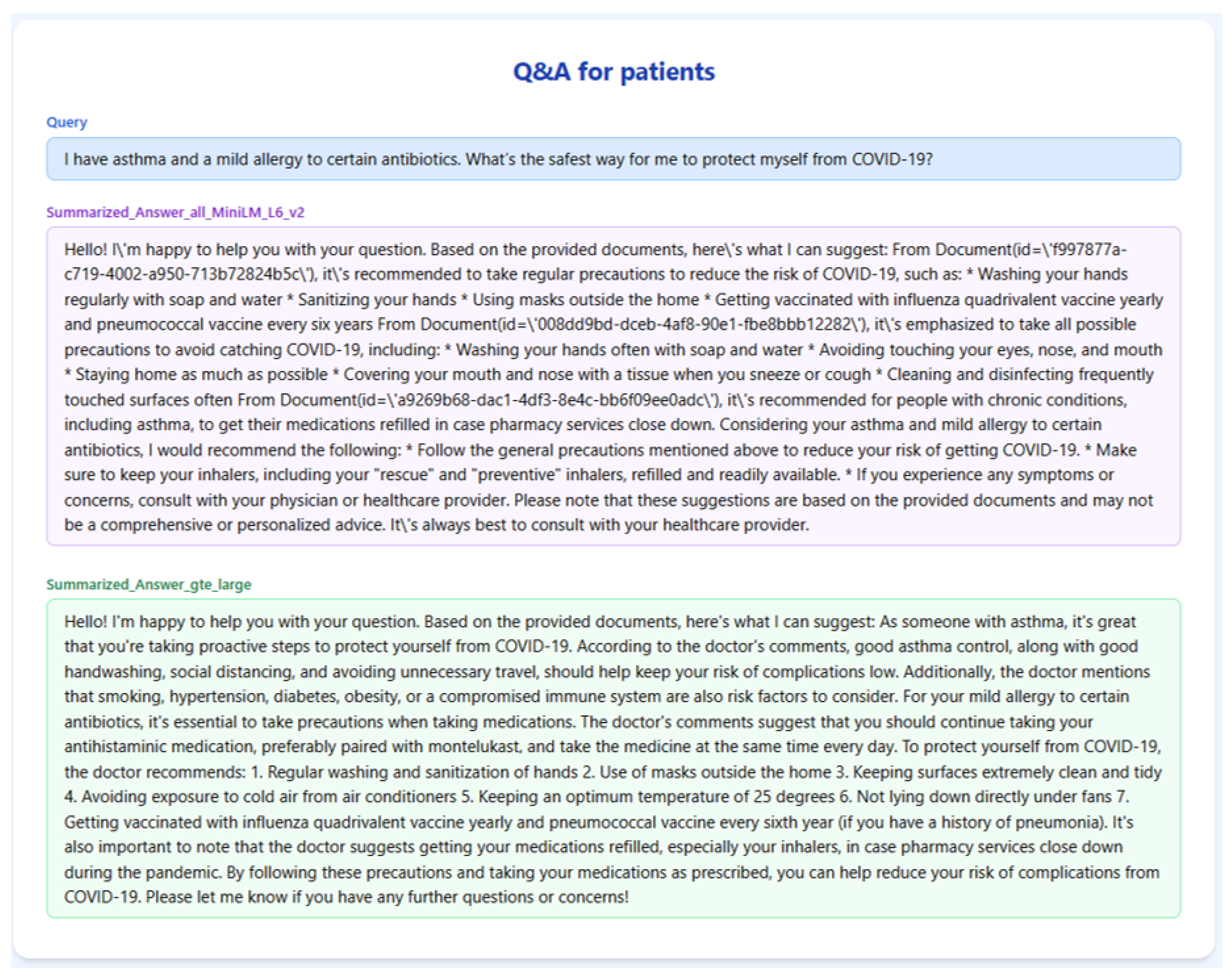
Figure 4 and
Table 1 illustrate the output generated by the RAGMed system in response to Query-1. gte-large retrieved better chunks specific to asthma and allergies, such as advice on inhaler use, keeping indoor air clean, and getting the right vaccines. Meanwhile, all-MiniLM-L6-v2 mostly repeated the general COVID-19 precautions without taking into account the users’ medical background history. As a result, gte-large is able to produce a more tailored and helpful response, offering detailed instructions that someone with asthma could follow.
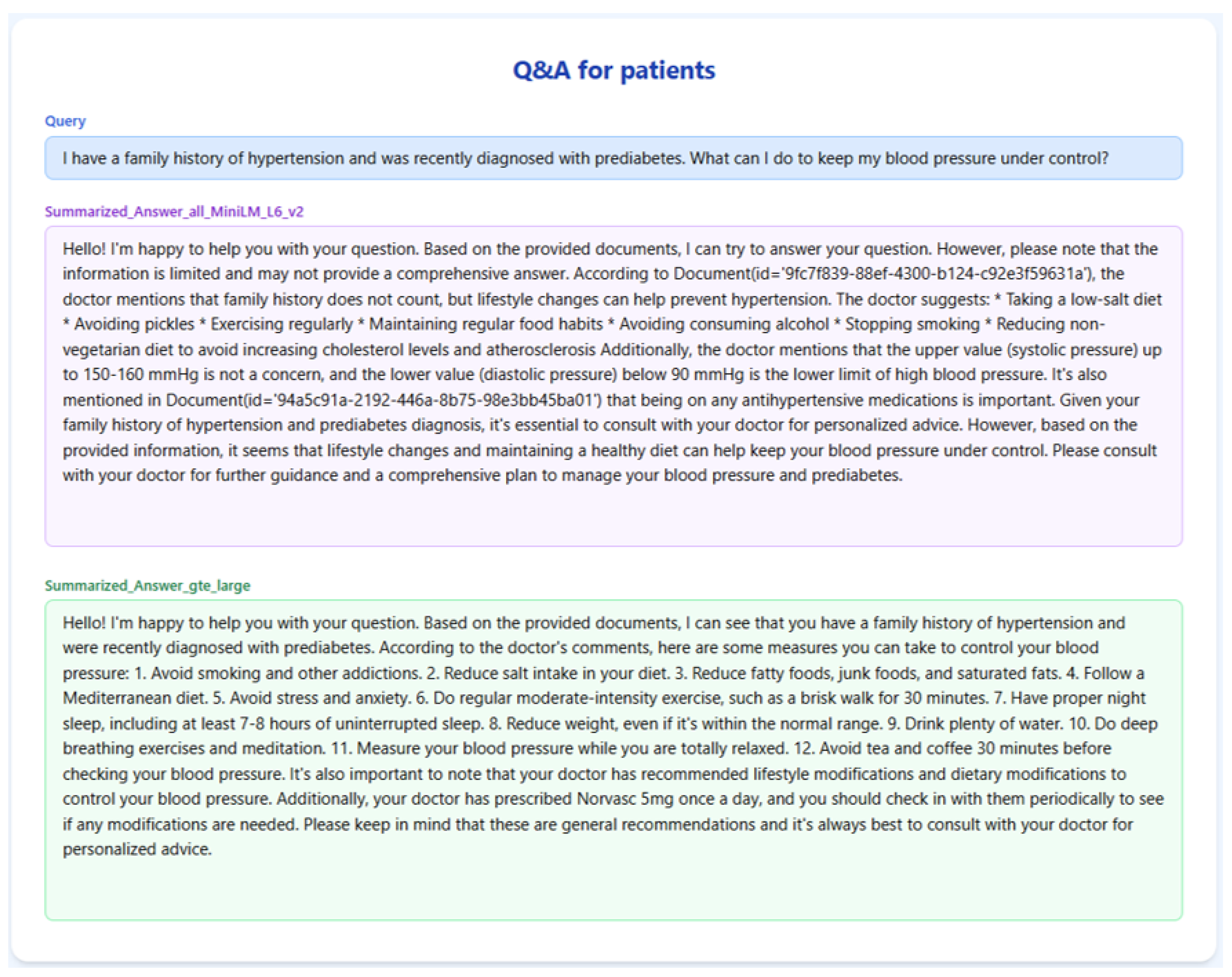
The results for Query-2 on the RAGMed system are presented in
Figure 5 and
Table 2. The final response of gte-large was more thorough and reliable since it extracted chunks that included specific lifestyle changes (such as the Mediterranean diet, sleep hygiene, and stress reduction) and referenced actual medication (Norvasc). Conversely, all-MiniLM-L6-v2 provided broad guidance that is unstructured and cited questionable blood pressure limits without providing context. As a result, gte-large generated a more structured and medically appropriate answer, providing the user with clear guidance to which to adhere.
Table 2.
RAG-TRIAD scores for Query-2.
Table 2.
RAG-TRIAD scores for Query-2.
| Embedding Model | Answer Relevance | Context Relevance | Groundedness |
|---|
| GTE-Large | 0.67 | 0.67 | 0.43 |
| all-MiniLM-L6-v2 | 0.58 | 0.63 | 0.25 |
Figure 5.
RAGMed output for Query-2.
Figure 5.
RAGMed output for Query-2.
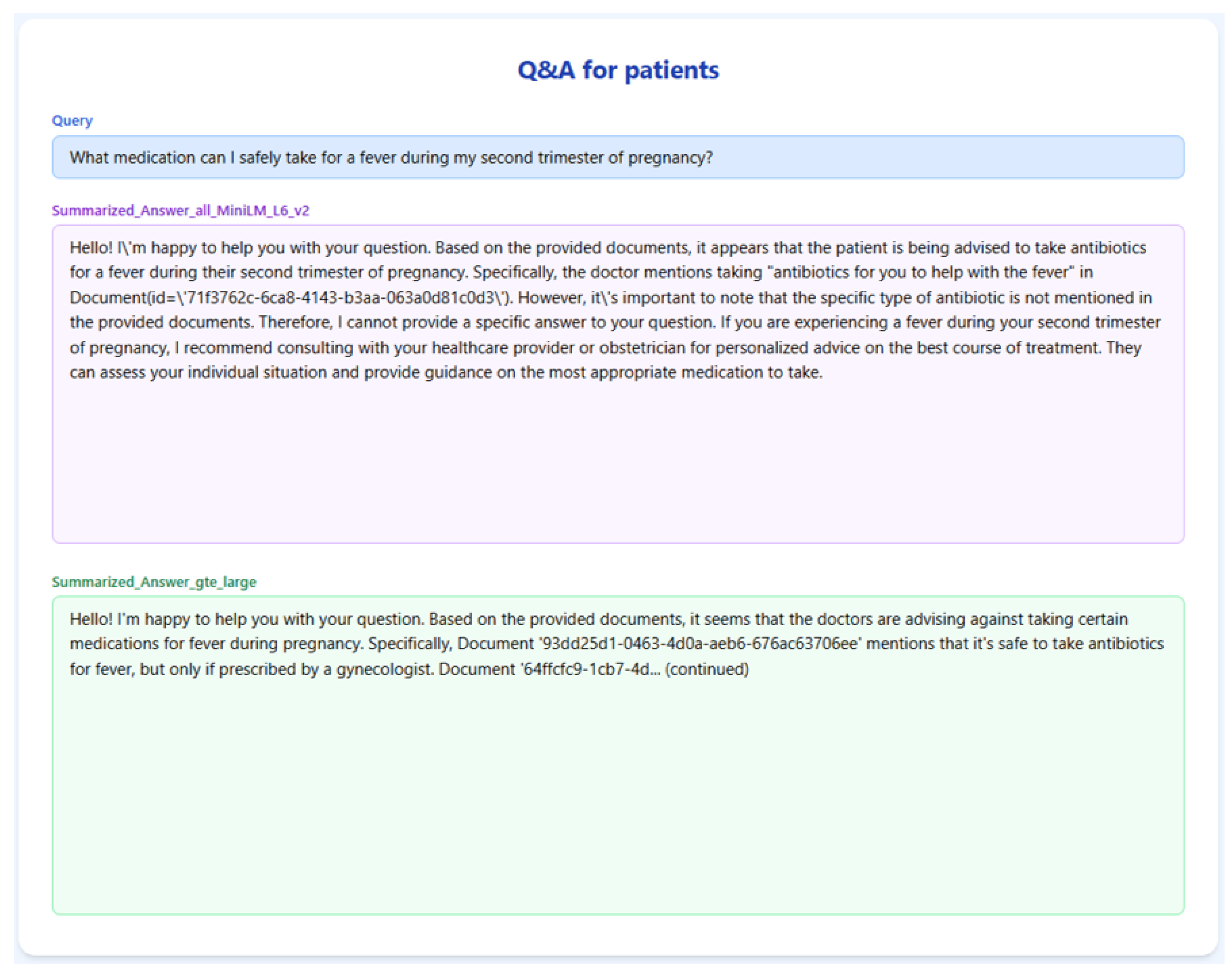
Figure 6 and
Table 3 showcase the RAGMed system’s response to Query-3. all-MiniLM-L6-v2 retrieved more chunks that mentioned specific medication names, but it did not clarify whether those drugs are safe during pregnancy, making it risky in this context. However, gte-large chunks are more cautious, mentioning antibiotics recommended by doctors and advising against using ibuprofen without a prescription. The final response from gte-large is less detailed but safer, better grounded, and more suitable for a medically sensitive query.
Across all 18 queries, the gte-large embedding model constantly produced better results when compared with the all-MiniLM-L6-v2 model. On average, GTE-large scored 0.72 in answer relevance, 0.70 in context relevance, and 0.47 in groundedness, reflecting its ability to grasp meaningful and clinically useful content. In comparison, all-MiniLM-L6-v2 scored lower in each category, with 0.53 for answer relevance, 0.61 for context relevance, and 0.31 for groundedness, indicating that it captured the general topic but often lacked the depth and clarity needed for high-quality responses. These results highlight the benefit of using semantically richer embeddings like GTE for retrieval augmented generation tasks in clinical applications.
Table 4 summarizes the average scores across all 18 queries:
Table 3.
RAG -TRIAD scores for Query-3.
Table 3.
RAG -TRIAD scores for Query-3.
| Embedding Model | Answer Relevance | Context Relevance | Groundedness |
|---|
| GTE-Large | 0.78 | 0.67 | 0.5 |
| all-MiniLM-L6-v2 | 0.56 | 0.5 | 0.19 |
Figure 6.
RAGMed output for Query-3.
Figure 6.
RAGMed output for Query-3.
Table 4.
Average RAG-TRIAD scores Across 18 queries.
Table 4.
Average RAG-TRIAD scores Across 18 queries.
| Embedding Model | Answer Relevance | Context Relevance | Groundedness |
|---|
| GTE-Large | 0.72 | 0.70 | 0.47 |
| all-MiniLM-L6-v2 | 0.53 | 0.61 | 0.31 |
5.3.1. Quantitative Evaluation
GTE-Large consistently outperformed all-MiniLM-L6-v2 across all dimensions. It demonstrated a stronger capacity to retrieve semantically rich and highly aligned content, which in turn enabled the language model to produce more informative and clinically sound responses. In contrast, while all-MiniLM-L6-v2 offered lower latency and faster inference, it occasionally retrieved context that was overly generic or tangential to the query’s intent.
5.3.2. Qualitative Observations
In addition to quantitative scoring, qualitative analysis revealed that
GTE-Large captured subtle semantic nuances better, particularly in queries involving multi-step reasoning or complex medical terminology.
MiniLM, while efficient, often missed specific contextual cues in longer or compound queries, leading to less precise responses.
Responses generated with GTE-Large context were more likely to cite specific clinical actions, findings, or terminology from the retrieved documents, thereby enhancing traceability and user trust.
5.3.3. Implications
These results underscore the critical role that embedding model selection plays in the performance of RAG-based systems for healthcare. While lightweight models like MiniLM are attractive for real-time systems due to their speed, high-dimensional embeddings from models like GTE-Large offer superior retrieval quality, which directly translates into better downstream response generation. The findings also validate the utility of the RAG-Triad framework as a multidimensional evaluation approach that captures not only linguistic fluency but also factual fidelity and contextual alignment—key concerns in clinical AI systems.
5.4. Appointment Scheduling
While traditional software systems can monitor and manage a physician’s calendar, they typically require structured inputs through predefined forms or drop-down menus. In contrast, our LLM-enabled assistant allows patients (and clinicians) to book or inquire about appointments by using simple English sentences. The system interprets this free-text request, extracts relevant details (physician, timeframe, reason), checks availability, and presents scheduling options. This offers several benefits over traditional systems, such as (1) improved accessibility, since patients who may not be tech-savvy can interact naturally without learning rigid interfaces, and (2) reduced administrative burden, as natural language scheduling decreases the need for staff to translate patient requests into structured calendar entries. By enabling natural, context-aware interaction, RAGMed goes beyond static rule-based tools and provides a more flexible, user-friendly scheduling experience.
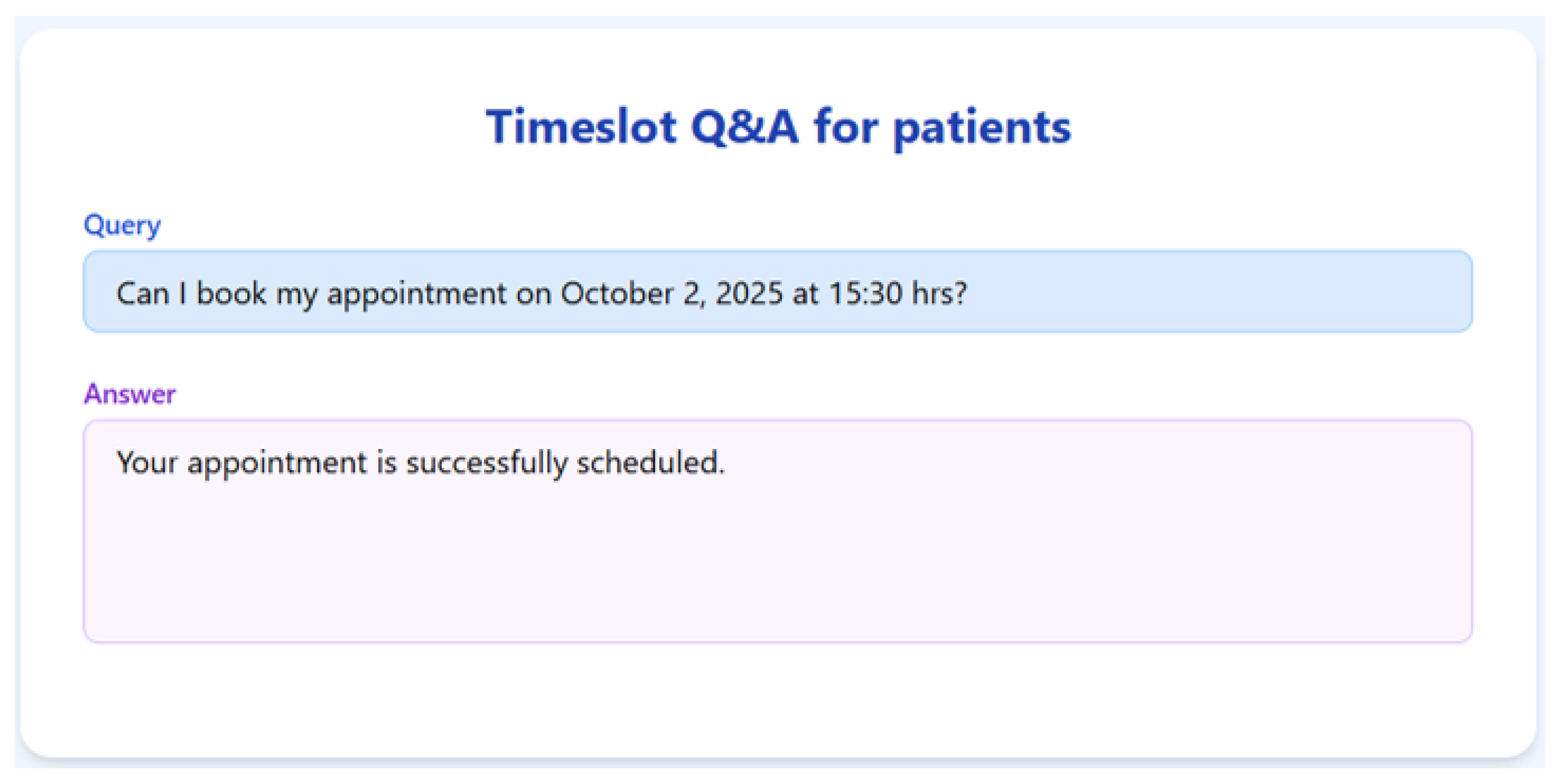
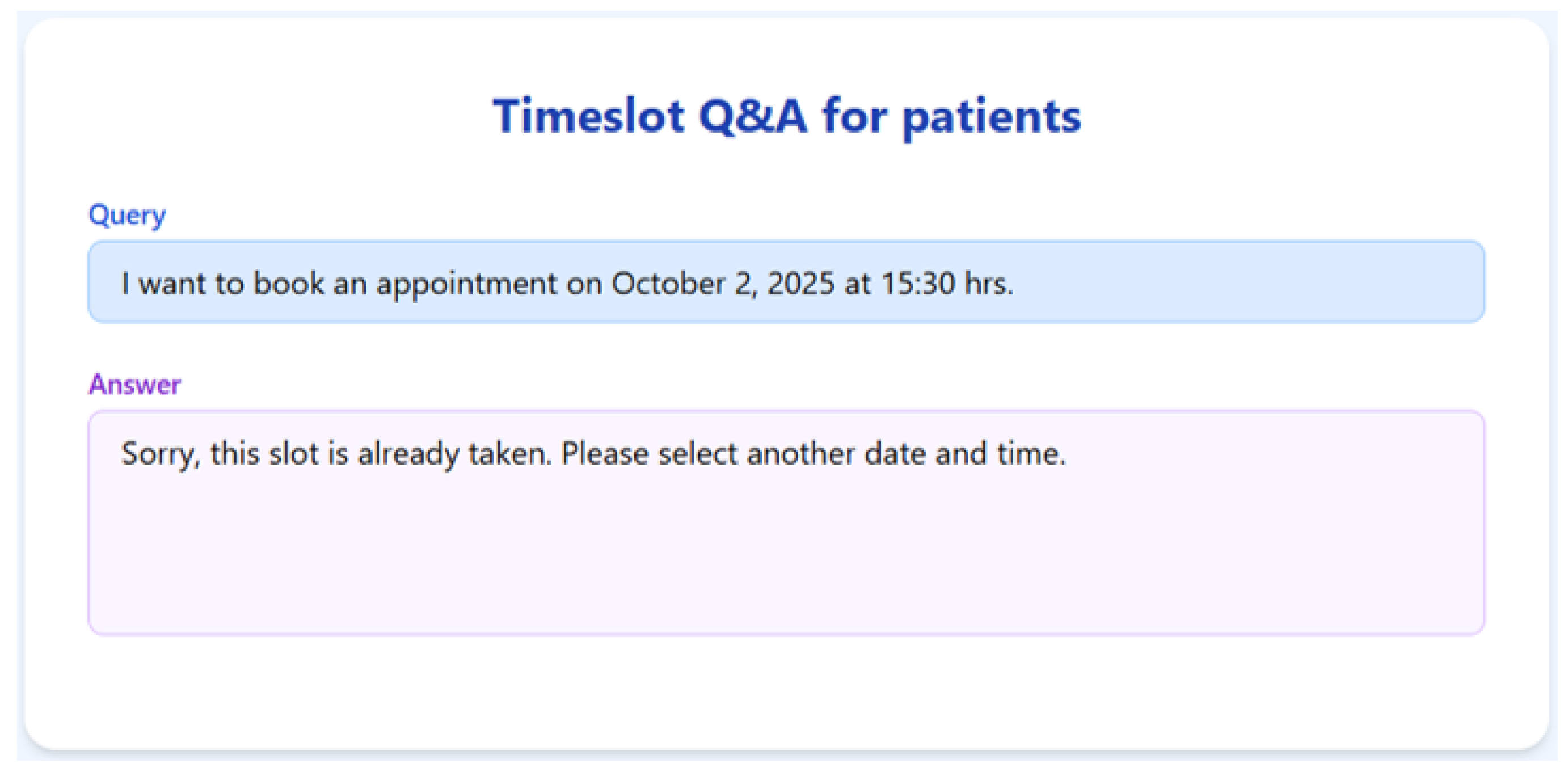
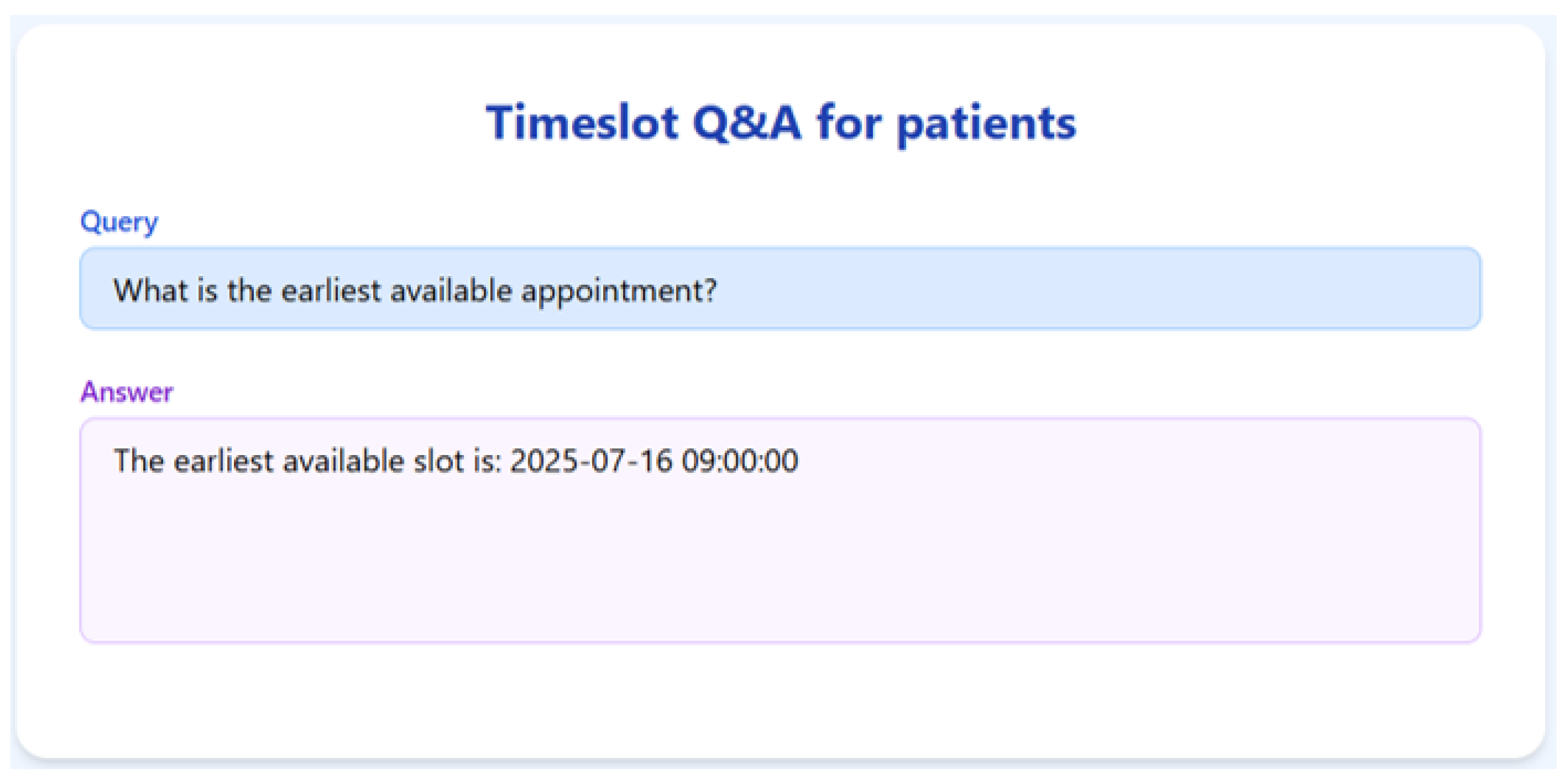
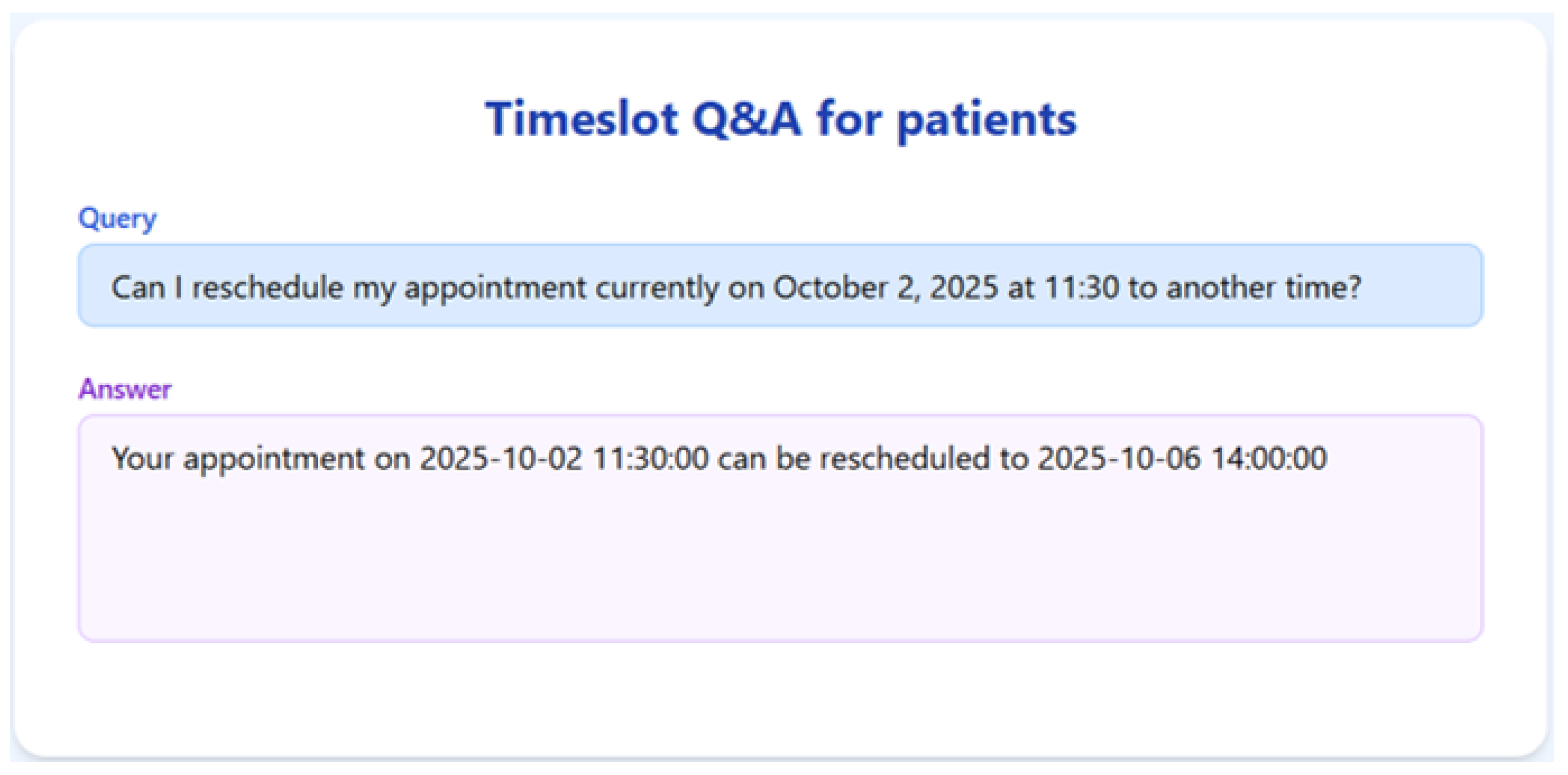
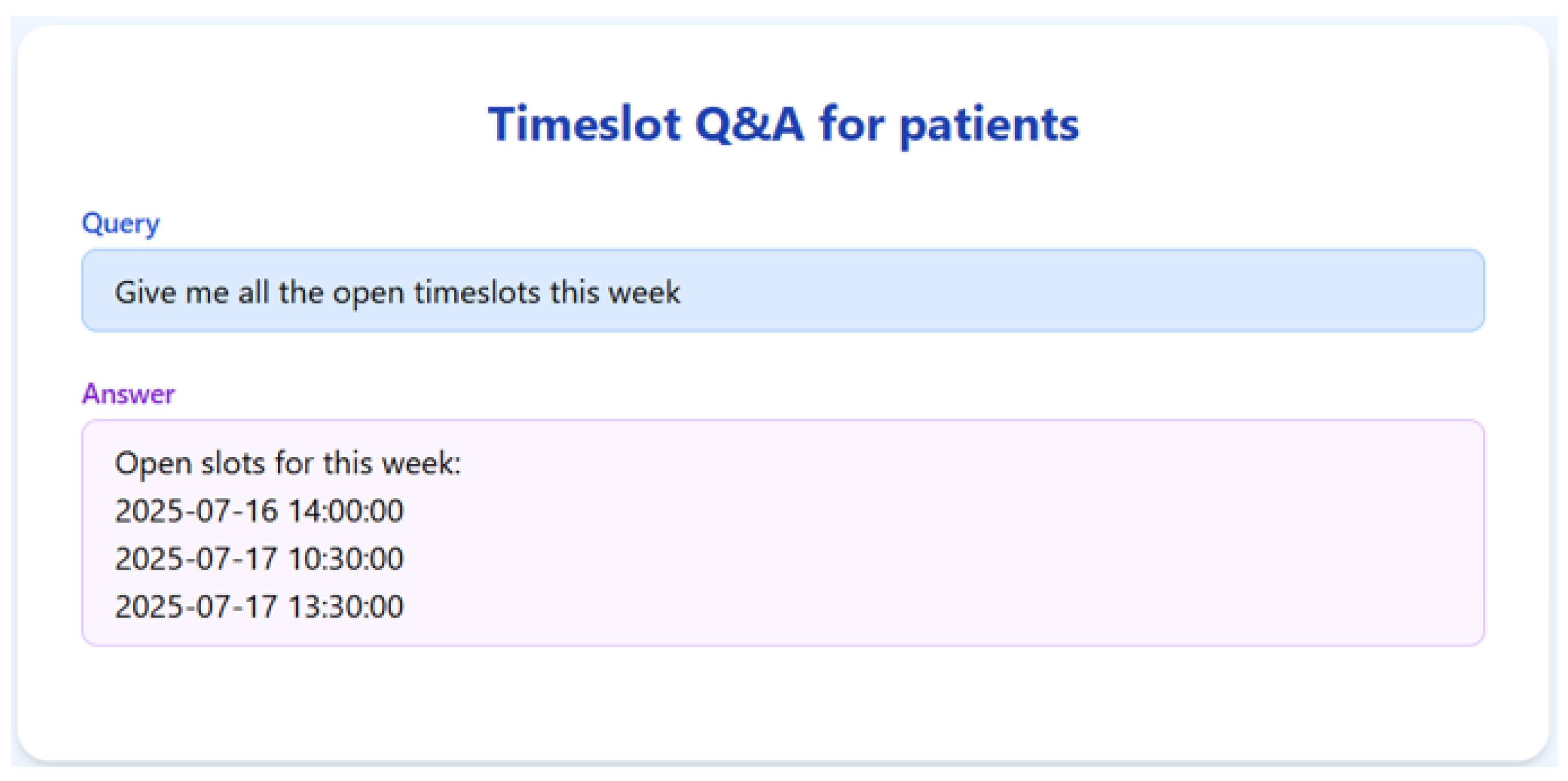
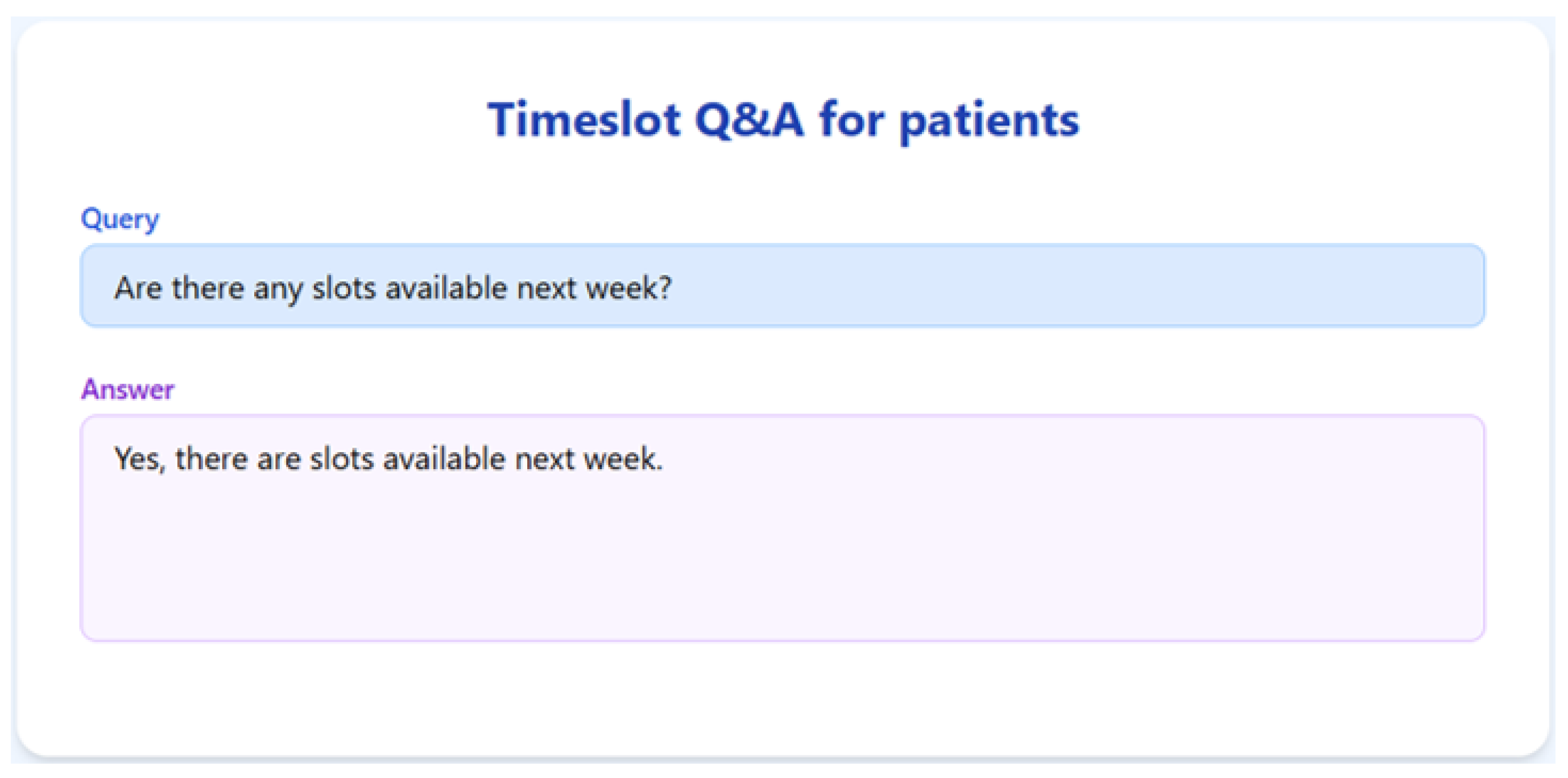
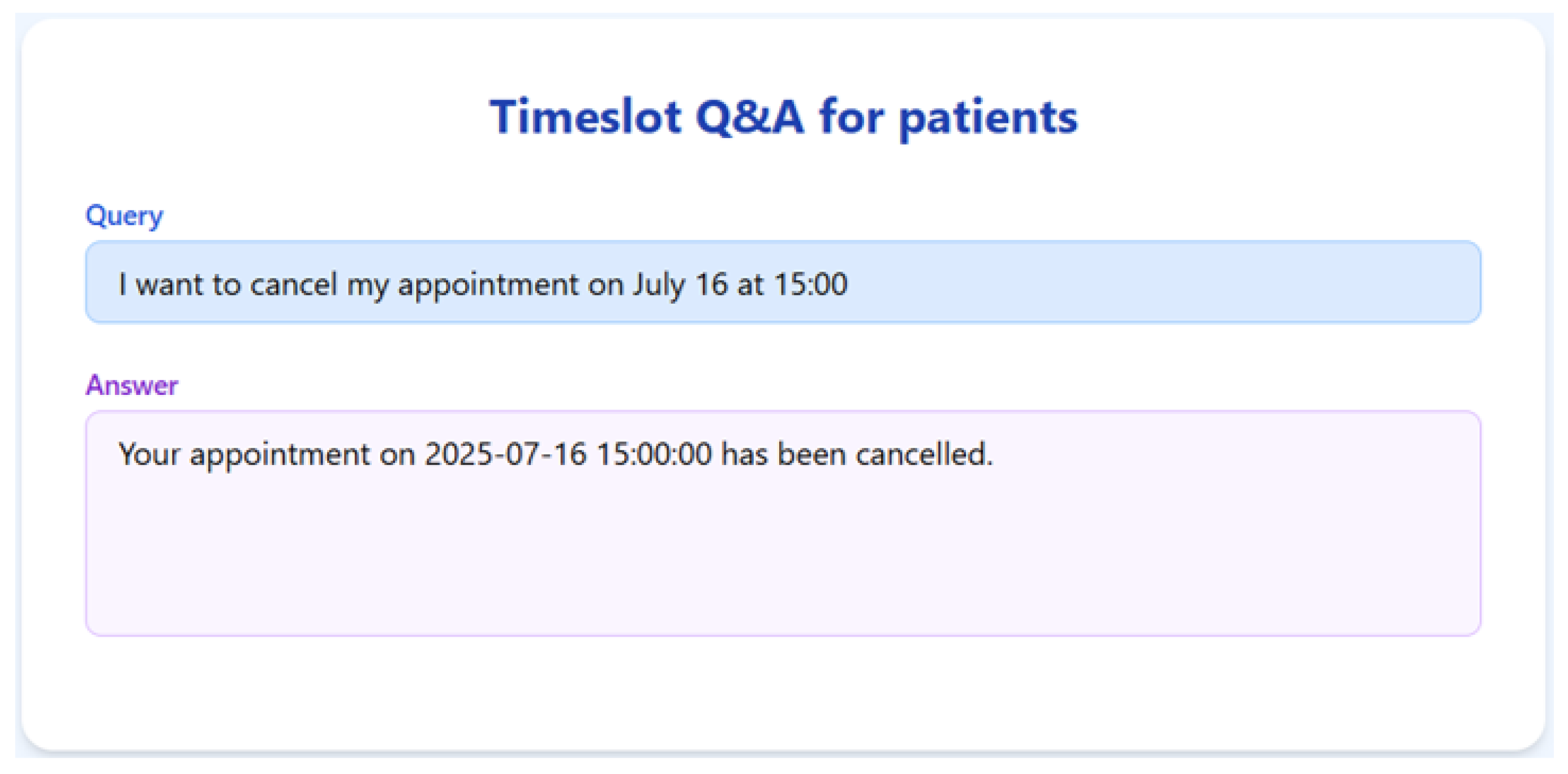
To validate the appointment scheduling functionality, we simulated a 3-month dataset of structured appointment records. The system was tested on several practical queries, and we demonstrate the results for seven of them below.
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11,
Figure 12 and
Figure 13 present the RAGMed output for those queries.
The RAGMed System was able to analyze existing appointments, identified open slots during business hours on Monday–Friday (9AM-5PM), and responded accordingly. It accurately distinguished between booked and available time slots. And it correctly interpreted temporal expressions such as “this week” or “next week” using prompt-based logic. Although, the system consistently generated accurate scheduling responses across most tested scenarios, demonstrating its potential for real-time deployment, occasional hallucinations were observed, indicating a need for future refinement.
Figure 7.
RAGMed output for Query-4.
Figure 7.
RAGMed output for Query-4.
Figure 8.
RAGMed output for Query-5.
Figure 8.
RAGMed output for Query-5.
Figure 9.
RAGMed output for Query-6.
Figure 9.
RAGMed output for Query-6.
Figure 10.
RAGMed output for Query-7.
Figure 10.
RAGMed output for Query-7.
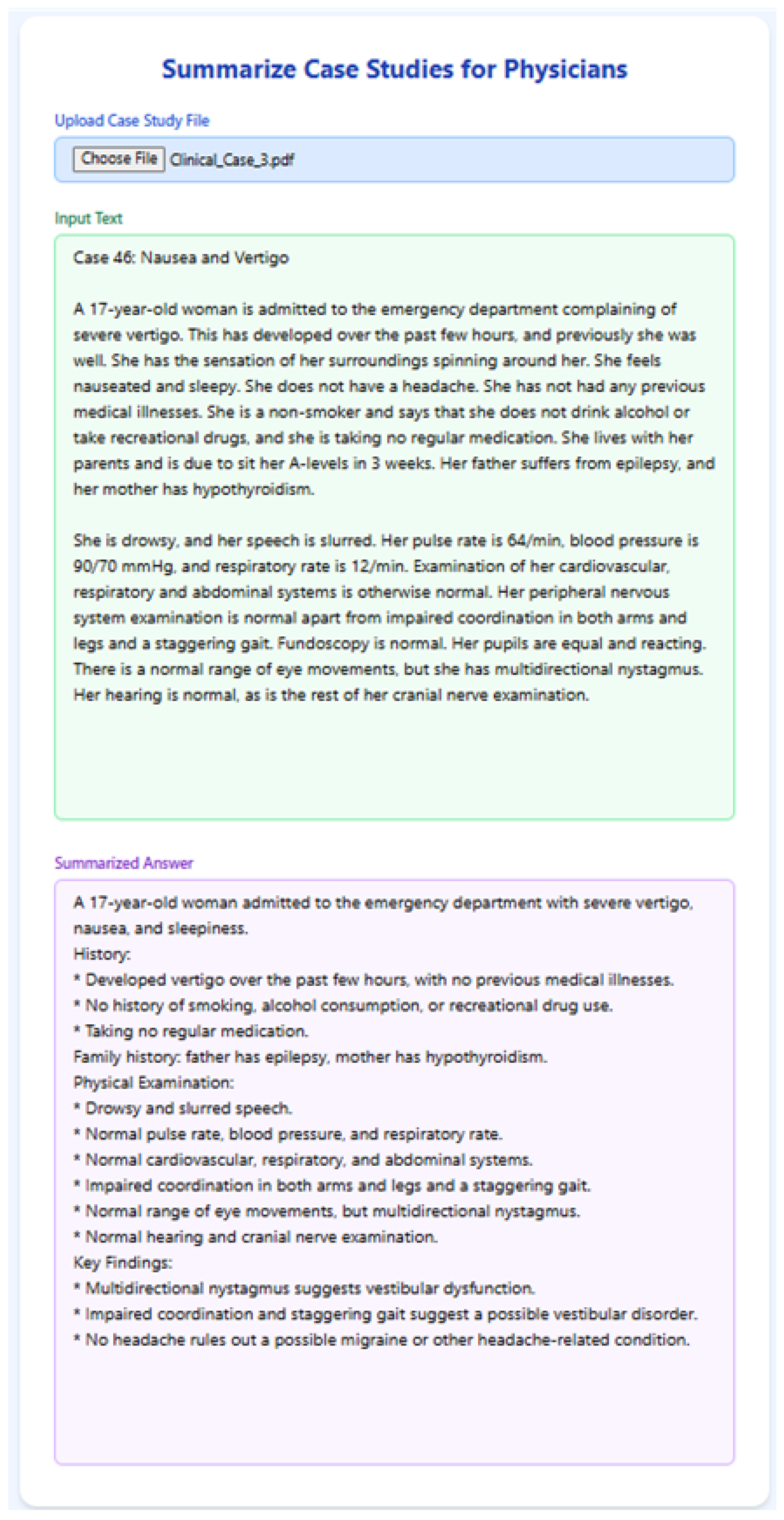
5.5. Clinical Note Summarization
To evaluate RAGMEd’s ability in summarizing structured clinical case studies, we implemented the following functionality, which allows physicians to upload a clinical case file and receive a clear bullet-point summary. The content is extracted from the file and passed to llama3-8b-8192 with a structured prompt designed for clinical summarization. The prompt instructs the model to organize the summary using bullet points and section headings, mimicking physician-style documentation.
Figure 14,
Figure 15 and
Figure 16 illustrate the system’s output for three selected clinical cases.
Figure 11.
RAGMed output for Query-8.
Figure 11.
RAGMed output for Query-8.
Figure 12.
RAGMed output for Query-9.
Figure 12.
RAGMed output for Query-9.
Figure 13.
RAGMed output for Query-10.
Figure 13.
RAGMed output for Query-10.
The RAGMed system consistently produced well-structured, clinically trustworthy summaries that aligned with standard clinical documentation practices. It captured key sections such as presenting complaints, medical history, and examination findings, resulting in outputs that physicians can review quickly and integrate into electronic health records. Importantly, the model demonstrated the ability to process long-form clinical input and distill it into concise, physician-friendly summaries. By grounding its outputs in retrieved context, RAGMed reduces the risk of unsupported statements or omissions, highlighting its potential as a practical tool for documentation assistance within real-world clinical workflows.
Figure 14.
RAGMed output for Clinical Case-1.
Figure 14.
RAGMed output for Clinical Case-1.
Figure 15.
RAGMed output for Clinical Case-2.
Figure 15.
RAGMed output for Clinical Case-2.
Figure 16.
RAGMed output for Clinical Case-3.
Figure 16.
RAGMed output for Clinical Case-3.
6. Related Work
Retrieval-Augmented Generation (RAG) has recently emerged as a promising approach to enhance the performance and reliability of generative models, particularly in domains requiring access to large and dynamic knowledge bases, such as healthcare. Unlike conventional large language models (LLMs) that rely solely on their internal parameters, RAG combines generative models with external retrieval systems, allowing the model to ground its outputs in up-to-date and domain-specific information [
10]. This feature is particularly critical in healthcare, where accuracy, explainability, and current knowledge are essential for clinical reasoning, patient education, and biomedical research. Retrieval-Augmented Generation (RAG) helps overcome key limitations of large language models (LLMs) in the medical domain—such as outdated clinical practice, hallucinations, and lack of transparency—by grounding responses in evidence-based, best practice external sources. The study in [
18] highlights how RAG improves the reliability of LLMs in healthcare tasks and surveys various RAG techniques (Naive, Advanced, Modular) applied to medical datasets.
Ref. [
19] introduced i-MedRAG, an iterative RAG approach where LLMs generate and refine follow-up queries across multiple rounds to build deeper understanding. This method significantly improves performance on challenging medical benchmarks like USMLE and MedQA, outperforming existing prompt engineering and fine-tuning strategies, and demonstrates strong potential for advancing medical question answering. The manuscript [
20] introduces MedSummRAG, a RAG framework tailored for medical text summarization, addressing limitations of large language models in domain-specific understanding. By integrating a fine-tuned dense retriever trained with contrastive learning, it enhances summary quality using relevant external knowledge. Experiments show notable ROUGE improvements across various settings.
Article [
21] presents CLI-RAG, a clinically informed retrieval-augmented generation framework for structured clinical text generation from unstructured EHR data. It introduces hierarchical chunking and dual-stage retrieval to handle the complexity and heterogeneity of clinical documentation. When evaluated on MIMIC-III, CLI-RAG outperforms baselines in semantic and temporal alignment, demonstrating potential for reliable and consistent clinical documentation. Ref. [
22]’s work introduces MIRAGE, a comprehensive benchmark for evaluating medical retrieval-augmented generation (RAG) systems, and MEDRAG, a toolkit enabling large-scale experimentation across various LLMs, retrievers, and corpora. Through extensive testing, the study shows that optimal RAG configurations significantly boost QA accuracy up to 18% and offers practical guidelines for deploying RAG in medical applications.
Manuscript [
23] presents SMARThealth GPT, a RAG-based system designed to support community health workers in low-resourced/socioeconomic settings with guideline-based maternal care education. Developed using Indian pregnancy guidelines, the model emphasizes traceability, scalability, and adaptability. The case study demonstrates the practical value of RAG and LLMs in improving healthcare education and offers a blueprint for similar applications in resource-limited contexts. Ref. [
24] introduces MedRAG, a RAG framework enhanced with knowledge graph-based reasoning to improve diagnostic accuracy and specificity from EHRs. By integrating hierarchical diagnostic KGs and dynamically retrieving similar cases, MedRAG supports precise, patient-centered recommendations and proactive diagnostic questioning. Evaluations on public and private datasets show that it outperforms existing RAG models, particularly in reducing misdiagnosis for clinically similar conditions.
The study in [
25] evaluates the effectiveness of combining fine-tuning and Retrieval-Augmented Generation (RAG) in open-source LLMs for medical question answering, particularly in resource-constrained settings. Among the tested models, Mistral-7B with fine-tuning and RAG showed the best performance, achieving strong accuracy and alignment between confidence and correctness. The work introduces a novel MCQ evaluation methodology and highlights the potential of such models for clinical reasoning and patient education. Article [
26] explores the use of zero-shot prompting with LLaMA 2 (13B) and RAG to extract and summarize malnutrition-related data from aged care EHRs. The combined approach achieved high accuracy (up to 99.25%) in generating structured summaries and extracting clinical risk factors. RAG improved summarization performance and reduced hallucinations, highlighting its value in enhancing data accessibility and improved quality of care in healthcare settings.
The scoping review in [
27] maps the current applications and challenges of retrieval-augmented generation (RAG) in healthcare, highlighting its use in clinical reasoning and clinical judgment, education, and pharmacovigilance. While RAG enhances accuracy and transparency over traditional LLMs, key issues such as data privacy, bias, and lack of standardized validation remain. The study emphasizes the need for ethical implementation and interdisciplinary collaboration to ensure safe and effective RAG deployment in clinical settings. Ref. [
28] evaluated LLM-RAG models for surgical fitness assessment and preoperative education generation using local and international guidelines. Among ten models tested, GPT-4 with RAG achieved the highest accuracy (96.4%), outperforming clinicians and maintaining low hallucination rates. The findings highlight its potential as a reliable, consistent, and scalable support tool in preoperative and broader clinical workflows.
Article [
29] introduces SelfRewardRAG, a RAG-based framework that dynamically integrates real-time medical data with LLMs to address knowledge obsolescence in healthcare AI. Demonstrating strong performance across benchmarks like PubMedQA and MedQA, it delivers accurate, timely medical responses, surpassing some state-of-the-art models. While promising, its reliance on external data quality and high computational demands highlight the need for further optimization and ethical integration into clinical workflows. Study [
30] presents a Retrieval-Augmented Generation (RAG) pipeline tailored for preoperative medicine, integrating guideline-based knowledge into LLMs to enhance clinical accuracy. Using 35 preoperative guidelines, the GPT-4.0-RAG model achieved 91.4% accuracy, outperforming base LLMs and showing non-inferiority to junior doctors (86.3%). The system delivered rapid, safe, and guideline-concordant responses with lower hallucination rates. These findings highlight the potential of LLM-RAG systems as scalable and upgradeable tools in clinical reasoning and clinical judgment.
Manuscript [
31]’s work introduces Self-BioRAG, a domain-specific RAG framework designed for biomedical and clinical tasks, integrating retrieval, generation, and self-reflection modules. Trained on 84k biomedical instructions, it outperforms prior open-source models (≤
7B) by 7.2% on average across major biomedical QA benchmarks and achieves an 8% improvement in Rouge-1 for long-form QA. The authors emphasize the need for tailored retrievers, corpora, and instruction tuning to enhance domain adherence and factual accuracy in medical NLP tasks. Article [
32] evaluates Almanac, a retrieval-augmented LLM designed for clinical applications, using a dataset of 314 clinical questions. Compared to standard LLMs like ChatGPT-4, Bing, and Bard, Almanac demonstrated superior performance in factual accuracy, completeness, user preference, and safety. The study highlights the promise of domain-specific LLMs in medical decision making while emphasizing the need for rigorous validation before deployment.
Ref. [
33] presents ReinRAG, a reinforced reasoning-augmented generation method that leverages medical knowledge graphs to guide LLMs in generating long-form clinical discharge instructions from limited pre-admission data. By optimizing retrieval quality with group-normalized rewards, ReinRAG improves reasoning depth and reduces clinical misinterpretation. Experiments on real-world data demonstrate its superior performance in clinical accuracy and language generation compared to baseline models. Paper [
34] presents MEDGPT, a healthcare chatbot built on the Retrieval-Augmented Generation (RAG) framework, integrating external data sources like PDFs, CSVs, and PubMed to improve response accuracy and user satisfaction. By combining specialized retrieval tools and reasoning agents, MEDGPT delivers contextually relevant and personalized medical information.
Collectively, these research studies underscore the effectiveness of Retrieval-Augmented Generation (RAG) in enhancing the performance and reliability of large language models (LLMs) in the medical domain. By grounding their responses with current evidence-based practices and domain-specific information, RAG allows LLMs to dynamically integrate medical data and standards, enabling more accurate and customized outputs for tasks like diagnoses, drug development, and personalized care. The RAGMed system proposed in this manuscript advances the state of the art by comparing two embedding models and evaluating their impact on the performance and output quality of the RAG framework.
7. Discussion and Limitations
7.1. Novelty and Contributions
Beyond the comparison of embedding models, this work makes several contributions. First, it demonstrates the application of retrieval-augmented generation in healthcare dialogue and workflow support, a safety-critical domain where factual grounding is essential. Second, by adopting the RAG-Triad framework, we provide an evaluation that goes beyond surface-level overlap metrics and directly measures answer relevance, context precision, and recall—dimensions critical for clinical decision support. Third, we present a working prototype that integrates question answering, appointment scheduling through natural language, and clinical note summarization, thereby illustrating how such assistants can reduce the documentation burden and improve workflow efficiency. Finally, we explicitly address compliance and responsible use considerations, outlining the safeguards required for deployment in healthcare environments. Collectively, these contributions extend the state of the art by positioning RAG not only as a research technique but as a practically oriented, domain-grounded assistant for digital healthcare. In addition to these contributions, our system also has the following limitations.
7.2. Sample Size
The evaluation was based on a relatively small sample size, which restricts the statistical power and may limit the generalizability of the findings. A larger and more diverse set of dialogues would be needed to better capture the variability of real-world patient–provider interactions. A further limitation is that we did not include well-known medical QA benchmarks such as BioASQ, MedQA, PubMedQA, i2b2/n2c2, or MIMIC. While these datasets are highly valuable for benchmarking, many are task-mismatched with our setting. For example, BioASQ, MedQA, and PubMedQA focus on factoid question answering from biomedical literature rather than dialogue-based consultation, whereas i2b2/n2c2 and MIMIC primarily contain EHRs, clinical notes, or structured data rather than conversational exchanges. In addition, benchmarks such as MIMIC and i2b2/n2c2 involve restricted-access clinical records that require IRB approval and data use agreements. As this study was an early-stage, proof-of-concept prototype, we focused on fully public, reproducible dialogue datasets. Future work will extend evaluation to these benchmarks to enable broader comparability and stronger validation.
7.3. Lack of Physician Validation
This research is currently in an early-stage, proof-of-concept phase, focused on developing and refining a workable prototype of a retrieval-augmented medical AI assistant. As such, no practicing physicians, clinical experts, or medical professionals were directly involved in validating the system’s responses at this stage. Our goal in this phase was to establish the technical feasibility of retrieval-augmented generation using publicly available datasets. Validation with physicians and other healthcare professionals is a critical next step to ensure clinical accuracy, safety, and usability, and will be incorporated into subsequent phases of this research once the system has matured beyond the prototype stage. While public datasets and automatic metrics (RAG-Triad) provide a useful initial benchmark, expert review is essential to confirm the clinical accuracy, safety, and usability of the assistant in practice. Future work will include qualitative evaluation with physicians and domain specialists.
7.4. Dataset Bias
The use of public datasets introduces a potential risk of bias. Many of these corpora (e.g., MedDialog) are curated or translated, and may not fully reflect authentic clinical language, workflows, or cultural context. This could lead to mismatches between system behavior and real clinical communication. Additionally, because rare conditions and complex multimorbidity are underrepresented in public datasets, performance may not generalize evenly across all patient scenarios.
Finally, as a proof of concept, our system did not incorporate statistical significance testing, confidence intervals, or comparative baselines with standard NLP metrics, which limits the rigor of the reported results. Future work will address these limitations by (i) expanding the evaluation dataset, (ii) involving medical professionals in validation, (iii) incorporating safeguards against dataset bias, and (iv) conducting more comprehensive statistical and comparative analyses.
7.5. Evaluation Rigor
We acknowledge that our evaluation did not include commonly reported text generation metrics such as BLEU, ROUGE, and F1. While these measures are widely used and enable comparability across studies, they may not fully reflect correctness in the medical domain, where multiple semantically valid answers may differ in wording. Our decision to prioritize RAG-Triad was guided by the need for clinically meaningful evaluation. Nonetheless, future work will incorporate both RAG-specific and general NLP metrics to balance domain faithfulness with broader benchmarking. Another limitation is the absence of statistical significance testing and confidence intervals, which are valuable for quantifying uncertainty and enabling more rigorous comparisons. As this study was intended as a proof-of-concept study, we focused on domain-specific evaluation, but future work will expand the analysis to include confidence intervals, hypothesis testing, and other quantitative measures alongside RAG-Triad scores.
7.6. Dataset Limitations
While public datasets such as MedDialog provide large-scale patient–doctor dialogues, they may not fully reflect the complexity of real-world clinical communication. For example, MedDialog may differ in linguistic style, cultural context, and clinical workflow compared to typical healthcare encounters. Similarly, other publicly available corpora often rely on curated or synthetic interactions rather than transcripts from authentic patient visits.
Despite these limitations, such datasets remain valuable for proof-of-concept development because they offer diverse coverage of medical topics, are ethically shareable, and enable reproducibility. To ensure eventual clinical applicability, however, future work should validate the system using de-identified real-world clinical dialogues obtained under Institutional Review Board (IRB) approval and HIPAA-compliant data-sharing agreements.
7.7. Data Privacy and Compliance
This study used only publicly available, de-identified datasets (e.g., MedDialog, ACI-Bench, Augmented Medical Dialogue, MTS Dialog, Patient–Doctor Conversations). As such, no protected health information (PHI) was accessed, and therefore HIPAA and GDPR compliance concerns were not directly applicable at this stage. Because the work is an early-stage, proof-of-concept prototype, all the experiments were conducted exclusively on open datasets that are widely used for research and do not contain identifiable patient data.
For future phases that involve real clinical data or physician validation, strict compliance with data protection regulations will be followed, including Institutional Review Board (IRB) approval, HIPAA adherence in the U.S., and GDPR alignment for international collaborations. All the patient data would be de-identified, securely stored, and accessed only under approved data use agreements
7.8. Responsible Use and Clinical Safety Considerations
Healthcare is a safety-critical domain, and the outputs of large language models (LLMs) cannot be relied upon in isolation for clinical decision making. Our prototype is intended as an early-stage, proof-of-concept research system to explore the feasibility of retrieval-augmented generation for medical dialogue, not as a tool for direct patient use. We acknowledge that LLMs can produce hallucinations, incomplete reasoning, or contextually inappropriate responses, which poses risks if applied without oversight.
For this reason, we emphasize that such systems should be deployed only as decision support aids under the supervision of qualified healthcare professionals, not as autonomous decision makers. Future work will incorporate expert physician validation, human-in-the-loop safeguards, and reliability mechanisms (e.g., confidence estimation, grounding verification) to ensure safety, correctness, and clinical usability before any real-world application.
7.9. Evaluation Limitations
We recognize that the question test set used in this proof-of-concept study is not sufficient for formal validation. This small set was intended only as an illustrative demonstration of system capabilities. The primary aim of including these questions was to show the baseline behavior of the system and to demonstrate how responses remain grounded in retrieved evidence rather than relying solely on model priors.
More clinically meaningful benefits of the RAG approach will emerge when the system is applied to patient-specific scenarios in which the vector database contains individualized health information (e.g., allergies, family history, comorbidities). In such cases, the assistant can tailor responses based on structured context, which a non-retrieval LLM would not reliably achieve. Future work will therefore involve evaluation on larger and more diverse question sets, including patient-specific queries linked to synthetic or de-identified health records, and will incorporate both automatic metrics (e.g., RAG-Triad, standard NLP measures) and domain expert validation to assess correctness, completeness, and safety at scale.
8. Conclusions and Future Work
8.1. Conclusions
In this study, we presented RAGMed, a Retrieval-Augmented Generation (RAG)-based AI assistant designed to support clinical workflows. A central focus was the comparative evaluation of two sentence embedding models—all-MiniLM-L6-v2 and GTE-Large—using the RAG-Triad framework. While prior studies have compared embeddings for general NLP or biomedical retrieval tasks, our work specifically evaluates their impact within a retrieval-augmented clinical assistant. The contribution lies not merely in confirming that GTE-Large outperforms MiniLM, but in showing how embedding dimensionality directly affects grounding quality in a safety-critical domain. By assessing answer relevance, context precision, and recall, we demonstrate that embedding choice has measurable consequences for retrieving clinically useful passages and generating factually grounded responses. To our knowledge, this is among the first evaluations to link embedding dimensionality with RAG performance in medical dialogue and workflow support, extending state-of-the-art retrieval analyses into a practical healthcare context.
Beyond question answering, RAGMed can also handle appointment scheduling via natural language and the summarization of lengthy clinical narratives, illustrating its potential to reduce the administrative burden and enhance workflow efficiency. Collectively, these contributions highlight the promise of retrieval-augmented assistants to improve the accessibility, responsiveness, and reliability of digital healthcare services.
8.2. Future Work
Looking ahead, we plan to extend the assistant’s capabilities in several directions. First, we will explore voice-based querying to support more natural interactions, integration with Electronic Health Record (EHR) systems for seamless clinical workflows, and advanced privacy-preserving mechanisms to safeguard sensitive health data. These developments are intended to ensure that RAG-based systems are not only technically robust but also operationally viable for real-world healthcare deployment. Furthermore, we will address the current prototype’s limitation in handling negative findings (e.g., absence of symptoms, normal results, non-diagnostic statements). Negative findings carry significant clinical value, yet our proof-of-concept system focused only on retrieval-grounded positive evidence. Future work will therefore incorporate clinical NLP methods for negation detection (e.g., rule-based tools such as NegEx or more recent neural approaches) into the RAG pipeline. This will enable the assistant to capture and highlight both positive and negative evidence, resulting in more balanced, clinically comprehensive outputs that better reflect real-world reasoning.
Equally important, future phases will directly address the limitations identified in this study. We will involve physicians in iterative validation and co-design; expand evaluation to widely used biomedical benchmarks such as BioASQ, PubMedQA, i2b2/n2c2, and MIMIC; and incorporate statistical rigor through confidence intervals, hypothesis testing, and effect size reporting. These steps, together with safeguards for privacy, dataset bias, and responsible deployment, will establish a more clinically credible foundation for RAGMed and ensure its readiness for real-world integration.