A Deep Learning Enabled Multi-Class Plant Disease Detection Model Based on Computer Vision
Abstract
:1. Introduction
2. The Proposed Network Structure of the Detection Model
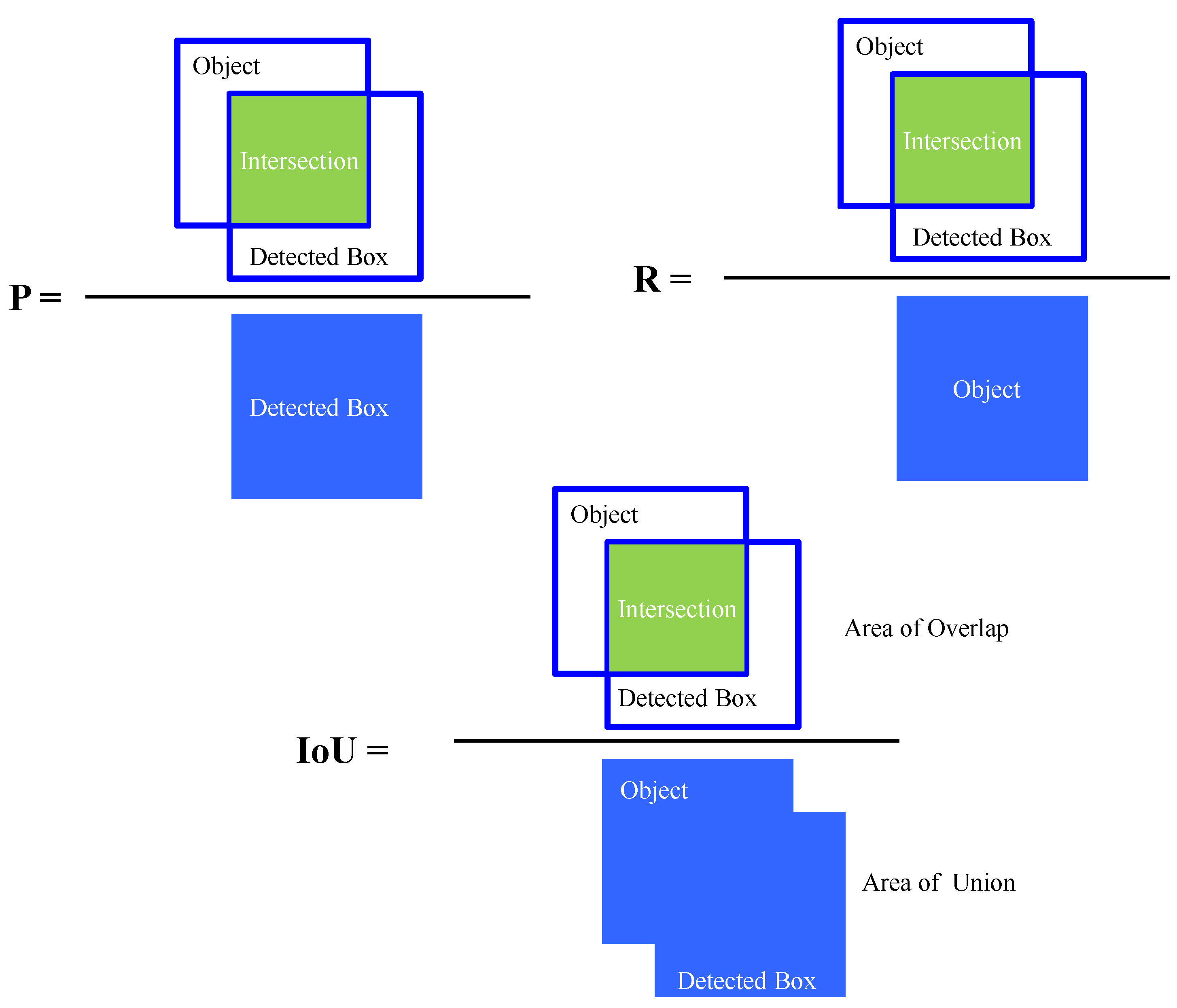
3. Performance Matrices of the Detection Model
4. Result and Discussion
4.1. Overall Performance of the Proposed Detection Model
4.2. Detection Results for Different Plant Disease Class
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tyagi, A.C. Towards a second green revolution. Irrig. Drain. 2016, 65, 388–389. [Google Scholar] [CrossRef]
- Vougioukas, S.G. Agricultural robotics. Annu. Rev. Control Robot. Auton. Syst. 2019, 2, 365–392. [Google Scholar] [CrossRef]
- Wang, Q.; Nuske, S.; Bergerman, M.; Singh, S. Automated crop yield estimation for apple orchards. Exp. Robot. 2013, 88, 745–758. [Google Scholar]
- Basnet, B.; Bang, J. The state-of-the-art of knowledge-intensive agriculture: A review on applied sensing systems and data analytics. J. Sens. 2018, 2018, 3528296. [Google Scholar]
- Fu, L.; Gao, F.; Wu, J.; Li, R.; Karkee, M.; Zhang, Q. Application of consumer RGB-D cameras for fruit detection and localization in field: A critical review. Comput. Electron. Agric. 2020, 177, 105687. [Google Scholar] [CrossRef]
- SepuLveda, D.; Fernández, R.; Navas, E.; Armada, M.; González-De-Santos, P. Robotic aubergine harvesting using dual-arm manipulation. IEEE Access 2020, 8, 121889–121904. [Google Scholar] [CrossRef]
- Peltoniemi, J.I.; Gritsevich, M.; Puttonen, E. Reflectance and polarization characteristics of various vegetation types. In Light Scattering Reviews; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9, pp. 257–294. [Google Scholar]
- Zhao, Y.; Gong, L.; Huang, Y.; Liu, C. A review of key techniques of vision-based control for harvesting robot. Comput. Electron. Agric. 2016, 127, 311–323. [Google Scholar] [CrossRef]
- Shamshiri, R.; Redmond, C.W.; Hameed, I.A.; Yule, I.J.; Grift, T.E.; Balasundram, S.K.; Pitonakova, L.; Ahmad, D.; Chowdhary, G. Research and development in agricultural robotics: A perspective of digital farming. Int. J. Agric. Biol. Eng. 2018, 11, 1–14. [Google Scholar] [CrossRef]
- Tang, Y.C.; Wang, C.; Luo, L.; Zou, X. Recognition and localization methods for vision-based fruit picking robots: A review. Front. Plant Sci. 2020, 11, 510. [Google Scholar] [CrossRef]
- Ling, X.; Zhao, Y.; Gong, L.; Liu, C.; Wang, T. Dual-arm cooperation and implementing for robotic harvesting tomato using binocular vision. Robot. Auton. Syst. 2019, 114, 134–143. [Google Scholar] [CrossRef]
- Qin, F.; Liu, D.; Sun, B.; Ruan, L.; Ma, Z.; Wang, H. Identification of alfalfa leaf diseases using image recognition technology. PLoS ONE 2016, 11, e0168274. [Google Scholar] [CrossRef] [Green Version]
- Chuanlei, Z.; Shanwen, Z.; Jucheng, Y.; Yancui, S.; Jia, C. Apple leaf disease identification using genetic algorithm and correlation based feature selection method. Int. J. Agric. Biol. Eng. 2017, 10, 74–83. [Google Scholar]
- Al Bashish, D.; Braik, M.; Bani-Ahmad, S. Detection and classification of leaf diseases using k-means-based segmentation and neural networks-based classification. Inf. Technol. J. 2011, 10, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Dhaygude, S.B.; Kumbhar, N.P. Agricultural plant leaf disease detection using image processing. Int. J. Adv. Res. Elect. 2013, 2, 599–602. [Google Scholar]
- Rajan, P.; Radhakrishnan, B.; Suresh, L.P. Detection and classification of pests from crop images using support vector machine. In Proceedings of the 2016 International Conference on Emerging Technological Trends (ICETT), Kollam, India, 21–22 October 2016; pp. 1–6. [Google Scholar]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early detection and classification of plant diseases with support vector machines based on hyperspectral rectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Islam, M.; Dinh, A.; Wahid, K.; Bhowmik, P. Detection of potato diseases using image segmentation and multiclass support vector machine. In Proceedings of the 2017 IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE), Windsor, ON, Canada, 30 April–3 May 2017; pp. 1–4. [Google Scholar]
- Wu, C.; Luo, C.; Xiong, N.; Zhang, W.; Kim, T. A greedy deep learning method for medical disease analysis. IEEE Access 2018, 6, 20021–20030. [Google Scholar] [CrossRef]
- Lu, H.; Li, Y.; Chen, M.; Kim, H.; Serikawa, S. Brain intelligence: Go beyond artificial intelligence. Mob. Netw. Appl. 2018, 23, 368–375. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, N.; Wang, Z.-H.; Li, H.; Chang, C.-C.; Wang, H. New secret sharing scheme based on faster R-CNNs image retrieval. IEEE Access 2018, 6, 49348–49357. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Chan, C.S.; Mayo, S.J.; Remagnino, P. How deep learning extracts and learns leaf features for plant classification. Pattern Recogn. 2017, 71, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.D.; Dong, Z.; Chen, X.; Jia, W.; Du, S.; Muhammad, K.; Wang, S.H. Image based fruit category classification by 13-layer deep convolutional neural network and data augmentation. Multimed. Tools Appl. 2017, 78, 3613–3632. [Google Scholar] [CrossRef]
- Tang, J.L.; Wang, D.; Zhang, Z.G.; He, L.J.; Xin, J.; Xu, Y. Weed identification based on K-means feature learning combined with convolutional neural network. Comput. Electron. Agric. 2017, 135, 63–70. [Google Scholar] [CrossRef]
- Arribas, J.I.; Sánchez-Ferrero, G.V.; Ruiz-Ruiz, G.; Gómez-Gil, J. Leaf classification in sunflower crops by computer vision and neural networks. Comput. Electron. Agric. 2011, 78, 9–18. [Google Scholar] [CrossRef]
- Dias, P.A.; Tabb, A.; Medeiros, H. Apple flower detection using deep convolutional networks. Comput. Ind. 2018, 99, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, K.; Guo, W.; Yoshioka, Y.; Ninomiya, S. On plant detection of intact tomato fruits using image analysis and machine learning methods. Sensors 2014, 14, 12191–12206. [Google Scholar] [CrossRef] [Green Version]
- Caglayan, A.; Can, A.B. Volumetric object recognition using 3-D CNNs on depth data. IEEE Access 2018, 6, 20058–20066. [Google Scholar] [CrossRef]
- Lu, H.; Li, Y.; Uemura, T.; Kim, H.; Serikawa, S. Low illumination underwater light field images reconstruction using deep convolutional neural networks. Future Gener. Comput. Syst. 2018, 82, 142–148. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, A.F.; Yoon, S.; Lee, J.; Park, D.S. High-performance deep neural network-based tomato plant diseases and pests diagnosis system with refinement filter bank. Front. Plant Sci. 2018, 9, 1162. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, A.F.; Yoon, S.; Park, D.S. Deep learning-based phenotyping system with glocal description of plant anomalies and symptoms. Front. Plant Sci. 2019, 10, 1321. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Hu, J.; Zhao, G.; Mei, F.; Zhang, C. An in-field automatic wheat disease diagnosis system. Comput. Electron. Agric. 2017, 142, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Yi, S.; Zeng, N.; Liu, Y.; Zhang, Y. Identification of Rice diseases using deep convolutional neural networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; He, D.J.; Li, Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 2017, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Qiao, Y.; Meng, F.; Fan, C.; Zhang, M. Identification of maize leaf diseases using improved deep convolutional neural networks. IEEE Access 2018, 6, 30370–30377. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Ross, G. Fast r-cnn. In Proceedings of the IEEE international Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 1137–1149. [Google Scholar] [CrossRef] [Green Version]
- Kaiming, H.; Georgia, G.; Piotr, D.; Ross, G. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Bargoti, S.; Underwood, J. Deep fruit detection in orchards. In Proceedings of the 2017 IEEE International Conference on Robotics and Automation (ICRA), Singapore, 29 May–3 June 2017; pp. 1–8. [Google Scholar]
- Inkyu, S.; Ge, Z.; Feras, D.; Ben, U.; Tristan, P.; Chris, M.C. DeepFruits: A fruit detection system using deep neural networks. Sensors 2016, 16, 1222. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, realtime object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6517–6525. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLOv3: An incremental improvement. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Misra, D. Mish: A self regularized non-monotonic neural activation function. arXiv 2019, arXiv:1908.08681. [Google Scholar]
- Huang, G.; Liu, Z.; Laurens, V.D.M.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for activation functions. arXiv 2017, arXiv:1710.05941. [Google Scholar]
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. In Proceedings of the ICML 2013, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. (TPAMI) 2015, 37, 1904–1916. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Qi, L.; Qin, H.F.; Shi, J.P.; Jia, J.Y. Path Aggregation Network for Instance Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8759–8768. [Google Scholar]
- Ghiasi, G.; Lin, T.-Y.; Le, Q.V. Dropblock: A regularization method for convolutional networks. In Proceedings of the Advances in Neural Information Processing Systems, Montréal, QC, USA, 3–8 December 2018; pp. 10727–10737. Available online: https://arxiv.org/pdf/1810.12890.pdf (accessed on 11 December 2020).
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU loss: Faster and better learning for bounding box regression. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–11 February 2020; Volume 34, No. 7. pp. 12993–13000. [Google Scholar]
- Kaggle PlantPathology Apple Dataset 2020. Available online: https://www.kaggle.com/piantic/plantpathology-apple-dataset (accessed on 9 March 2021).
- LabelImg 2021. Available online: https://github.com/tzutalin/labelImg (accessed on 24 March 2021).
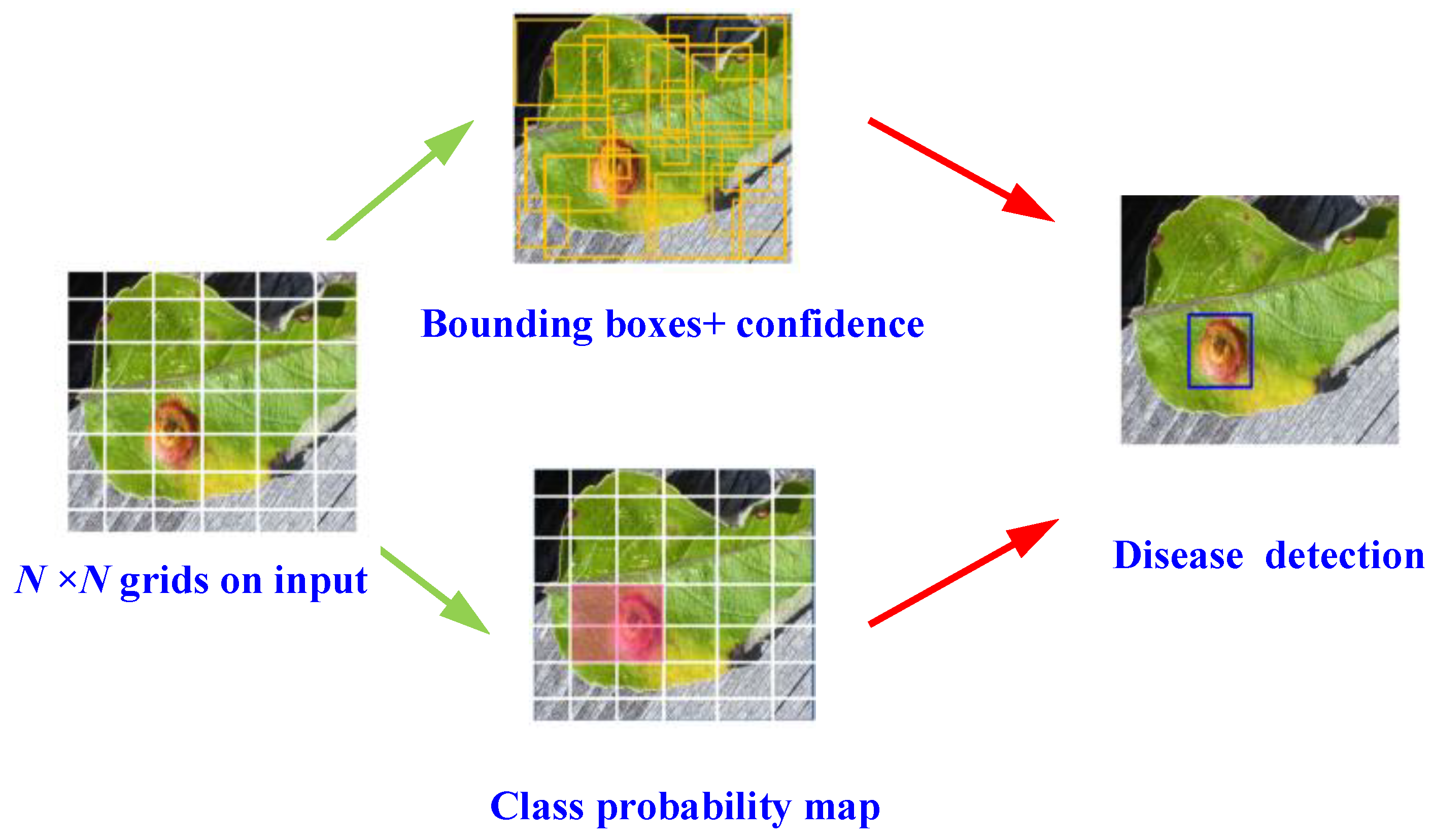
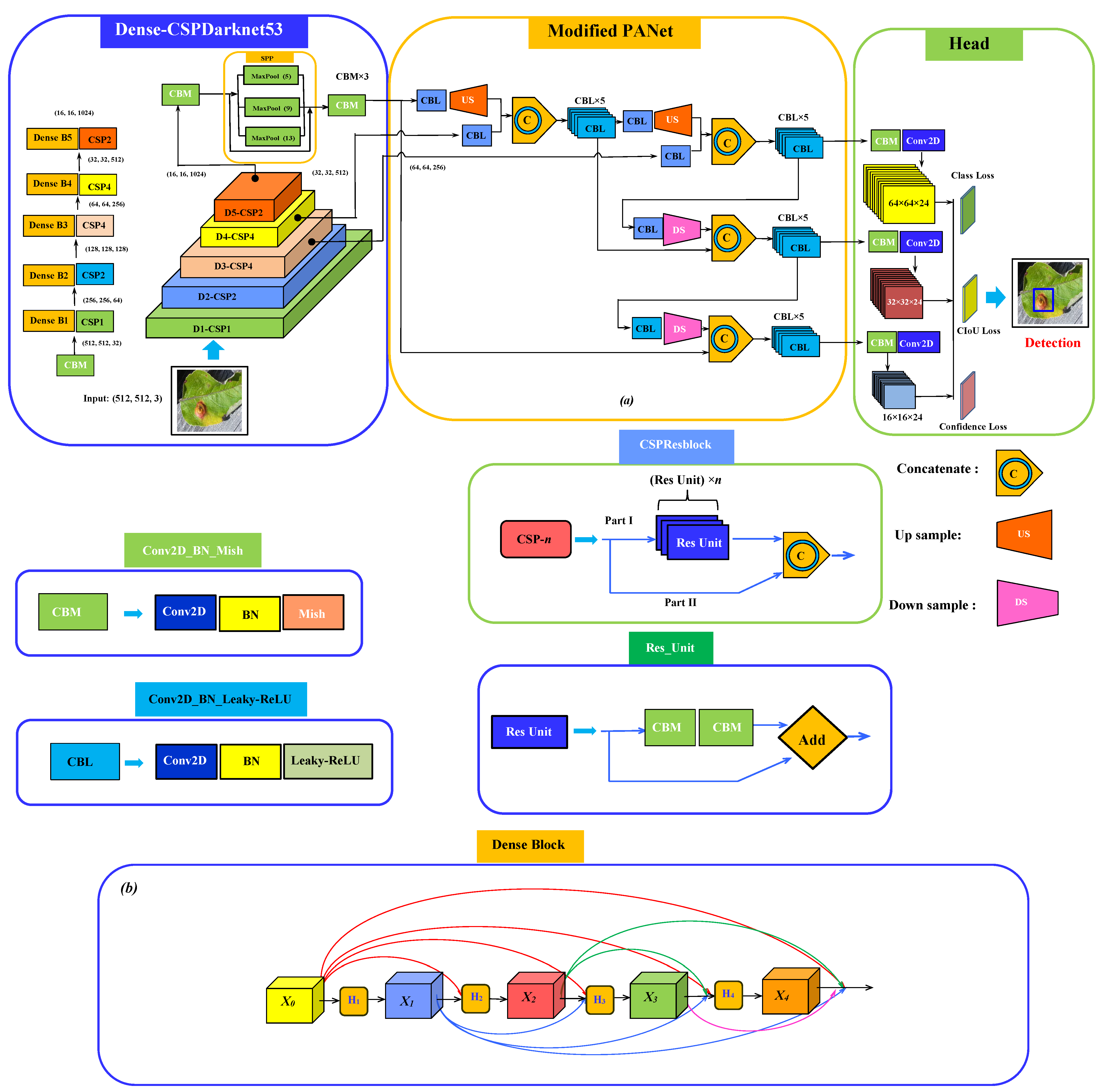
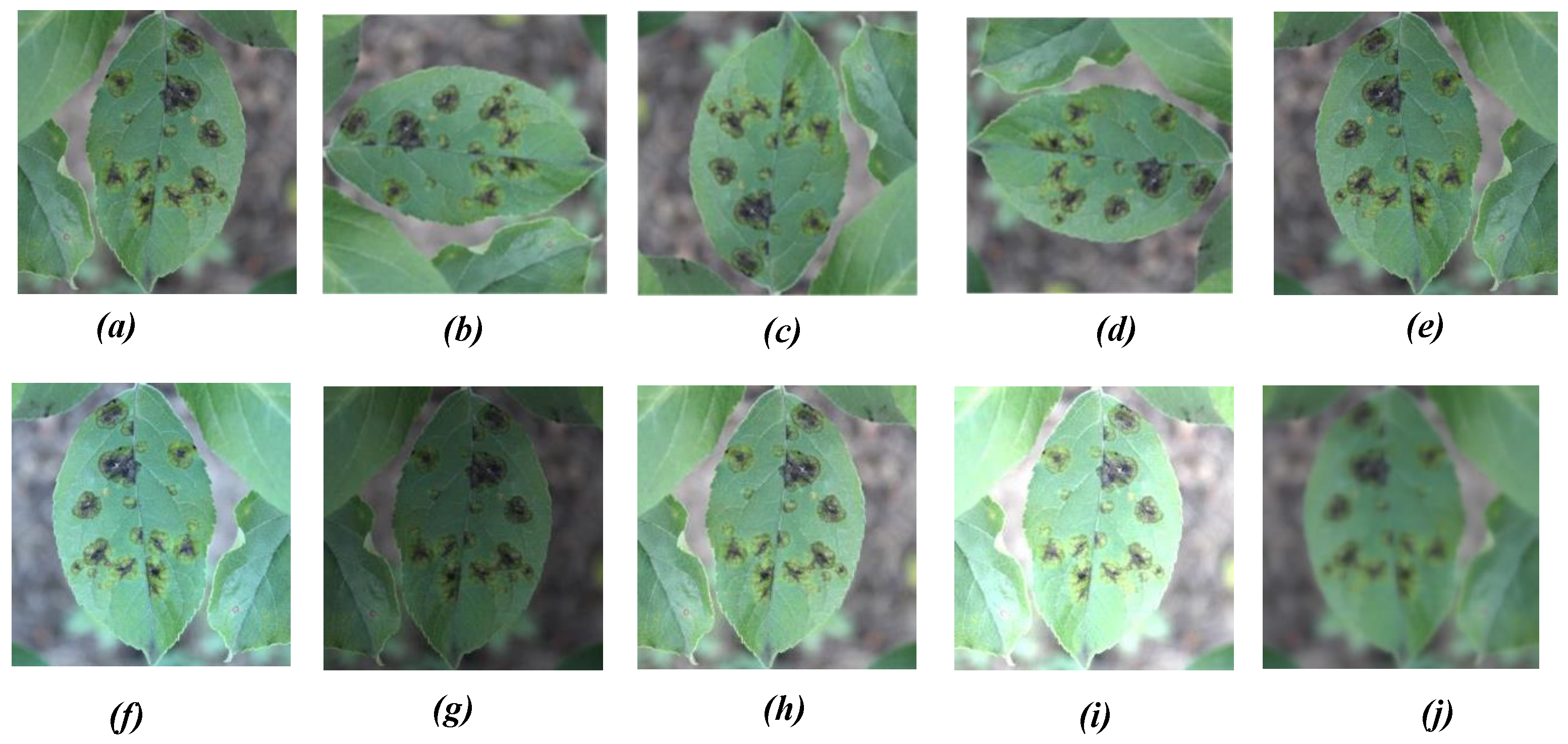

| Object Class | Scab | Rust | Mix (Scab and Rust) |
|---|---|---|---|
| Class identifier | 1 | 2 | 1, 2 |
| Original images | 200 | 200 | 200 |
| Rotation | 800 | 800 | 800 |
| Color balancing | 200 | 200 | 200 |
| Brightness transform | 600 | 600 | 600 |
| Blur processing | 200 | 200 | 200 |
| Total number of images/class | 2000 | 2000 | 2000 |
| Testing Environment | Configuration Parameter |
|---|---|
| OS | Windows 10 Pro 64 |
| CPU | Intel Core i5-10210U |
| RAM | 8 GB DDR4 |
| GPU | NVIDIA GeForce RTX 2080 |
| GPU acceleration env. | CUDA 10.2.89 |
| GPU acclerated DNN lib. | cuDNN 10.2 v7.6.5 |
| Int. development env. | Visual Studio comm. v15.9 (2017) |
| Comp. Vision lib. | OpenCV 4.5.1-vc14 |
| Input Size of Image | Batch | Subdivision | Channels | Momentum |
| 16 | 4 | 3 | 0.9 | |
| Initial Learning Rate | Decay | Classes | Filters | Training Steps |
| 0.001 | 0.005 | 4 | 27 | 85,000 |
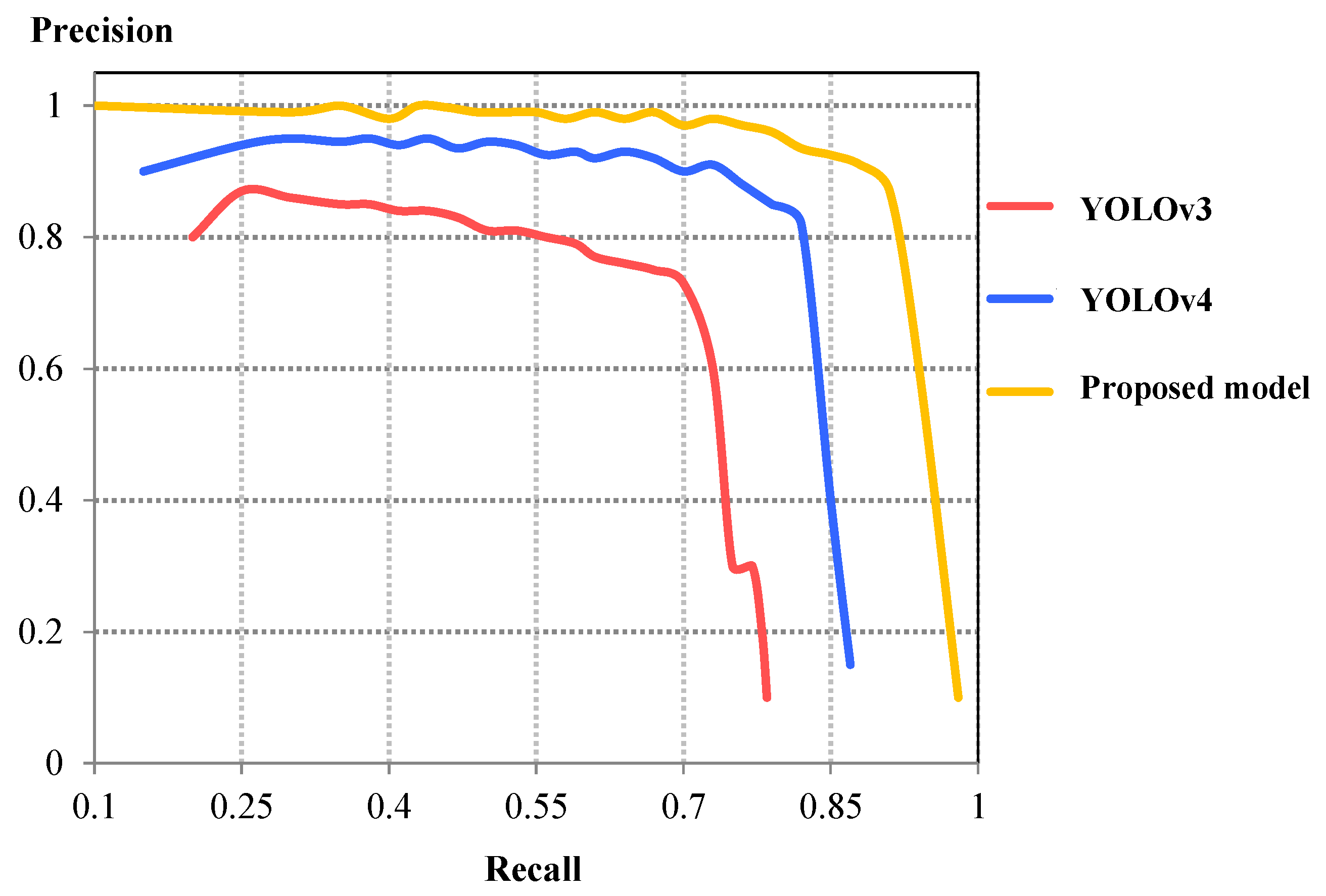
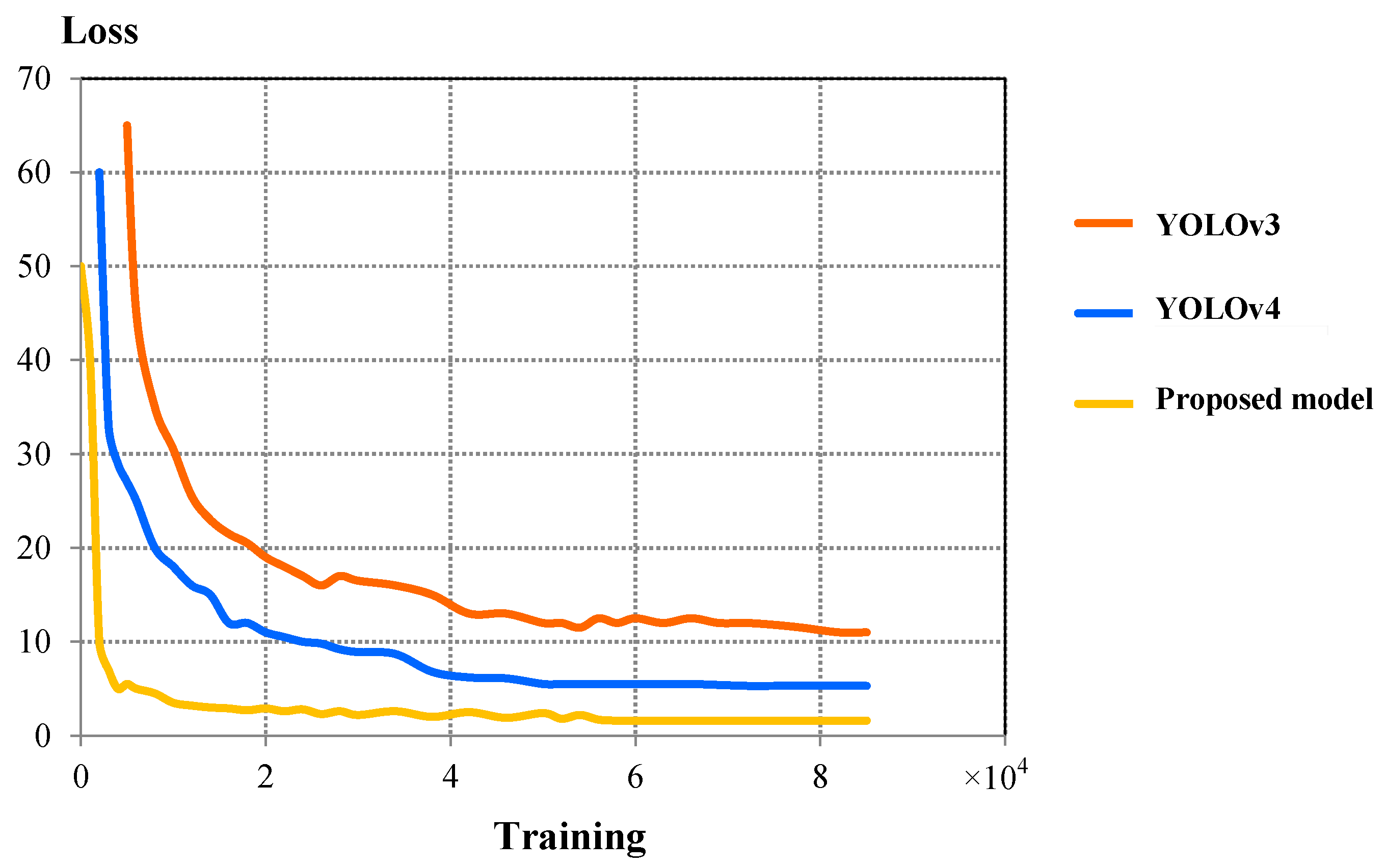
| Detection Model | IoU | F1-Score | mAP | Validation Loss | Detection Time (ms) | Detection Speed (FPS) |
|---|---|---|---|---|---|---|
| YOLOv3 | 0.787 | 0.822 | 0.781 | 11.12 | 16.254 | 61.52 |
| YOLOv4 | 0.861 | 0.883 | 0.839 | 4.31 | 15.301 | 65.22 |
| Proposed model | 0.922 | 0.959 | 0.912 | 1.65 | 17.577 | 56.89 |
| Model | Class | Objects | TP | FP | FN | P (%) | R (%) | F1-Score |
|---|---|---|---|---|---|---|---|---|
| YOLOv3 | All | 3517 | 2688 | 750 | 408 | 78.18 | 86.82 | 82.27 |
| Scab | 1975 | 1501 | 398 | 187 | 79.04 | 88.92 | 83.69 | |
| Rust | 1542 | 1187 | 352 | 221 | 77.12 | 84.30 | 80.56 | |
| YOLOv4 | All | 3517 | 2944 | 525 | 248 | 84.86 | 92.23 | 88.39 |
| Scab | 1975 | 1643 | 286 | 137 | 85.17 | 92.30 | 88.59 | |
| Rust | 1542 | 1301 | 239 | 111 | 84.48 | 92.13 | 88.14 | |
| Proposed Model | All | 3517 | 3272 | 212 | 62 | 93.91 | 98.14 | 95.98 |
| Scab | 1975 | 1845 | 127 | 39 | 93.55 | 97.93 | 95.69 | |
| Rust | 1542 | 1427 | 85 | 23 | 94.37 | 98.41 | 96.35 |
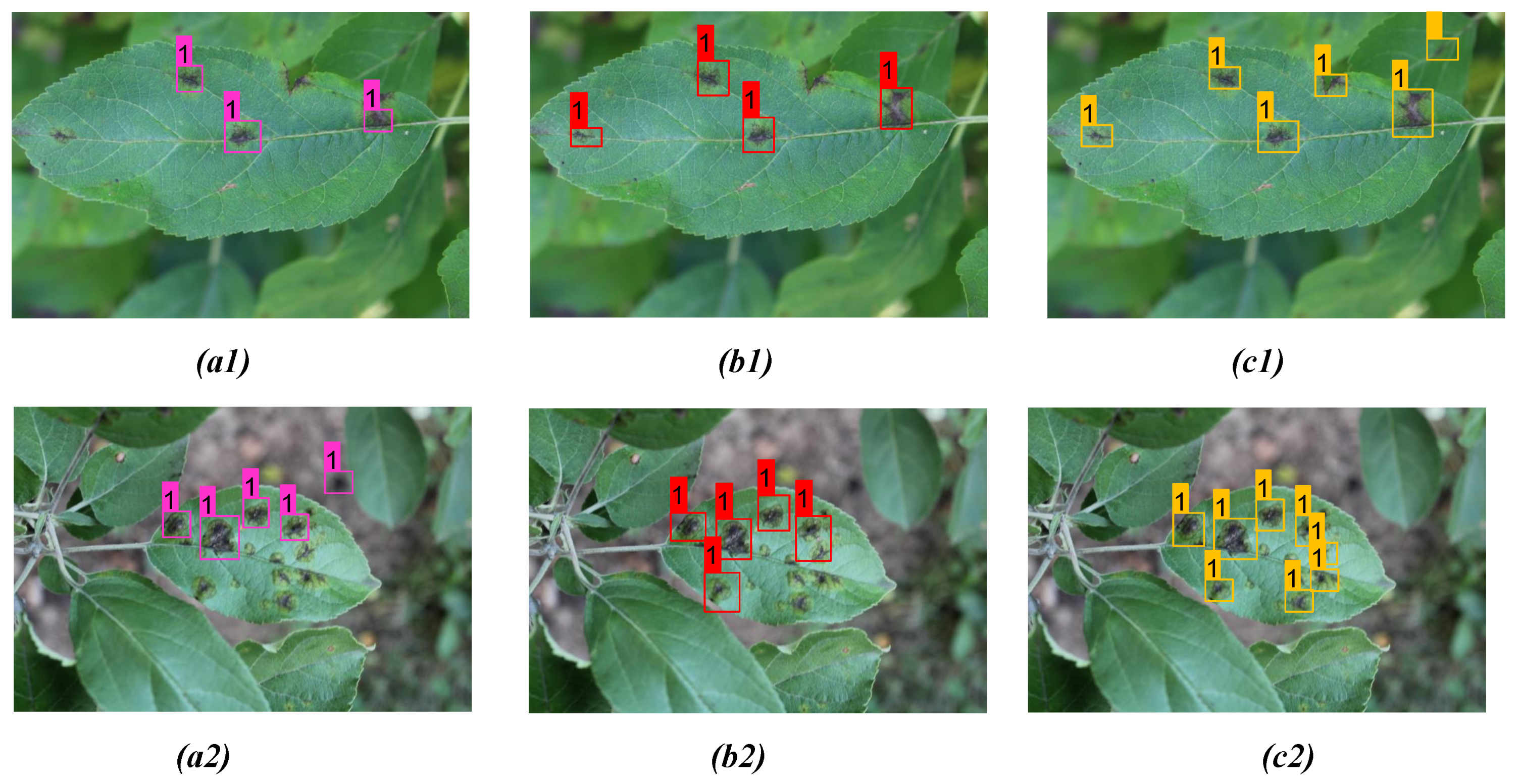
| Figs. No | Model | Detc. | Undetc. | Misdetc. | Confidence Scores |
|---|---|---|---|---|---|
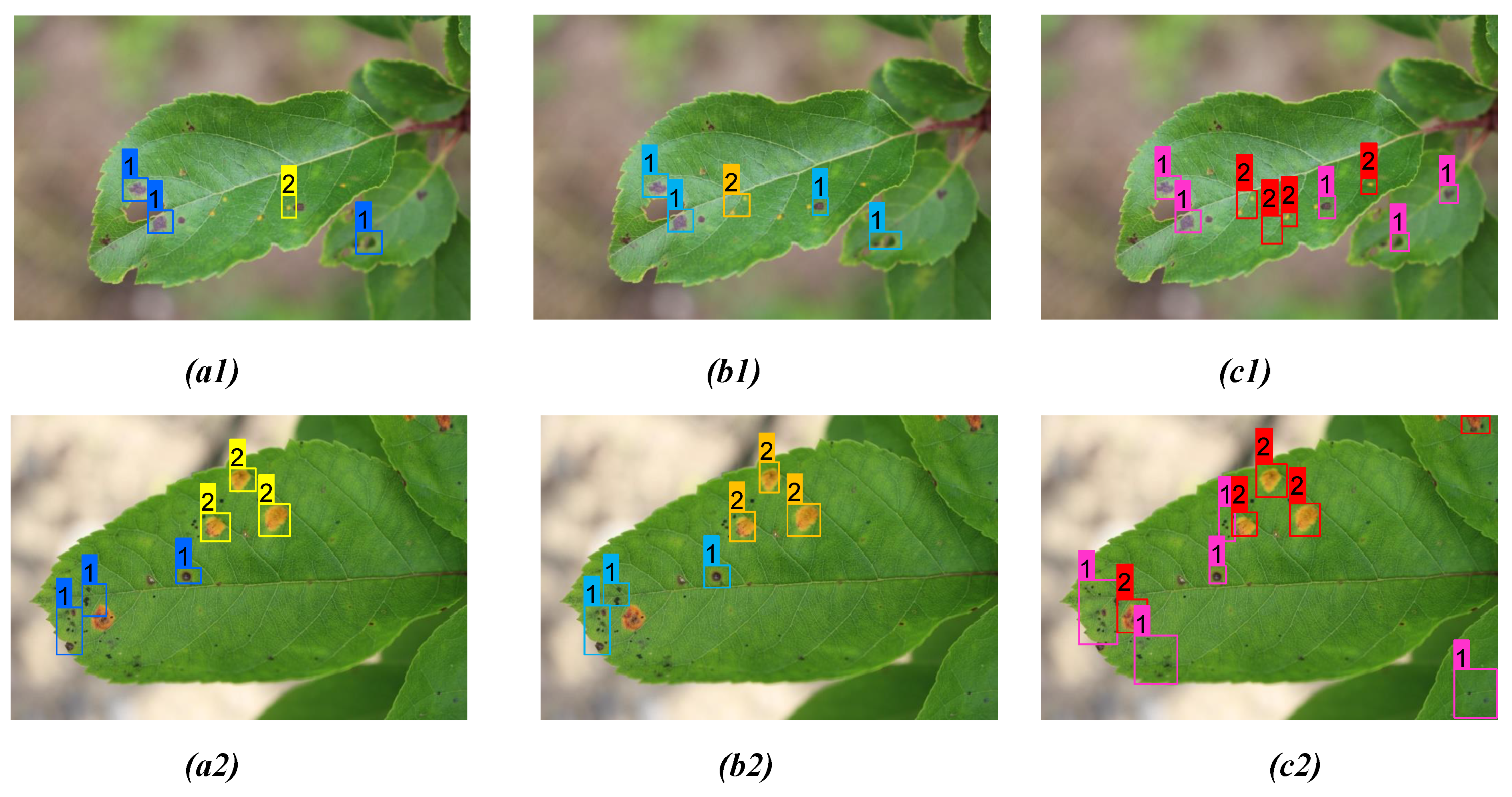
| Figure 8(a1) | YOLOv3 | 3 | 3 | 0 | 0.84, 0.93, 0.98 |
| Figure 8(b1) | YOLOv4 | 4 | 2 | 0 | 0.94, 1.00, 1.00, 0.98 |
| Figure 8(c1) | Proposed model | 6 | 0 | 0 | 0.98, 1.00, 1.00, 0.97, 1.00, 1.00 |
| Figure 8(a2) | YOLOv3 | 3 | 5 | 1 | 0.81, 0.94, 0.77 |
| Figure 8(b2) | YOLOv4 | 4 | 4 | 1 | 0.97, 0.81, 1.00, 0.78 |
| Figure 8(c2) | Proposed model | 8 | 1 | 0 | 1.00, 1.00, 0.92, 1.00 1.00, 1.00, 1.00, 0.97 |
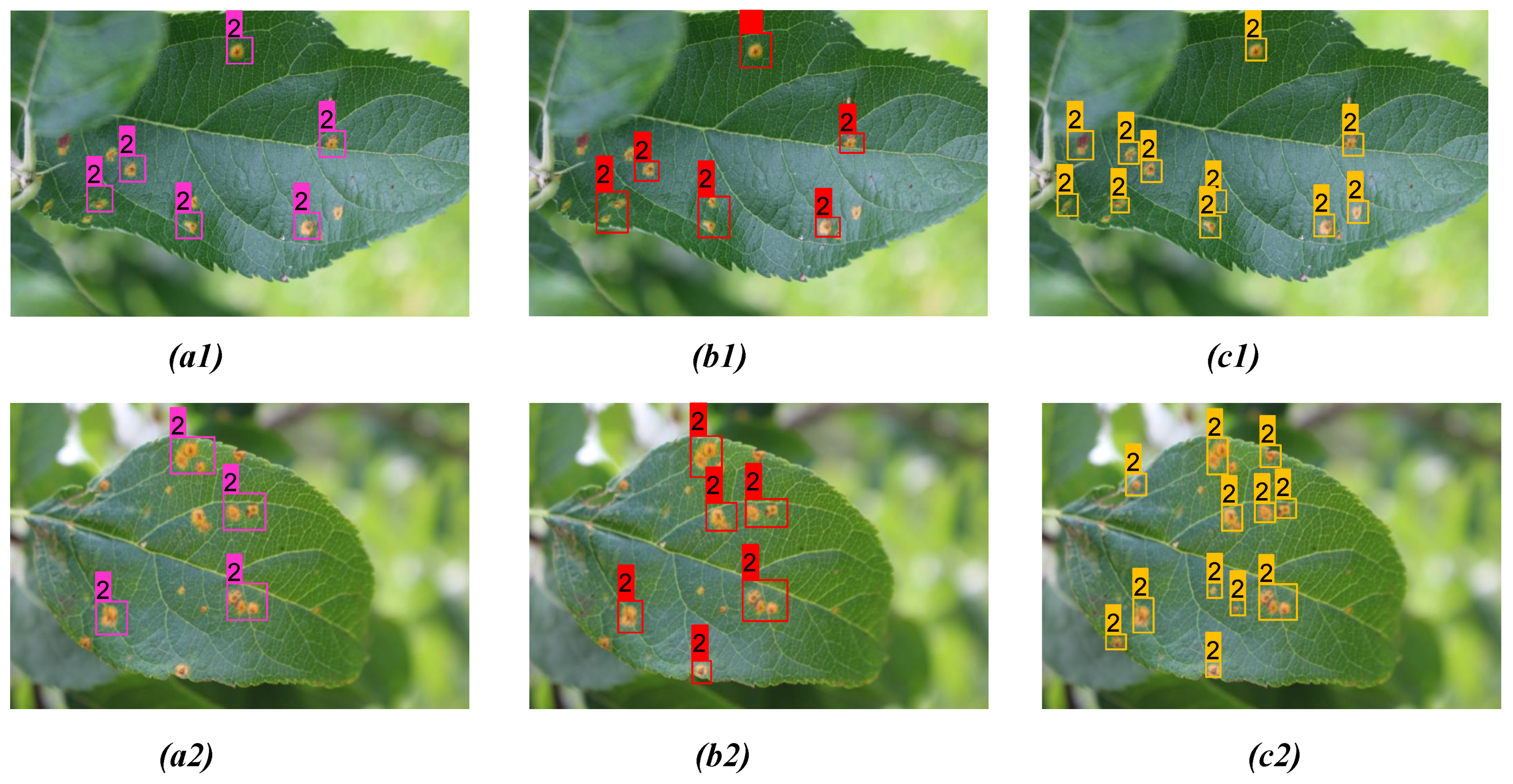
| Figs. No | Model | Detc. | Undetc. | Misdetc. | Confidence Scores |
|---|---|---|---|---|---|
| Figure 9(a1) | YOLOv3 | 6 | 5 | 0 | 0.84, 0.93, 0.79, 0.93, 0.89, 0.94 |
| Figure 9(b1) | YOLOv4 | 8 | 3 | 0 | 0.88, 0.91, 0.87, 0.91, 0.78, 0.83 0.99, 0.89 |
| Figure 9(c1) | Proposed model | 11 | 0 | 0 | 0.96, 0.91, 1.00, 1.00, 0.92, 1.00 1.00, 1.00, 0.98, 1.00, 0.97 |
| Figure 9(a2) | YOLOv3 | 7 | 7 | 1 | 0.91, 0.78, 0.98, 0.76, 0.79, 0.92, 0.86 |
| Figure 9(b2) | YOLOv4 | 8 | 6 | 1 | 0.92, 0.91, 0.83, 1.00, 0.92, 0.87 0.91, 0.83 |
| Figure 9(c2) | Proposed model | 12 | 2 | 0 | 0.95, 0.99, 0.87, 1.00, 0.92, 1.00 0.94, 1.00, 0.98, 1.00, 0.83, 0.97 |
| Figs. No | Model | Detc. | Undetc. | Misdetc. | Confidence Scores |
|---|---|---|---|---|---|
| Figure 10(a1) | YOLOv3 | 6 | 5 | 0 | 0.84, 0.88, 0.76, 0.79, 1.00, 0.98 |
| Figure 10(b1) | YOLOv4 | 7 | 4 | 0 | 0.82, 0.77, 1.00, 0.93, 1.00 0.83, 0.94 |
| Figure 10(c1) | Proposed model | 10 | 1 | 0 | 0.90, 0.87, 1.00, 0.92, 1.00 0.94, 1.00, 1.00, 0.83, 0.97 |
| Figure 10(a2) | YOLOv3 | 6 | 6 | 1 | 0.91, 0.67, 0.81 0.94, 0.77, 0.79 |
| Figure 10(b2) | YOLOv4 | 6 | 6 | 1 | 0.97, 0.86, 1.00, 0.77, 0.85, 0.67 |
| Figure 10(c2) | Proposed model | 9 | 3 | 0 | 0.72, 0.90, 1.00, 0.92, 1.00 0.83, 0.95, 0.94, 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roy, A.M.; Bhaduri, J. A Deep Learning Enabled Multi-Class Plant Disease Detection Model Based on Computer Vision. AI 2021, 2, 413-428. https://doi.org/10.3390/ai2030026
Roy AM, Bhaduri J. A Deep Learning Enabled Multi-Class Plant Disease Detection Model Based on Computer Vision. AI. 2021; 2(3):413-428. https://doi.org/10.3390/ai2030026
Chicago/Turabian StyleRoy, Arunabha M., and Jayabrata Bhaduri. 2021. "A Deep Learning Enabled Multi-Class Plant Disease Detection Model Based on Computer Vision" AI 2, no. 3: 413-428. https://doi.org/10.3390/ai2030026
APA StyleRoy, A. M., & Bhaduri, J. (2021). A Deep Learning Enabled Multi-Class Plant Disease Detection Model Based on Computer Vision. AI, 2(3), 413-428. https://doi.org/10.3390/ai2030026







