Enhancing Genomic Selection in Dairy Cattle Through Artificial Intelligence: Integrating Advanced Phenotyping and Predictive Models to Advance Health, Climate Resilience, and Sustainability
Abstract
1. Introduction
2. Influence of Genomic Selection on Dairy Cattle Health
2.1. Impact on Disease Resistance and Prevalence
2.2. Adaptation in Heat-Stressed Dairy Cattle
2.3. Harnessing Genetics to Reduce Enteric Methane Emissions in Livestock
3. Enhanced Productivity
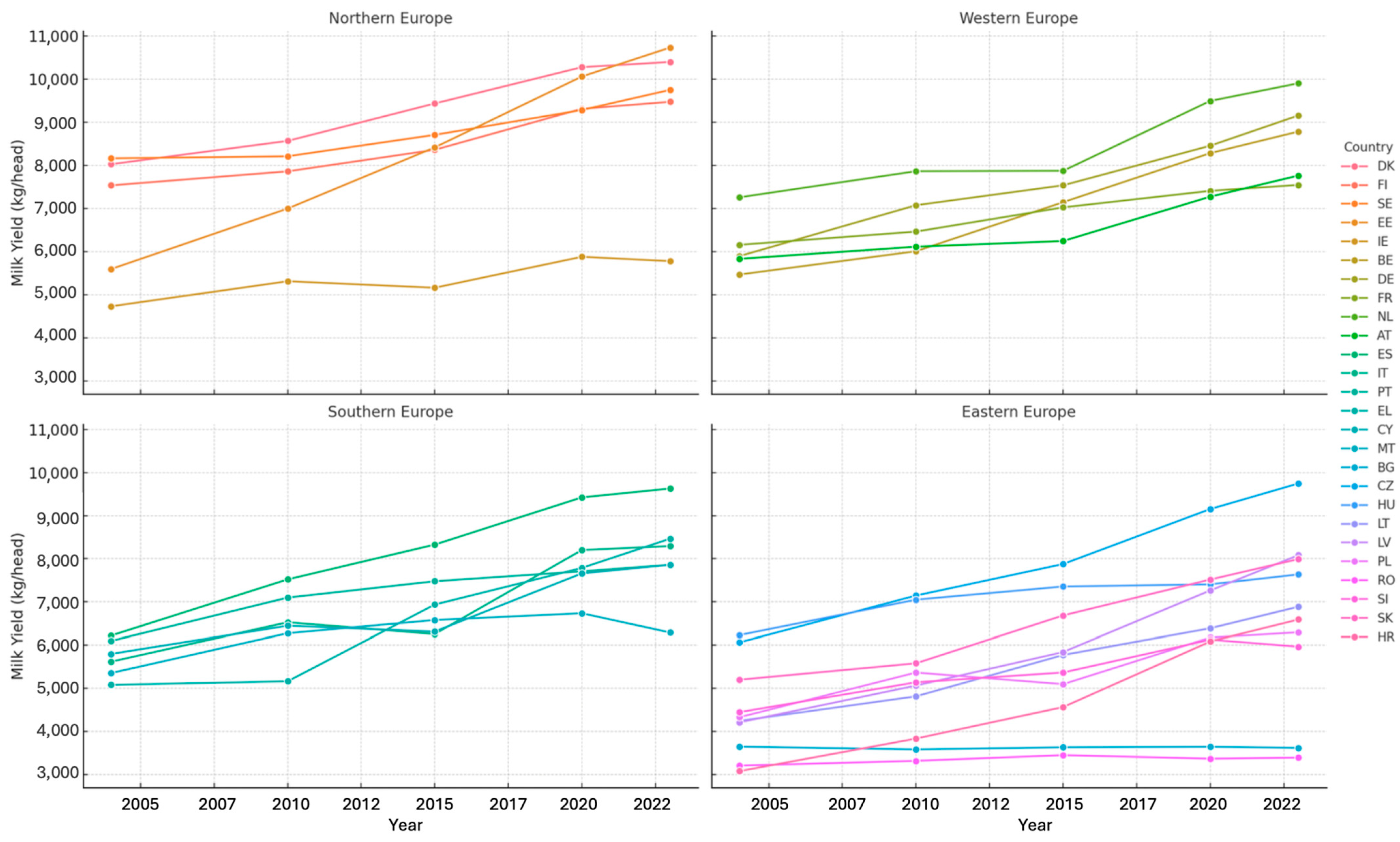
3.1. Milk Yield
3.2. Fertility
4. Artificial Intelligence in Genomic Selection: Opportunities and Challenges in Dairy Cattle Breeding
4.1. Machine Learning and Deep Learning Approaches in Genomic Selection
4.2. Milk Production and Reproductive Efficiency
4.3. Health Monitoring and Early Disease Prediction
4.4. Machine Learning Applications in Heat Stress and Environmental Adaptation
5. Challenges and Considerations
5.1. Data Integration and Interpretation
5.2. Future Issues
6. Conclusions
7. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association studies |
| GEBV | Genomic breeding values |
| DNA | Deoxyribonucleic acid |
| GS | Genomic selection |
| AI | Artificial intelligence |
| ML | Machine learning |
| DL | Deep learning |
| PCR | Polymerase chain reaction |
| SCC | Somatic cell count |
| HSF | Heat shock transcription factors |
| HSP | Heat shock protein genes |
| HS | Heat stress |
| SNP | Single nucleotide polymorphism |
| PRLP | Prolactin receptor gene |
| QTL | Quantitative trait loci |
| GHG | Greenhouse gas |
| CH4 | Methane |
| CO2 | Carbon dioxide |
| H2 | Hydrogen |
| DMI | Dry matter intake |
| AMS | Automatic milking systems |
| GBV | Genomic breeding values |
| TAI | Timed artificial insemination |
| GDRR | Genomic daughter pregnancy rate |
| DSS | Decision support systems |
| SHAP | Shapley additive explanations |
| XGBoost | Extreme gradient boosting |
| AUC | Area under the receiver operating characteristic curve |
| XAI | Explainable artificial intelligence |
| BLUP | Best linear unbiased prediction |
| LASSO | Least absolute shrinkage and selection operator |
References
- Peñagaricano, F. Chapter 6-Genetics and Genomics of Dairy Cattle. In Animal Agriculture; Bazer, F.W., Lamb, G.C., Wu, G., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 101–119. ISBN 978-0-12-817052-6. [Google Scholar]
- Cole, J.B.; VanRaden, P.M. Possibilities in an Age of Genomics: The Future of Selection Indices1. J. Dairy Sci. 2018, 101, 3686–3701. [Google Scholar] [CrossRef]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; Cabezas, J.; Rodriguez-Alvarez, L.; Garcia-Herreros, M. Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits. Animals 2020, 10, 1704. [Google Scholar] [CrossRef]
- Gutierrez-Reinoso, M.A.; Aponte, P.M.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.J.; Goddard, M.E. Invited Review: Genomic Selection in Dairy Cattle: Progress and Challenges. J. Dairy Sci. 2009, 92, 433–443. [Google Scholar] [CrossRef]
- Seno, L.d.O.; Guidolin, D.G.F.; Aspilcueta-Borquis, R.R.; do Nascimento, G.B.; da Silva, T.B.R.; de Oliveira, H.N.; Munari, D.P. Genomic Selection in Dairy Cattle Simulated Populations. J. Dairy Res. 2018, 85, 125–132. [Google Scholar] [CrossRef]
- Mueller, M.L.; Van Eenennaam, A.L. Synergistic Power of Genomic Selection, Assisted Reproductive Technologies, and Gene Editing to Drive Genetic Improvement of Cattle. CABI Agric. Biosci. 2022, 3, 13. [Google Scholar] [CrossRef]
- Tade, B.; Melesse, A. A Review on the Application of Genomic Selection in the Improvement of Dairy Cattle Productivity. Ecol. Genet. Genom. 2024, 31, 100257. [Google Scholar] [CrossRef]
- Kasimanickam, R.; Ferreira, J.C.P.; Kastelic, J.; Kasimanickam, V. Application of Genomic Selection in Beef Cattle Disease Prevention. Animals 2025, 15, 277. [Google Scholar] [CrossRef]
- Pedrosa, V.B.; Chen, S.-Y.; Gloria, L.S.; Doucette, J.S.; Boerman, J.P.; Rosa, G.J.M.; Brito, L.F. Machine Learning Methods for Genomic Prediction of Cow Behavioral Traits Measured by Automatic Milking Systems in North American Holstein Cattle. J. Dairy Sci. 2024, 107, 4758–4771. [Google Scholar] [CrossRef] [PubMed]
- Chafai, N.; Hayah, I.; Houaga, I.; Badaoui, B. A Review of Machine Learning Models Applied to Genomic Prediction in Animal Breeding. Front. Genet. 2023, 14, 1150596. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, N.; Wang, Y.-G.; George, A.W.; Reverter, A.; Li, Y. Genomic Prediction of Breeding Values Using a Subset of SNPs Identified by Three Machine Learning Methods. Front. Genet. 2018, 9, 237. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A Review of Deep Learning Applications for Genomic Selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review. Animals 2023, 13, 1588. [Google Scholar] [CrossRef]
- Parker Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. Genomic Selection for Producer-Recorded Health Event Data in US Dairy Cattle. J. Dairy Sci. 2014, 97, 3190–3199. [Google Scholar] [CrossRef]
- Morris, C.A. A Review of Genetic Resistance to Disease in Bos Taurus Cattle. Vet. J. 2007, 174, 481–491. [Google Scholar] [CrossRef]
- Abdelsayed, M.; Haile-Mariam, M.; Pryce, J.E. Genetic Parameters for Health Traits Using Data Collected from Genomic Information Nucleus Herds. J. Dairy Sci. 2017, 100, 9643–9655. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Cui, N.; Liang, Y. Cadherins Associate with Distinct Stem Cell-Related Transcription Factors to Coordinate the Maintenance of Stemness in Triple-Negative Breast Cancer. Stem Cells Int. 2017, 2017, 5091541. [Google Scholar] [CrossRef]
- Li, Y.; Han, B.; Liu, L.; Zhao, F.; Liang, W.; Jiang, J.; Yang, Y.; Ma, Z.; Sun, D. Genetic Association of 3, 23A, 2 and 4A1 Genes with Milk Yield and Composition in Dairy Cattle. Anim. Genet. 2019, 50, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.L.; Vinson, W.E.; Pearson, R.E.; Dickinson, F.N.; Johnson, L.P. Relationships between Linear Type Scores, Objective Type Measures, and Indicators of Mastitis. J. Dairy Sci. 1984, 67, 1281–1292. [Google Scholar] [CrossRef]
- Talebi, E.; Nezhad, M.K. Revolutionizing Animal Sciences: Multifaceted Solutions and Transformative Impact of AI Technologies. CABI Rev. 2024, 19. [Google Scholar] [CrossRef]
- Siva Kiran, R.R.; Dhamodhar, P. The Role of Artificial Intelligence and Machine Learning in Advancing Animal Biotechnology: A Review. Arch. Razi Inst. 2025, 80, 819–832. [Google Scholar] [CrossRef]
- Moser, D.W.; Miller, S.P.; Retallick, K.J.; Lu, D.; Kuehn, L.A. 52 Genomic Selection in the Beef Industry: Current Achievements and Future Directions. J. Anim. Sci. 2019, 97, 54–55. [Google Scholar] [CrossRef]
- Schneider, M.J.; Tait, R.G.; Ruble, M.V.; Busby, W.D.; Reecy, J.M. Evaluation of Fixed Sources of Variation and Estimation of Genetic Parameters for Incidence of Bovine Respiratory Disease in Preweaned Calves and Feedlot Cattle. J. Anim. Sci. 2010, 88, 1220–1228. [Google Scholar] [CrossRef]
- Snowder, G.D.; Van Vleck, L.D.; Cundiff, L.V.; Bennett, G.L. Influence of Breed, Heterozygosity, and Disease Incidence on Estimates of Variance Components of Respiratory Disease in Preweaned Beef Calves. J. Anim. Sci. 2005, 83, 1247–1261. [Google Scholar] [CrossRef]
- Berry, D.P.; Bermingham, M.L.; Good, M.; More, S.J. Genetics of Animal Health and Disease in Cattle. Ir. Vet. J. 2011, 64, 5. [Google Scholar] [CrossRef]
- Gonda, M.G.; Chang, Y.M.; Shook, G.E.; Collins, M.T.; Kirkpatrick, B.W. Genetic Variation of Mycobacterium Avium Ssp. Paratuberculosis Infection in US Holsteins. J. Dairy Sci. 2006, 89, 1804–1812. [Google Scholar] [CrossRef]
- Mortensen, H.; Nielsen, S.S.; Berg, P. Genetic Variation and Heritability of the Antibody Response to Mycobacterium Avium Subspecies Paratuberculosis in Danish Holstein Cows. J. Dairy Sci. 2004, 87, 2108–2113. [Google Scholar] [CrossRef] [PubMed]
- Uribe, H.A.; Kennedy, B.W.; Martin, S.W.; Kelton, D.F. Genetic Parameters for Common Health Disorders of Holstein Cows. J. Dairy Sci. 1995, 78, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B. Genetic Analysis of Fertility-Related Diseases and Disorders in Norwegian Red Cows. J. Dairy Sci. 2010, 93, 2751–2756. [Google Scholar] [CrossRef]
- Kadarmideen, H.N.; Thompson, R.; Simm, G. Linear and threshold model genetic parameters for disease, fertility and milk production in dairy cattle. Anim. Sci. 2000, 71, 411–419. [Google Scholar] [CrossRef]
- Heringstad, B.; Chang, Y.M.; Gianola, D.; Klemetsdal, G. Genetic Analysis of Clinical Mastitis, Milk Fever, Ketosis, and Retained Placenta in Three Lactations of Norwegian Red Cows. J. Dairy Sci. 2005, 88, 3273–3281. [Google Scholar] [CrossRef]
- Pryce, J.E.; Veerkamp, R.F.; Thompson, R.; Hill, W.G.; Simm, G. Genetic Aspects of Common Health Disorders and Measures of Fertility in Holstein Friesian Dairy Cattle. Anim. Sci. 1997, 65, 353–360. [Google Scholar] [CrossRef]
- Guarini, A.R.; Lourenco, D.a.L.; Brito, L.F.; Sargolzaei, M.; Baes, C.F.; Miglior, F.; Misztal, I.; Schenkel, F.S. Genetics and Genomics of Reproductive Disorders in Canadian Holstein Cattle. J. Dairy Sci. 2019, 102, 1341–1353. [Google Scholar] [CrossRef]
- van der Drift, S.G.A.; van Hulzen, K.J.E.; Teweldemedhn, T.G.; Jorritsma, R.; Nielen, M.; Heuven, H.C.M. Genetic and Nongenetic Variation in Plasma and Milk β-Hydroxybutyrate and Milk Acetone Concentrations of Early-Lactation Dairy Cows. J. Dairy Sci. 2012, 95, 6781–6787. [Google Scholar] [CrossRef] [PubMed]
- Tsiamadis, V.; Banos, G.; Panousis, N.; Kritsepi-Konstantinou, M.; Arsenos, G.; Valergakis, G.E. Genetic Parameters of Subclinical Macromineral Disorders and Major Clinical Diseases in Postparturient Holstein Cows. J. Dairy Sci. 2016, 99, 8901–8914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, Y.; Liu, G.; Ning, Y.; Li, J. Dairy Cow Mastitis Detection by Thermal Infrared Images Based on CLE-UNet. Animals 2023, 13, 2211. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, S. Genomic Prediction: Enhancing Breeding Strategies for Complex Traits in Livestock. Anim. Mol. Breed. 2024, 14, 95–105. [Google Scholar] [CrossRef]
- Kašná, E.; Zavadilová, L.; Vařeka, J. Genetic Evaluation of Resilience Indicators in Holstein Cows. Animals 2025, 15, 667. [Google Scholar] [CrossRef]
- Kotlarz, K.; Mielczarek, M.; Biecek, P.; Wojdak-Maksymiec, K.; Suchocki, T.; Topolski, P.; Jagusiak, W.; Szyda, J. An Explainable Deep Learning Classifier of Bovine Mastitis Based on Whole-Genome Sequence Data—Circumventing the p >> n Problem. Int. J. Mol. Sci. 2024, 25, 4715. [Google Scholar] [CrossRef]
- Cheng, M.; McCarl, B.; Fei, C. Climate Change and Livestock Production: A Literature Review. Atmosphere 2022, 13, 140. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Bowman, P.J.; Haile-Mariam, M.; Pryce, J.E.; Hayes, B.J. Genomic Selection for Tolerance to Heat Stress in Australian Dairy Cattle. J. Dairy Sci. 2016, 99, 2849–2862. [Google Scholar] [CrossRef]
- Cheruiyot, E.K.; Haile-Mariam, M.; Cocks, B.G.; MacLeod, I.M.; Xiang, R.; Pryce, J.E. New Loci and Neuronal Pathways for Resilience to Heat Stress in Cattle. Sci. Rep. 2021, 11, 16619. [Google Scholar] [CrossRef] [PubMed]
- Antanaitis, R.; Džermeikaitė, K.; Bespalovaitė, A.; Ribelytė, I.; Rutkauskas, A.; Japertas, S.; Baumgartner, W. Assessment of Ruminating, Eating, and Locomotion Behavior during Heat Stress in Dairy Cattle by Using Advanced Technological Monitoring. Animals 2023, 13, 2825. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; Collier, J.L.; Rhoads, R.P.; Baumgard, L.H. Invited Review: Genes Involved in the Bovine Heat Stress Response1. J. Dairy Sci. 2008, 91, 445–454. [Google Scholar] [CrossRef]
- Olson, T.A.; Lucena, C.; Chase, C.C., Jr.; Hammond, A.C. Evidence of a Major Gene Influencing Hair Length and Heat Tolerance in Bos Taurus Cattle1,2,3. J. Anim. Sci. 2003, 81, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Carabaño, M.J. The Challenge of Genetic Selection for Heat Tolerance: The Dairy Cattle Example. Adv. Anim. Biosci. 2016, 7, 218–222. [Google Scholar] [CrossRef]
- Sonna, L.A.; Fujita, J.; Gaffin, S.L.; Lilly, C.M. Invited Review: Effects of Heat and Cold Stress on Mammalian Gene Expression. J. Appl. Physiol. 2002, 92, 1725–1742. [Google Scholar] [CrossRef]
- Garner, J.B.; Douglas, M.L.; Williams, S.R.O.; Wales, W.J.; Marett, L.C.; Nguyen, T.T.T.; Reich, C.M.; Hayes, B.J. Genomic Selection Improves Heat Tolerance in Dairy Cattle. Sci. Rep. 2016, 6, 34114. [Google Scholar] [CrossRef]
- Worku, D.; Hussen, J.; De Matteis, G.; Schusser, B.; Alhussien, M.N. Candidate Genes Associated with Heat Stress and Breeding Strategies to Relieve Its Effects in Dairy Cattle: A Deeper Insight into the Genetic Architecture and Immune Response to Heat Stress. Front. Vet. Sci. 2023, 10, 1151241. [Google Scholar] [CrossRef]
- Otto, P.I.; Guimarães, S.E.F.; Verardo, L.L.; Azevedo, A.L.S.; Vandenplas, J.; Sevillano, C.A.; Marques, D.B.D.; Pires, M.d.F.A.; de Freitas, C.; Verneque, R.S.; et al. Genome-Wide Association Studies for Heat Stress Response in Bos taurus × Bos indicus Crossbred Cattle. J. Dairy Sci. 2019, 102, 8148–8158. [Google Scholar] [CrossRef]
- Hariyono, D.N.H.; Prihandini, P.W. Association of selected gene polymorphisms with thermotolerance traits in cattle–A review. Anim. Biosci. 2022, 35, 1635. [Google Scholar] [CrossRef] [PubMed]
- Zamorano-Algandar, R.; Medrano, J.F.; Thomas, M.G.; Enns, R.M.; Speidel, S.E.; Sánchez-Castro, M.A.; Luna-Nevárez, G.; Leyva-Corona, J.C.; Luna-Nevárez, P. Genetic Markers Associated with Milk Production and Thermotolerance in Holstein Dairy Cows Managed in a Heat-Stressed Environment. Biology 2023, 12, 679. [Google Scholar] [CrossRef] [PubMed]
- Del Corvo, M.; Lazzari, B.; Capra, E.; Zavarez, L.; Milanesi, M.; Utsunomiya, Y.T.; Utsunomiya, A.T.H.; Stella, A.; de Paula Nogueira, G.; Garcia, J.F.; et al. Methylome Patterns of Cattle Adaptation to Heat Stress. Front. Genet. 2021, 12, 633132. [Google Scholar] [CrossRef]
- Silpa, M.V.; König, S.; Sejian, V.; Malik, P.K.; Nair, M.R.R.; Fonseca, V.F.C.; Maia, A.S.C.; Bhatta, R. Climate-Resilient Dairy Cattle Production: Applications of Genomic Tools and Statistical Models. Front. Vet. Sci. 2021, 8, 625189. [Google Scholar] [CrossRef] [PubMed]
- Dikmen, S.; Wang, X.-Z.; Ortega, M.S.; Cole, J.B.; Null, D.J.; Hansen, P.J. Single Nucleotide Polymorphisms Associated with Thermoregulation in Lactating Dairy Cows Exposed to Heat Stress. J. Anim. Breed. Genet. 2015, 132, 409–419. [Google Scholar] [CrossRef]
- Luo, H.; Li, X.; Hu, L.; Xu, W.; Chu, Q.; Liu, A.; Guo, G.; Liu, L.; Brito, L.F.; Wang, Y. Genomic Analyses and Biological Validation of Candidate Genes for Rectal Temperature as an Indicator of Heat Stress in Holstein Cattle. J. Dairy Sci. 2021, 104, 4441–4451. [Google Scholar] [CrossRef]
- Bang, N.N.; Hayes, B.J.; Lyons, R.E.; Randhawa, I.A.S.; Gaughan, J.B.; Trach, N.X.; McNeill, D.M. Genomic Prediction and Genome-Wide Association Studies for Productivity, Conformation and Heat Tolerance Traits in Tropical Smallholder Dairy Cows. J. Anim. Breed. Genet. 2025, 142, 322–341. [Google Scholar] [CrossRef]
- Luo, H.; Hu, L.; Brito, L.F.; Dou, J.; Sammad, A.; Chang, Y.; Ma, L.; Guo, G.; Liu, L.; Zhai, L.; et al. Weighted Single-Step GWAS and RNA Sequencing Reveals Key Candidate Genes Associated with Physiological Indicators of Heat Stress in Holstein Cattle. J. Anim. Sci. Biotechnol. 2022, 13, 108. [Google Scholar] [CrossRef]
- Luo, H.; Xu, W.; Liu, A.; Li, X.; Liu, L.; Wang, Y. Genome-Wide Association Study for Rectal Temperature in Chinese Holstein Population. In Proceedings of the 11th World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, 11–16 February 2018. [Google Scholar]
- Sigdel, A.; Abdollahi-Arpanahi, R.; Aguilar, I.; Peñagaricano, F. Whole Genome Mapping Reveals Novel Genes and Pathways Involved in Milk Production Under Heat Stress in US Holstein Cows. Front. Genet. 2019, 10, 928. [Google Scholar] [CrossRef]
- Bohlouli, M.; Halli, K.; Yin, T.; Gengler, N.; König, S. Genome-Wide Associations for Heat Stress Response Suggest Potential Candidate Genes Underlying Milk Fatty Acid Composition in Dairy Cattle. J. Dairy Sci. 2022, 105, 3323–3340. [Google Scholar] [CrossRef]
- Bohlouli, M.; Yin, T.; Hammami, H.; Gengler, N.; König, S. Climate Sensitivity of Milk Production Traits and Milk Fatty Acids in Genotyped Holstein Dairy Cows. J. Dairy Sci. 2021, 104, 6847–6860. [Google Scholar] [CrossRef]
- Habeeb, A.A.; Osman, S.F.; Teama, F.E.I.; Gad, A.E. The Detrimental Impact of High Environmental Temperature on Physiological Response, Growth, Milk Production, and Reproductive Efficiency of Ruminants. Trop. Anim. Health Prod. 2023, 55, 388. [Google Scholar] [CrossRef]
- Cheruiyot, E.K.; Haile-Mariam, M.; Cocks, B.G.; Pryce, J.E. Improving Genomic Selection for Heat Tolerance in Dairy Cattle: Current Opportunities and Future Directions. Front. Genet. 2022, 13, 894067. [Google Scholar] [CrossRef]
- Džermeikaitė, K.; Krištolaitytė, J.; Antanaitis, R. Relationship between Dairy Cow Health and Intensity of Greenhouse Gas Emissions. Animals 2024, 14, 829. [Google Scholar] [CrossRef]
- Bačėninaitė, D.; Džermeikaitė, K.; Antanaitis, R. Global Warming and Dairy Cattle: How to Control and Reduce Methane Emission. Animals 2022, 12, 2687. [Google Scholar] [CrossRef] [PubMed]
- Colombi, D.; Perini, F.; Bettini, S.; Mastrangelo, S.; Abeni, F.; Conte, G.; Marletta, D.; Cassandro, M.; Bernabucci, U.; Ciampolini, R.; et al. Genomic Responses to Climatic Challenges in Beef Cattle: A Review. Anim. Genet. 2024, 55, 854–870. [Google Scholar] [CrossRef]
- Breider, I.S.; Wall, E.; Garnsworthy, P.C. Short Communication: Heritability of Methane Production and Genetic Correlations with Milk Yield and Body Weight in Holstein-Friesian Dairy Cows. J. Dairy Sci. 2019, 102, 7277–7281. [Google Scholar] [CrossRef]
- Difford, G.F.; Plichta, D.R.; Løvendahl, P.; Lassen, J.; Noel, S.J.; Højberg, O.; Wright, A.-D.G.; Zhu, Z.; Kristensen, L.; Nielsen, H.B.; et al. Host Genetics and the Rumen Microbiome Jointly Associate with Methane Emissions in Dairy Cows. PLoS Genet. 2018, 14, e1007580. [Google Scholar] [CrossRef]
- Binsulong, B.; Gunha, T.; Kongphitee, K.; Maeda, K.; Sommart, K. Enteric Methane Emissions, Rumen Fermentation Characteristics, and Energetic Efficiency of Holstein Crossbred Bulls Fed Total Mixed Ration Silage with Cassava Instead of Rice Straw. Fermentation 2023, 9, 850. [Google Scholar] [CrossRef]
- Dressler, E.A.; Bormann, J.M.; Weaber, R.L.; Rolf, M.M. Use of Methane Production Data for Genetic Prediction in Beef Cattle: A Review. Transl. Anim. Sci. 2024, 8, txae014. [Google Scholar] [CrossRef] [PubMed]
- Kooverjee, B.B.; Soma, P.; Van Der Nest, M.A.; Scholtz, M.M.; Neser, F.W.C. Selection Signatures in South African Nguni and Bonsmara Cattle Populations Reveal Genes Relating to Environmental Adaptation. Front. Genet. 2022, 13, 909012. [Google Scholar] [CrossRef]
- Le Gloux, F.; Duvaleix, S.; Dupraz, P. Taking the Diet of Cows into Consideration in Designing Payments to Reduce Enteric Methane Emissions on Dairy Farms. J. Dairy Sci. 2023, 106, 6961–6985. [Google Scholar] [CrossRef]
- Pickering, N.K.; Chagunda, M.G.G.; Banos, G.; Mrode, R.; McEwan, J.C.; Wall, E. Genetic parameters for predicted methane production and laser methane detector measurements1. J. Anim. Sci. 2015, 93, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Lassen, J.; Løvendahl, P. Heritability Estimates for Enteric Methane Emissions from Holstein Cattle Measured Using Noninvasive Methods. J. Dairy Sci. 2016, 99, 1959–1967. [Google Scholar] [CrossRef]
- Pszczola, M.; Rzewuska, K.; Mucha, S.; Strabel, T. Heritability of Methane Emissions from Dairy Cows over a Lactation Measured on Commercial Farms1. J. Anim. Sci. 2017, 95, 4813–4819. [Google Scholar] [CrossRef]
- Arthur, P.F.; Archer, J.A.; Johnston, D.J.; Herd, R.M.; Richardson, E.C.; Parnell, P.F. Genetic and Phenotypic Variance and Covariance Components for Feed Intake, Feed Efficiency, and Other Postweaning Traits in Angus Cattle. J. Anim. Sci. 2001, 79, 2805–2811. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Júnior, G.A.; Peripolli, E.; Schmidt, P.I.; Campos, G.S.; Mota, L.F.M.; Mercadante, M.E.Z.; Baldi, F.; Carvalheiro, R.; de Albuquerque, L.G. Current Applications and Perspectives of Genomic Selection in Bos indicus (Nellore) Cattle. Livest. Sci. 2022, 263, 105001. [Google Scholar] [CrossRef]
- Schwarz, L.; Križanac, A.-M.; Schneider, H.; Falker-Gieske, C.; Heise, J.; Liu, Z.; Bennewitz, J.; Thaller, G.; Tetens, J. Genetic and Genomic Analysis of Reproduction Traits in Holstein Cattle Using SNP Chip Data and Imputed Sequence Level Genotypes. BMC Genom. 2024, 25, 880. [Google Scholar] [CrossRef]
- Vergani, A.M.; Bagnato, A.; Masseroli, M. Predicting Bovine Daily Milk Yield by Leveraging Genomic Breeding Values. Comput. Electron. Agric. 2024, 219, 108777. [Google Scholar] [CrossRef]
- Miglior, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.F. A 100-Year Review: Identification and Genetic Selection of Economically Important Traits in Dairy Cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef]
- Milk and Milk Product Statistics. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Milk_and_milk_product_statistics (accessed on 5 April 2025).
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.K.; Kumar, A.; Pradhan, A.K.; Kumar, U.; Kumar, R.R.; Juliana, P.; Mishra, D.C.; et al. Genomic Selection: A Tool for Accelerating the Efficiency of Molecular Breeding for Development of Climate-Resilient Crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic Selection of High-Yielding Dairy Cattle toward Sustainable Farming Systems in a Rapidly Changing World. Animal 2021, 15, 100292. [Google Scholar] [CrossRef] [PubMed]
- Kemel, C.; Salamone, M.; Aernouts, B.; Adriaens, I.; Opsomer, G.; Hut, P.; Hostens, M. Association of Artificial Intelligence-Predicted Milk Yield Residuals to Behavioral Patterns and Transition Success in Multiparous Dairy Cows. J. Dairy Sci. 2025, 108, 8859–8876. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-M.; Dang, C.-G.; Alam, M.Z.; Kim, Y.-S.; Cho, K.-H.; Park, K.-D.; Kim, J.-J. The Effectiveness of Genomic Selection for Milk Production Traits of Holstein Dairy Cattle. Asian-Australas. J. Anim. Sci. 2019, 33, 382–389. [Google Scholar] [CrossRef]
- Bednarski, M.; Kupczyński, R. Factors Affecting Milk Productivity, Milk Quality and Dairy Cow Health. Animals 2024, 14, 3707. [Google Scholar] [CrossRef]
- Schneider, H.; Krizanac, A.-M.; Falker-Gieske, C.; Heise, J.; Tetens, J.; Thaller, G.; Bennewitz, J. Genomic Dissection of the Correlation between Milk Yield and Various Health Traits Using Functional and Evolutionary Information about Imputed Sequence Variants of 34,497 German Holstein Cows. BMC Genom. 2024, 25, 265. [Google Scholar] [CrossRef]
- Huang, W.; Peñagaricano, F.; Ahmad, K.R.; Lucey, J.A.; Weigel, K.A.; Khatib, H. Association between Milk Protein Gene Variants and Protein Composition Traits in Dairy Cattle. J. Dairy Sci. 2012, 95, 440–449. [Google Scholar] [CrossRef]
- Iung, L.H.S.; Petrini, J.; Ramírez-Díaz, J.; Salvian, M.; Rovadoscki, G.A.; Pilonetto, F.; Dauria, B.D.; Machado, P.F.; Coutinho, L.L.; Wiggans, G.R.; et al. Genome-Wide Association Study for Milk Production Traits in a Brazilian Holstein Population. J. Dairy Sci. 2019, 102, 5305–5314. [Google Scholar] [CrossRef]
- Mahmoudi, P.; Rashidi, A. Strong Evidence for Association between K232A Polymorphism of the DGAT1 Gene and Milk Fat and Protein Contents: A Meta-Analysis. J. Dairy Sci. 2023, 106, 2573–2587. [Google Scholar] [CrossRef]
- Erdoğan, M.; Çinkaya, S.; Brenig, B.; Çelikeloğlu, K.; Demirtaş, M.; Sarıibrahimoğlu, S.; Tekerli, M. Genome-Wide Association Studies for Milk Production Traits and Persistency of First Calving Holstein Cattle in Türkiye. Front. Vet. Sci. 2024, 11, 1461075. [Google Scholar] [CrossRef]
- CORDIS. European Commission. Available online: https://cordis.europa.eu/project/id/613689/reporting#:~:text=The%20combined%20approach%20will%20promote%20expansion%20of,based%20on%20easily%20accessible%20data%20and%20samples (accessed on 5 May 2025).
- Mckimmie, C.; Forutan, M.; Tajet, H.M.; Ehsani, A.; Hickford, J.; Amirpour, H. Impact of Implementing Female Genomic Selection and the Use of Sex-Selected Semen Technology on Genetic Gain in a Dairy Herd in New Zealand. Int. J. Mol. Sci. 2025, 26, 990. [Google Scholar] [CrossRef]
- Chen, Z.; Brito, L.F.; Luo, H.; Shi, R.; Chang, Y.; Liu, L.; Guo, G.; Wang, Y. Genetic and Genomic Analyses of Service Sire Effect on Female Reproductive Traits in Holstein Cattle. Front. Genet. 2021, 12, 713575. [Google Scholar] [CrossRef]
- Ayantoye, J.O.; Kolachi, H.A.; Zhang, X.; Shahzad, M.; Kandil, O.M.T.; Wan, P.; Zhao, X. Advances in Timed Artificial Insemination: Integrating Omics Technologies for Enhanced Reproductive Efficiency in Dairy Cattle. Animals 2025, 15, 816. [Google Scholar] [CrossRef]
- Rahman, M.A.; Juyena, N.S.; Shamsuddin, M.; Bhuiyan, M.M.U. Genomic Tools and Genetic Improvement of Crossbred Friesian Cattle. Res. Agric. Livest. Fish. 2021, 8, 89–107. [Google Scholar] [CrossRef]
- Pryce, J.E.; Egger-Danner, C.; Simm, G. Strategies and Tools for Genetic Selection in Dairy Cattle and Their Application to Improving Animal Welfare. In Cattle Welfare in Dairy and Beef Systems; Springer: Cham, Switzerland, 2023; pp. 323–348. ISBN 978-3-031-21020-4. [Google Scholar]
- Walsh, D.P.; Fahey, A.G.; Lonergan, P.; Wallace, M. Economics of Timed Artificial Insemination with Unsorted or Sexed Semen in a High-Producing, Pasture-Based Dairy Production System. J. Dairy Sci. 2022, 105, 3192–3208. [Google Scholar] [CrossRef] [PubMed]
- Madureira, A.M.L.; Plenio, J.L.; Vasconcelos, J.L.M.; Guida, T.G.; Cerri, R.L.A.; Borchardt, S. Association between Genomic Daughter Pregnancy Rate and Expected Milk Production on the Resumption of Estrus Behavior in Holstein Cattle. J. Dairy Sci. 2024, 107, 1592–1602. [Google Scholar] [CrossRef]
- Lima, F.S.; Silvestre, F.T.; Peñagaricano, F.; Thatcher, W.W. Early Genomic Prediction of Daughter Pregnancy Rate Is Associated with Improved Reproductive Performance in Holstein Dairy Cows. J. Dairy Sci. 2020, 103, 3312–3324. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.R.; de Almeida, J.V.N.; Oliveira, A.C.; Paim, T.d.P.; Marques, T.C.; Leão, K.M. Artificial Insemination Timing on Pregnancy Rate of Holstein Cows Using an Automated Activity Monitoring. Ciênc. Rural 2023, 54, e20220557. [Google Scholar] [CrossRef]
- Laplacette, A.L.; Rial, C.; Sitko, E.; Perez, M.M.; Tompkins, S.; Stangaferro, M.L.; Thomas, M.J.; Giordano, J.O. Delaying Induction of Ovulation and Timed Artificial Insemination in a Double-Ovsynch Protocol Increased Expression of Estrus and Altered First-Service Reproductive Outcomes of Lactating Dairy Cows. J. Dairy Sci. 2025, 108, 1103–1124. [Google Scholar] [CrossRef]
- Fleming, A.; Abdalla, E.A.; Maltecca, C.; Baes, C.F. Invited Review: Reproductive and Genomic Technologies to Optimize Breeding Strategies for Genetic Progress in Dairy Cattle. Arch. Anim. Breed. 2018, 61, 43–57. [Google Scholar] [CrossRef]
- Wadood, A.A.; Bordbar, F.; Zhang, X. Integrating Omics Approaches in Livestock Biotechnology: Innovations in Production and Reproductive Efficiency. Front. Anim. Sci. 2025, 6, 1551244. [Google Scholar] [CrossRef]
- Santana, T.E.Z.; Silva, J.C.F.; da Silva, L.O.C.; Alvarenga, A.B.; Menezes, G.R.d.O.; Torres, R.A.A.; Duarte, M.d.S.; e Silva, F.F. Genome-Enabled Classification of Stayability in Nellore Cattle under a Machine Learning Framework. Livest. Sci. 2022, 260, 104935. [Google Scholar] [CrossRef]
- Becker, C.A.; Aghalari, A.; Marufuzzaman, M.; Stone, A.E. Predicting Dairy Cattle Heat Stress Using Machine Learning Techniques. J. Dairy Sci. 2021, 104, 501–524. [Google Scholar] [CrossRef] [PubMed]
- Cockburn, M. Review: Application and Prospective Discussion of Machine Learning for the Management of Dairy Farms. Animals 2020, 10, 1690. [Google Scholar] [CrossRef] [PubMed]
- Shahinfar, S.; Khansefid, M.; Haile-Mariam, M.; Pryce, J.E. Machine Learning Approaches for the Prediction of Lameness in Dairy Cows. Animal 2021, 15, 100391. [Google Scholar] [CrossRef]
- Ogutu, J.O.; Piepho, H.-P.; Schulz-Streeck, T. A Comparison of Random Forests, Boosting and Support Vector Machines for Genomic Selection. BMC Proc. 2011, 5, S11. [Google Scholar] [CrossRef]
- Li, M.; Wang, L.; Zhao, F. Research Progress on Machine Learning for Genomic Selection in Animals. Sci. Agric. Sin. 2023, 56, 3682–3692. [Google Scholar] [CrossRef]
- Nayeri, S.; Sargolzaei, M.; Tulpan, D. A Review of Traditional and Machine Learning Methods Applied to Animal Breeding. Anim. Health Res. Rev. 2019, 20, 31–46. [Google Scholar] [CrossRef]
- Grzesiak, W.; Zaborski, D.; Pluciński, M.; Jędrzejczak-Silicka, M.; Pilarczyk, R.; Sablik, P. The Use of Selected Machine Learning Methods in Dairy Cattle Farming: A Review. Animals 2025, 15, 2033. [Google Scholar] [CrossRef]
- Palma, O.; Plà-Aragonés, L.M.; Mac Cawley, A.; Albornoz, V.M. AI and Data Analytics in the Dairy Farms: A Scoping Review. Animals 2025, 15, 1291. [Google Scholar] [CrossRef] [PubMed]
- Billah, M.; Bermann, M.; Hollifield, M.K.; Tsuruta, S.; Chen, C.Y.; Psota, E.; Holl, J.; Misztal, I.; Lourenco, D. Review: Genomic Selection in the Era of Phenotyping Based on Digital Images. Animal 2025, 101486, in press. [Google Scholar] [CrossRef]
- Gocheva-Ilieva, S.; Yordanova, A.; Kulina, H. Predicting the 305-Day Milk Yield of Holstein-Friesian Cows Depending on the Conformation Traits and Farm Using Simplified Selective Ensembles. Mathematics 2022, 10, 1254. [Google Scholar] [CrossRef]
- Santana, M.L.; Bignardi, A.B.; Pereira, R.J.; Oliveira Junior, G.A.; Freitas, A.P.; Carvalheiro, R.; Eler, J.P.; Ferraz, J.B.S.; Cyrillo, J.N.S.G.; Mercadante, M.E.Z. Genotype by Prenatal Environment Interaction for Postnatal Growth of Nelore Beef Cattle Raised under Tropical Grazing Conditions. Animals 2023, 13, 2321. [Google Scholar] [CrossRef]
- Alemu, S.W.; Bilton, T.P.; Johnson, P.L.; Perry, B.J.; Henry, H.; Dodds, K.G.; McEwan, J.C.; Rowe, S.J. Improving Genomic Prediction Accuracy for Methane Emission and Feed Efficiency in Sheep: Integrating Rumen Microbial PCA with Host Genomic Variation Using Neural Network GBLUP (NN-GBLUP). Genet. Sel. Evol. 2025, 57, 41. [Google Scholar] [CrossRef]
- Wang, X.; Shi, S.; Ali Khan, M.Y.; Zhang, Z.; Zhang, Y. Improving the Accuracy of Genomic Prediction in Dairy Cattle Using the Biologically Annotated Neural Networks Framework. J. Anim. Sci. Biotechnol. 2024, 15, 87. [Google Scholar] [CrossRef]
- Hempstalk, K.; McParland, S.; Berry, D.P. Machine Learning Algorithms for the Prediction of Conception Success to a given Insemination in Lactating Dairy Cows. J. Dairy Sci. 2015, 98, 5262–5273. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bell, M.; Liu, X.; Liu, G. Machine-Learning Techniques Can Enhance Dairy Cow Estrus Detection Using Location and Acceleration Data. Animals 2020, 10, 1160. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi-Arpanahi, R.; Morota, G.; Peñagaricano, F. Predicting Bull Fertility Using Genomic Data and Biological Information. J. Dairy Sci. 2017, 100, 9656–9666. [Google Scholar] [CrossRef] [PubMed]
- Marques, T.C.; Marques, L.R.; Fernandes, P.B.; de Lima, F.S.; do Prado Paim, T.; Leão, K.M. Machine Learning to Predict Pregnancy in Dairy Cows: An Approach Integrating Automated Activity Monitoring and On-Farm Data. Animals 2024, 14, 1567. [Google Scholar] [CrossRef]
- Lucy, M.C. Symposium Review: Selection for Fertility in the Modern Dairy Cow—Current Status and Future Direction for Genetic Selection. J. Dairy Sci. 2019, 102, 3706–3721. [Google Scholar] [CrossRef]
- Ferreira, R.E.P.; Dórea, J.R.R. International Symposium on Ruminant Physiology: Leveraging Computer Vision, Large Language Models, and Multimodal Machine Learning for Optimal Decision Making in Dairy Farming. J. Dairy Sci. 2025, 108, 7493–7510. [Google Scholar] [CrossRef]
- Saarela, M.; Podgorelec, V. Recent Applications of Explainable AI (XAI): A Systematic Literature Review. Appl. Sci. 2024, 14, 8884. [Google Scholar] [CrossRef]
- Michelena, Á.; Fontenla-Romero, Ó.; Luis Calvo-Rolle, J. A Review and Future Trends of Precision Livestock over Dairy and Beef Cow Cattle with Artificial Intelligence. Log. J. IGPL 2024, 33, jzae111. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, C.; Wang, H.; Xu, W.; Zhao, Z.; Chen, M.; Jia, B.; Huang, B. The Early Prediction of Common Disorders in Dairy Cows Monitored by Automatic Systems with Machine Learning Algorithms. Animals 2022, 12, 1251. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; van Knegsel, A.T.M.; Vervoort, J.J.M.; Bruckmaier, R.M.; van Hoeij, R.J.; Kemp, B.; Saccenti, E. Prediction of Metabolic Status of Dairy Cows in Early Lactation with On-Farm Cow Data and Machine Learning Algorithms. J. Dairy Sci. 2019, 102, 10186–10201. [Google Scholar] [CrossRef]
- Gerakari, M.; Katsileros, A.; Kleftogianni, K.; Tani, E.; Bebeli, P.J.; Papasotiropoulos, V. Breeding of Solanaceous Crops Using AI: Machine Learning and Deep Learning Approaches—A Critical Review. Agronomy 2025, 15, 757. [Google Scholar] [CrossRef]
- Rao, Y.; Zhang, L.; Gao, L.; Wang, S.; Yang, L. ExAutoGP: Enhancing Genomic Prediction Stability and Interpretability with Automated Machine Learning and SHAP. Animals 2025, 15, 1172. [Google Scholar] [CrossRef] [PubMed]
- Gengler, N. Symposium Review: Challenges and Opportunities for Evaluating and Using the Genetic Potential of Dairy Cattle in the New Era of Sensor Data from Automation. J. Dairy Sci. 2019, 102, 5756–5763. [Google Scholar] [CrossRef]
- Sahana, G.; Cai, Z.; Sanchez, M.P.; Bouwman, A.C.; Boichard, D. Invited Review: Good Practices in Genome-Wide Association Studies to Identify Candidate Sequence Variants in Dairy Cattle. J. Dairy Sci. 2023, 106, 5218–5241. [Google Scholar] [CrossRef]
- Distante, D.; Albanello, C.; Zaffar, H.; Faralli, S.; Amalfitano, D. Artificial Intelligence Applied to Precision Livestock Farming: A Tertiary Study. Smart Agric. Technol. 2025, 11, 100889. [Google Scholar] [CrossRef]
- Mienye, I.D.; Swart, T.G. A Comprehensive Review of Deep Learning: Architectures, Recent Advances, and Applications. Information 2024, 15, 755. [Google Scholar] [CrossRef]
- Suminda, G.G.D.; Ghosh, M.; Son, Y.-O. The Innovative Informatics Approaches of High-Throughput Technologies in Livestock: Spearheading the Sustainability and Resiliency of Agrigenomics Research. Life 2022, 12, 1893. [Google Scholar] [CrossRef]
- Wise, J.; de Barron, A.G.; Splendiani, A.; Balali-Mood, B.; Vasant, D.; Little, E.; Mellino, G.; Harrow, I.; Smith, I.; Taubert, J.; et al. Implementation and Relevance of FAIR Data Principles in Biopharmaceutical R&D. Drug Discov. Today 2019, 24, 933–938. [Google Scholar] [CrossRef]
- Pryce, J.E.; Nguyen, T.T.T.; Axford, M.; Nieuwhof, G.; Shaffer, M. Symposium Review: Building a Better Cow—The Australian Experience and Future Perspectives1. J. Dairy Sci. 2018, 101, 3702–3713. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Oliveira, H.R.; McConn, B.R.; Schinckel, A.P.; Arrazola, A.; Marchant-Forde, J.N.; Johnson, J.S. Large-Scale Phenotyping of Livestock Welfare in Commercial Production Systems: A New Frontier in Animal Breeding. Front. Genet. 2020, 11, 793. [Google Scholar] [CrossRef]
- Berghof, T.V.L.; Poppe, M.; Mulder, H.A. Opportunities to Improve Resilience in Animal Breeding Programs. Front. Genet. 2019, 9, 692. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.J.; Tzimiropoulos, G. Novel Monitoring Systems to Obtain Dairy Cattle Phenotypes Associated with Sustainable Production. Front. Sustain. Food Syst. 2018, 2, 31. [Google Scholar] [CrossRef]
- Wójcik-Gront, E.; Zieniuk, B.; Pawełkowicz, M. Harnessing AI-Powered Genomic Research for Sustainable Crop Improvement. Agriculture 2024, 14, 2299. [Google Scholar] [CrossRef]
- Zhan, S.; Huang, L.; Luo, G.; Zheng, S.; Gao, Z.; Chao, H.-C. A Review on Federated Learning Architectures for Privacy-Preserving AI: Lightweight and Secure Cloud–Edge–End Collaboration. Electronics 2025, 14, 2512. [Google Scholar] [CrossRef]
- Alkhanbouli, R.; Matar Abdulla Almadhaani, H.; Alhosani, F.; Simsekler, M.C.E. The Role of Explainable Artificial Intelligence in Disease Prediction: A Systematic Literature Review and Future Research Directions. BMC Med. Inform. Decis. Mak. 2025, 25, 110. [Google Scholar] [CrossRef]
- Elrashedy, A.; Mousa, W.; Nayel, M.; Salama, A.; Zaghawa, A.; Elsify, A.; Hasan, M.E. Advances in Bioinformatics and Multi-Omics Integration: Transforming Viral Infectious Disease Research in Veterinary Medicine. Virol. J. 2025, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Rebez, E.B.; Sejian, V.; Silpa, M.V.; Kalaignazhal, G.; Thirunavukkarasu, D.; Devaraj, C.; Nikhil, K.T.; Ninan, J.; Sahoo, A.; Lacetera, N.; et al. Applications of Artificial Intelligence for Heat Stress Management in Ruminant Livestock. Sensors 2024, 24, 5890. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Mun, H.; Koo, Y.; Park, S.; Kim, J.; Yu, S.; Shin, J.; Lee, J.; Son, J.; Park, C.; et al. Enhancing Genomic Prediction Accuracy for Body Conformation Traits in Korean Holstein Cattle. Animals 2024, 14, 1052. [Google Scholar] [CrossRef] [PubMed]
| Genetic Resistance to Disease | Range in Heritability Estimate | Reference |
|---|---|---|
| Respiratory disease in pre-weaning calves | 0.11 | [24] |
| Respiratory disease in post-weaning calves | 0.07 | [24] |
| Bovine respiratory disease | 0.07 and 0.29 | [25] |
| Mastitis in Irish Holstein-Friesian dairy cows | 0.05 | [26] |
| Lameness in Irish Holstein-Friesian dairy cattle | 0.04 | [26] |
| Lameness | 0.00 to 0.02 | [17] |
| Metabolic disorders | 0.00 to 0.06 | [17] |
| Johne’s disease | 0.05 to 0.15 | [27,28] |
| Displaced abomasum | 0.15 to 0.31 | [29,30] |
| Hypomagnesaemia | 0.004 | [31] |
| Ketosis | 0.01 to 0.16 | [29,30] |
| Hypocalcaemia | 0.01 to 0.13 | [32,33] |
| Retained placenta | 0.02 | [34] |
| Metritis | 0.01 | [34] |
| Cystic ovaries | 0.02 | [34] |
| Concentrations of plasma β-hydroxybutyrate | 0.17 | [35] |
| Concentrations of milk β-hydroxybutyrate | 0.16 | [35] |
| Concentrations of milk acetone | 0.10 | [35] |
| Milk fever | 0.07–0.11 | [36] |
| Mastitis | 0.01 to 0.03 | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Džermeikaitė, K.; Šidlauskaitė, M.; Antanaitis, R.; Anskienė, L. Enhancing Genomic Selection in Dairy Cattle Through Artificial Intelligence: Integrating Advanced Phenotyping and Predictive Models to Advance Health, Climate Resilience, and Sustainability. Dairy 2025, 6, 50. https://doi.org/10.3390/dairy6050050
Džermeikaitė K, Šidlauskaitė M, Antanaitis R, Anskienė L. Enhancing Genomic Selection in Dairy Cattle Through Artificial Intelligence: Integrating Advanced Phenotyping and Predictive Models to Advance Health, Climate Resilience, and Sustainability. Dairy. 2025; 6(5):50. https://doi.org/10.3390/dairy6050050
Chicago/Turabian StyleDžermeikaitė, Karina, Monika Šidlauskaitė, Ramūnas Antanaitis, and Lina Anskienė. 2025. "Enhancing Genomic Selection in Dairy Cattle Through Artificial Intelligence: Integrating Advanced Phenotyping and Predictive Models to Advance Health, Climate Resilience, and Sustainability" Dairy 6, no. 5: 50. https://doi.org/10.3390/dairy6050050
APA StyleDžermeikaitė, K., Šidlauskaitė, M., Antanaitis, R., & Anskienė, L. (2025). Enhancing Genomic Selection in Dairy Cattle Through Artificial Intelligence: Integrating Advanced Phenotyping and Predictive Models to Advance Health, Climate Resilience, and Sustainability. Dairy, 6(5), 50. https://doi.org/10.3390/dairy6050050









