1. Introduction
One of the most damaging problems in all agriculture is the citrus huanglongbing (HLB) [
1], an incurable disease that is caused by the bacterium
Liberibacter spp.) [
2]. The main vector for this disease is the psyllid species
, a two to three-millimeter light-brown insect that is most often found in citrus shoots. The constant monitoring of psyllid infestation is thus essential for timely preventive actions to avoid substantial economic losses.
Psyllids can be sampled in the field using a variety of methods [
3], among which sticky traps are the most used for monitoring infestations due to their reliability [
4]. However, accurate population estimates have to rely on a very time-consuming sorting and counting process. In addition, fatigue and well-known cognitive phenomena tend to decrease the accuracy of the visual assessment [
5]. As a result, infestation estimates may become too inaccurate to properly support decisions. This is a problem that affects not only psyllids, but almost all methods of insect infestation estimation, which has given rise to many research efforts towards the automation of the process using imaging devices [
6].
Depending on the type of infestation, the detection of insects directly on leaves [
7,
8,
9,
10,
11,
12], stems [
13], and flowers [
14] can be appropriate, but in most cases counting insects caught by traps is a better proxy for the real situation in a given region [
15]. The literature has many examples of image-based insect monitoring that employ traps for sample collection [
5,
16,
17,
18,
19,
20,
21,
22]. In most cases, traps capture not only the specimens of interest, but also other invertebrate species, dust, and other types of debris. Detecting specific objects in images captured under this type of condition can be challenging. This is particularly true in the case of psyllids, as these tend to be among the smallest objects usually found in the traps. Pattern recognition in a busy environment is the type of task that can, in many cases, be suitably handled by machine learning algorithms. Thus, it is no surprise that this type of technique has been predominant in this context.
Traditional machine learning techniques such as multi-layered perceptron (MLP) neural networks [
20], Support Vector Machines (SVM) [
10,
13,
14,
23], and K-means [
21] have been applied to insect counting in sticky traps, with various degrees of success. However, an important limitation associated with those techniques is that they either require complex feature engineering to work properly, or are not capable of correctly modelling all the nuances and nonlinearities associated with pattern recognition with busy backgrounds. This may explain in part why deep learning techniques, and CNNs in particular, have gained so much popularity in the last few years for image recognition tasks.
The use of deep learning for insect recognition has mostly concentrated on the classification of samples into species and subspecies, with the insects of interest appearing prominently in the images captured either under natural [
11,
24] or controlled [
25] conditions. Some investigations have also addressed pest detection amidst other disorders [
26]. To the best of the authors’ knowledge, the only work found in the literature that uses deep learning for monitoring psyllids was proposed by Partel et al. [
27], achieving precision and recall values of 80% and 95%, respectively. Another study with similar characteristics, but dedicated to moth detection in sticky traps, was carried out by Ding and Taylor [
19]. In any case, a problem that is common to the vast majority of the investigations based on deep learning found in the literature is that training and tests are done with the datasets generated using the same camera or imaging device. As a result, the model will be trained using data that lack the variability that can be found in practice, reducing its reliability. This difference between the distribution of the data used to train the model and the data on which the model is to be applied in the real world is commonly called covariate shift [
28], and is one of the main reasons for models to fail under real world conditions.
In order to investigate how severe is the covariate shift problem when CNNs are used to detect and count psyllids in yellow sticky traps, nine distinct datasets were built using different imaging devices. Extensive experiments were conducted in order to determine the impact that the lack of variability has on the ability of the trained model to handle image data acquired by different people using different devices. This study is a follow-up to a previous investigation about the influence of image quality on the detection of psyllids [
29].
3. Results and Discussion
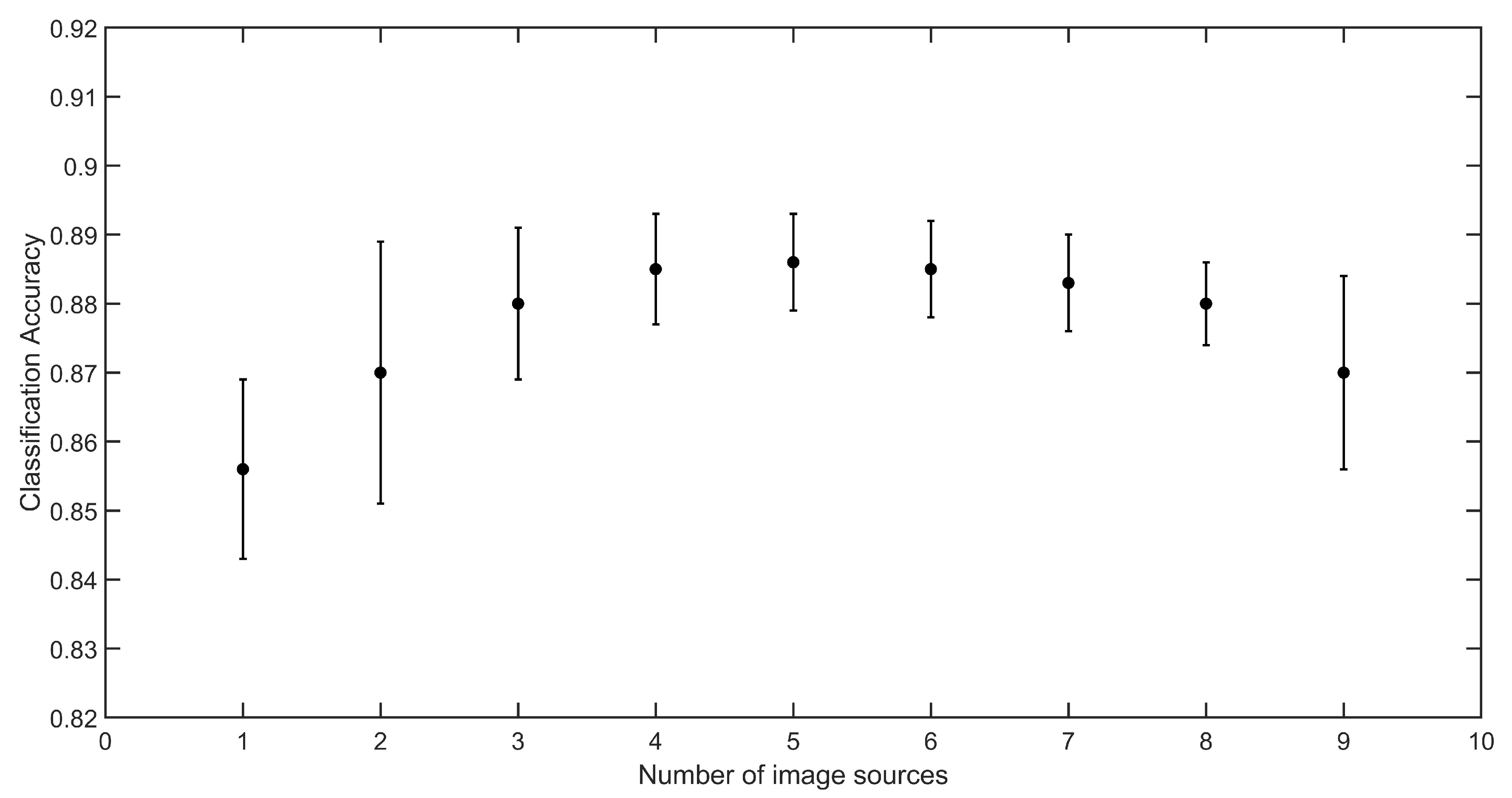
The results obtained in experiment 1 (
Figure 2 and
Table 4) clearly indicate that increasing the variability of the samples used for training the model, while keeping the number of images fixed, indeed improves the results, as error rates decreased by almost 40% (from 15.1% using a single source to 9.4% using all sources). The standard deviations were quite low, indicating that there was not much variation between repetitions and between different source combinations. However, when only a few sources were used, the inclusion of certain devices, especially the Xiaomi Redmi Note 3 and the iPhone 6s, led to slightly higher accuracies. On the other hand, the inclusion of the scanner images was not helpful in any case, confirming the conclusions reached in [
29]. The explanation for this is that the scanner images, despite having a much higher quality in terms of resolution and illumination homogeneity, are too different from the smartphone images to cause a positive impact on the model training. Thus, with the number of sources fixed, there was not much difference when different devices were included in the training, except in the case of the scanner. This indicates that the quality variation of the samples acquired with different smartphones is important to better prepare the model to deal with the variability expected in practice. In addition, since each device covers part of that variability, their replacement does not cause a significant change in the accuracy (but the samples that are misclassified may be different).
While the number of images used for training was above 300 (less than five sources included in the training set), the results obtained in experiment 2 were similar to those achieved in experiment 1 (
Figure 3 and
Table 5). As the number of images used for training fell below 300 and the number of sources increased, the accuracy plateaued, but did not fall significantly even when as few as 180 images (nine sources) were used. This indicates that data variability is more important than data quantity. Although it is evident that the more images are included, the higher the probability is that more variability is included, and a carefully selected training set may be effective even with a relatively low number of samples. This is very relevant considering that obtaining large amounts of labelled data is usually a challenge.
The results obtained in experiment 3 (model training using a single source) were mostly consistent with those yielded by the previous two experiments (
Table 6), but some relevant differences were observed as the source for the training samples was changed. The best training dataset was the first Motorola Moto Z2 Play, which resulted in an average accuracy of 0.876. A possible explanation for this is that this smartphone has an intermediate resolution and optical quality in comparison with the other devices, so the characteristics of its images tend to not be too dissimilar to those yielded by the other smartphones. In addition, the Motorola Moto Z2 Play was the only device operated by two people (thus generating two separate datasets), and very high accuracies (>0.92) were achieved when the model trained using the images captured by one person was used to classify the other set. Despite this, the accuracy associated with this smartphone would still be the highest if the second database was removed from the experiments. It is also worth noting that the two Motorola Moto Z2 Play models yielded relatively dissimilar results when used to classify images from other sources, with differences surpassing 5% when the trained models were applied to the Asus ZenFone 5 and Huawei Mate SE test sets. This is a clear indication that differences in the way people acquire images may have a sizeable impact on the characteristics of the dataset generated, even when the exact same imaging device is used.
As expected, accuracies dropped significantly when scanner images were used for training (0.708 in average), confirming the results obtained in the other experiments. However, models trained with smartphone images performed relatively well classifying scanner images (0.826). This unexpected asymmetry seems to indicate that, while CNN models trained with high quality images have problems dealing with samples of lower quality, the opposite is not necessarily true.
All models trained with smartphone images yielded classification accuracies above 0.89 when applied to the iPhone 6s dataset, a result that was considerably better than those obtained for the other test sets. A suitable explanation for this was only found upon a careful analysis of all images used in the experiments. Because all smartphone images were captured indoors with a relatively low light intensity, problems related to focus and motion blur were a relatively common occurrence. Images severely blurred or out-of-focus were not used, but those with mild distortions were included to better represent situations that can occur in practice. The iPhone 6s dataset was the least affected by distortions and, as observed in the case of the scanner images, high quality images tend to be suitably classified by models trained with more degraded images. It is not clear if this dataset was less distorted because of the operator dexterity, the effectiveness of the device’s post-processing, or both. Analyzing the image capture process and how to decrease distortions was beyond the scope of this work, but these may be interesting topics for future research.
The test dataset with the worst performance was the Huawei Mate SE (average accuracy of 0.786). This dataset was built for the previous research [
29], so the traps that were imaged were not the same as the ones used with the other devices. One important characteristic of the traps imaged by the Huawei Mate SE is that most of them contained a very large amount of psyllids and other objects. As a result, images were very busy, and image crops containing psyllids usually also contained other objects. Since this kind of situation may happen in practice, the inclusion of such busy image was warranted, but it caused this dataset to be more difficult to be correctly classified. Interestingly, the results when this dataset was used for training were not nearly as poor (average accuracy of 0.847), which again indicates that including images with distortions or spurious elements actually helps to increase classification accuracies.
The vast majority of the false positive errors (non-psyllid images classified as psyllid) were due to the presence of insects with features similar to those of psyllids (
Table 7). Because the features that distinguish different species of insects are often minor, it can be difficult for the model to properly learn those distinctive features if only a few representative samples are available for training. It is likely that one of the reasons for the steady model improvement as more data sources were included, even when the number of training samples was decreased, was that problematic cases were represented in a more diverse way in the training set. As a result, the model was better prepared to discriminate species under different circumstances.
Finally, it is worth mentioning that testing CNNs with deeper and more complex architectures was beyond the objectives of this work, but experiments like this have been carried out previously under similar conditions [
29]. In that study, the ResNet-101 architecture [
33] yielded only slightly higher accuracies (between 1% and 4%) at the cost of a prediction time almost 10 times larger.
4. Conclusions
This article presented a study on the impact of using different data sources to train and validate CNN models, for the specific purpose of psyllid detection and counting using images of sticky traps. Relatively good results were achieved even when only samples from a single smartphone were used for training. This indicates that reasonably accurate estimates for the psyllid infestation may be possible even with models trained with limited datasets, as long as the characteristics of the training samples do not depart too much from those commonly found in practice. However, for more accurate estimates, or if a large number of devices with significantly different characteristics are expected to be employed in practice, the CNN model should be trained and tested using data that is as diverse as possible, otherwise results will not reflect reality and practical failure will become inevitable. This is not an easy task because data collection can be expensive and time consuming. Incremental evolution, in which the model is continuously improved as more data become available, may be a viable option, especially if a network of early adopters is already available.
Some factors that are relevant in the context of psyllid detection were only superficially investigated in this study and should receive more attention in the future, such as the influence of illumination, the consistency of the manual annotation of the reference material, the impact of unfocused and blurred images on training and testing, the importance of having multiple different people acquiring the images used to develop the models, and the consequences of having different psyllid densities in the traps, among others.