Organic Farm Bedded Pack System Microbiomes: A Case Study with Comparisons to Similar and Different Bedded Packs
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.1.1. Micobiomes
2.1.2. Isolations
2.2. Part 1: Amplicon Sequencing
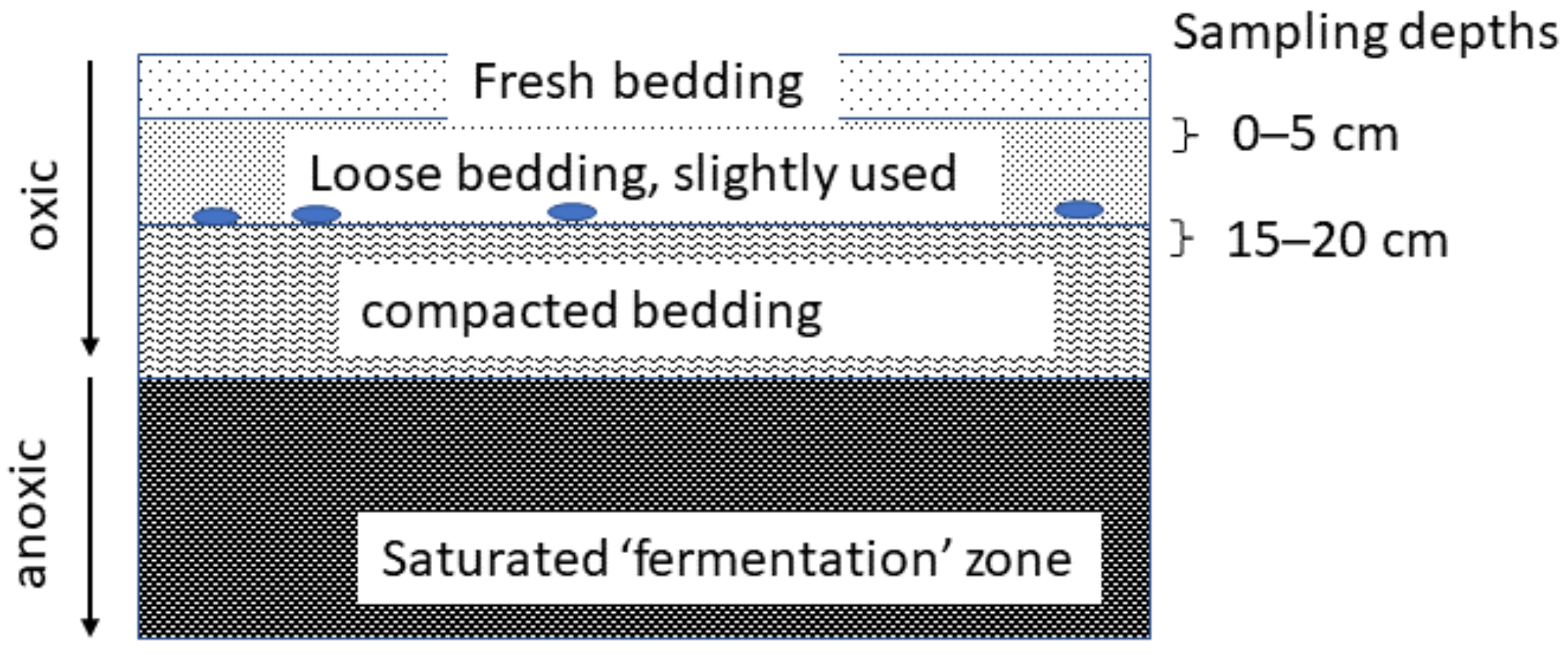
2.2.1. Sampling
2.2.2. DNA Extraction and Sequencing
2.2.3. Statistical Analysis
2.3. Part 2: Ecology
2.3.1. Culturing and Characterization by Morphology
2.3.2. Carbon Utilization
2.3.3. Indicators of Colonization and Pathogenicity
2.3.4. DNA Extraction, PCR and Sanger Sequencing
3. Results
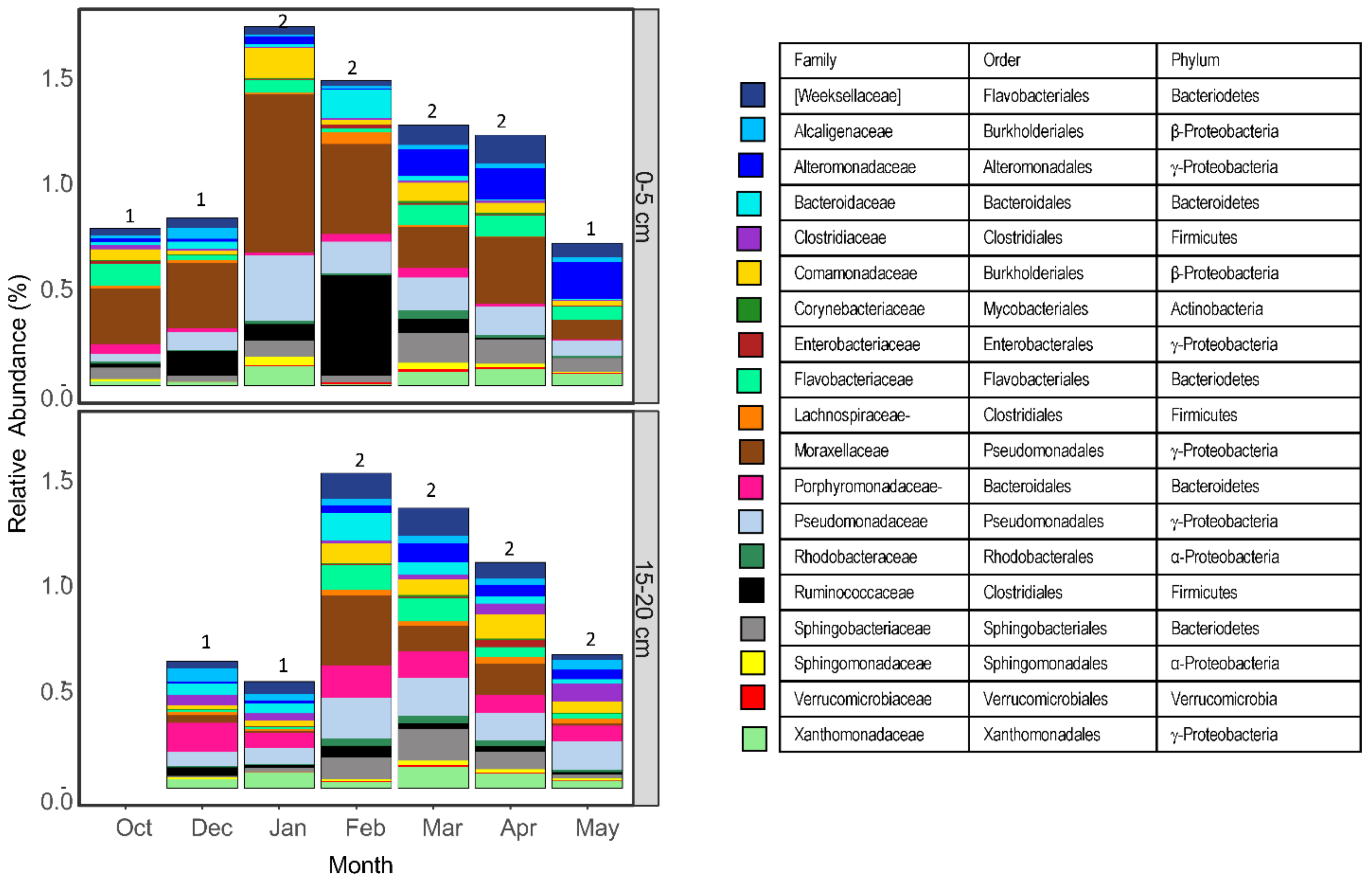
3.1. Most Common Taxa at Farm C and Their Distribution with Season and Depth
3.2. Cultured Filamentous Fungi and Yeast Isolates
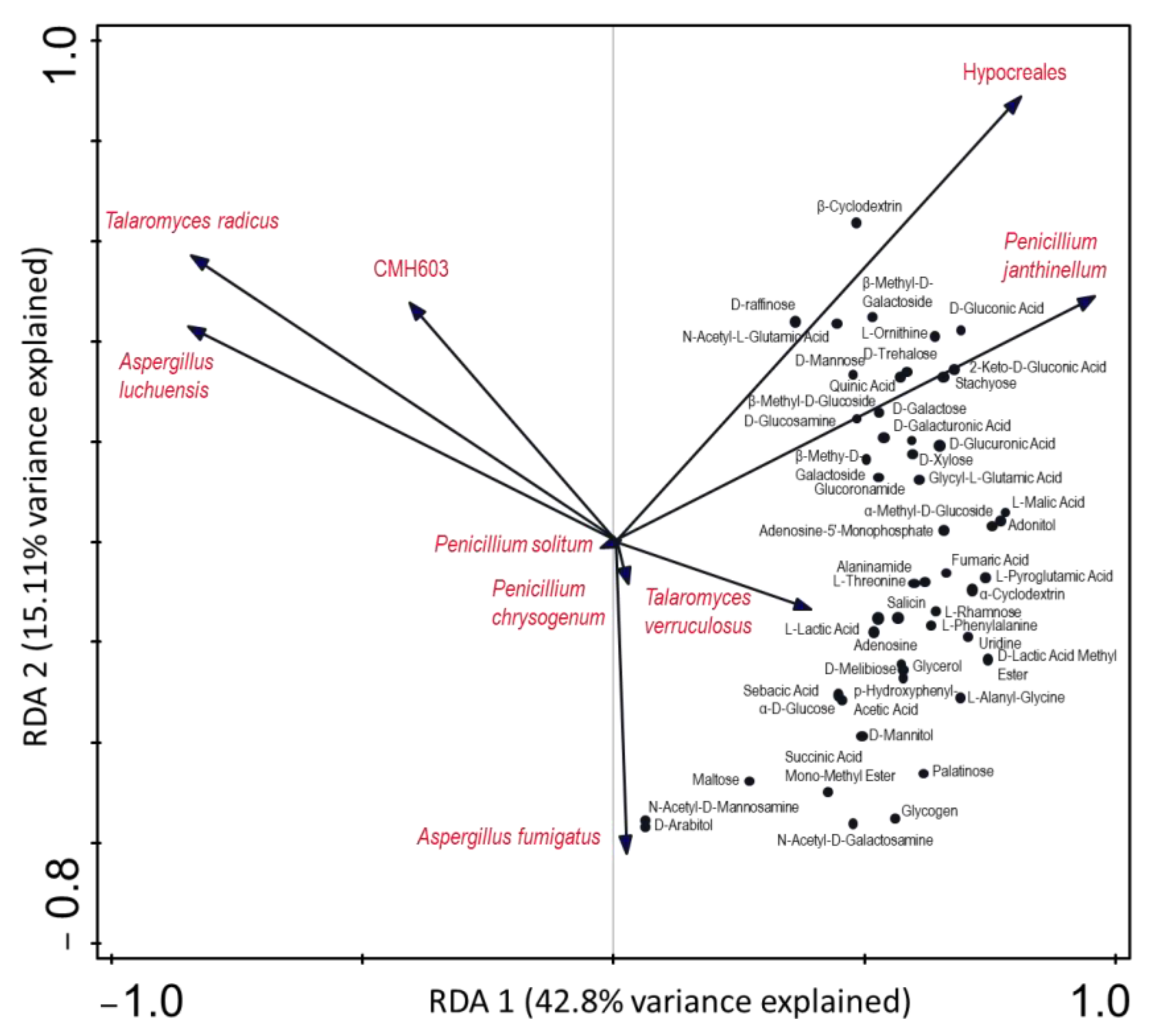
3.3. Indicators of Carbon Utilization and Pathogenicity
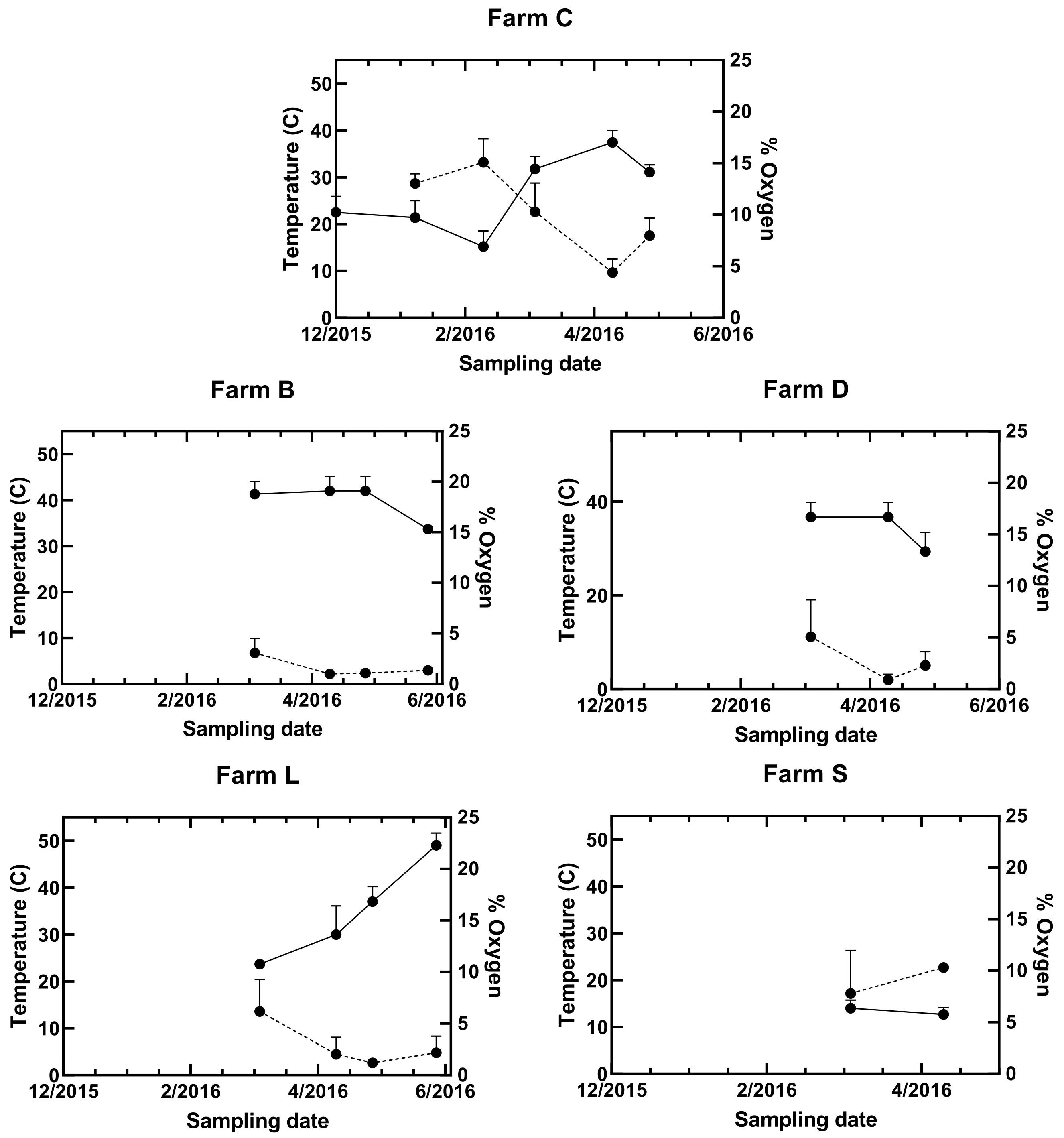
3.4. Comparison among Farms
4. Discussion
4.1. Core Bedding Microbiome
4.2. Trichocomaceae
4.3. Yeasts
4.4. Indicators of Pathogenicity
4.5. Source of Yeasts
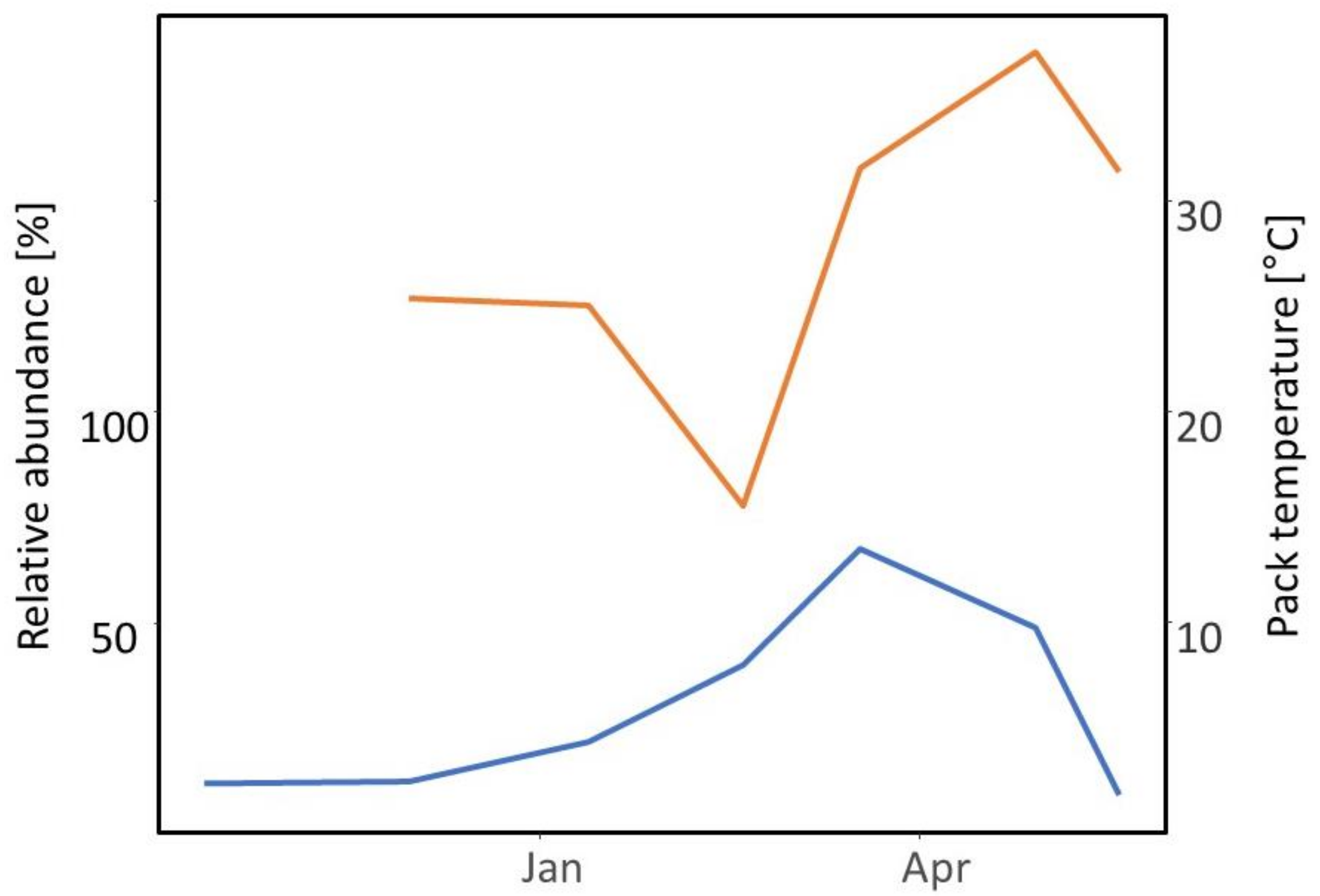
4.6. Bedding Management and the Microbiome
4.7. Bedding Material and the Microbiome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bewley, J.M.; Robertson, L.M.; Eckelkamp, E.A. A 100-Year Review: Lactating dairy cattle housing management. J. Dairy Sci. 2017, 100, 10418–10431. [Google Scholar] [CrossRef] [PubMed]
- USDA. Dairy Cattle Management Practices in the United States. 2014. Available online: https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy14/Dairy14_dr_PartI_1.pdf (accessed on 2 August 2022).
- Leso, L.; Barbari, M.; Lopes, M.A.; Damasceno, F.A.; Galama, P.; Taraba, J.L.; Kuipers, A. Invited review: Compost-bedded pack barns for dairy cows. J. Dairy Sci. 2020, 103, 1072–1099. [Google Scholar] [CrossRef] [PubMed]
- Black, R.A.; Taraba, J.L.; Day, G.B.; Damasceno, F.A.; Bewley, J.M. Compost bedded pack dairy barn management, performance, and producer satisfaction. J. Dairy Sci. 2013, 96, 8060–8074. [Google Scholar] [CrossRef] [PubMed]
- Barberg, A.E.; Endres, M.I.; Salfer, J.A.; Reneau, J.K. Performance and welfare of dairy cows in an alternative housing system in Minnesota. J. Dairy Sci. 2007, 90, 1575–1583. [Google Scholar] [CrossRef]
- Benson, A.F. Consider Deep Pack Barns for Cow Comfort and Manure Management. Available online: https://smallfarms.cornell.edu/2012/04/consider-deep-pack-barns-for-cow-comfort-and-manure-management/ (accessed on 2 August 2022).
- Ray, T.; Gaire, T.N.; Dean, C.J.; Rowe, S.; Godden, S.M.; Noyes, N.R. The microbiome of common bedding materials before and after use on commercial dairy farms. Anim. Microbiome 2022, 4, 18. [Google Scholar] [CrossRef]
- Bewley, J.M.; Taraba, J.; Day, G.; Black, R.A.; Damasceno, F. Compost Bedded Pack Barn Design: Features and Management Considerations. Available online: http://www2.ca.uky.edu/agcomm/pubs/ID/ID206/ID206.pdf (accessed on 2 August 2022).
- Neher, D.A.; Weicht, T.R.; Bates, S.T.; Leff, J.W.; Fierer, N. Changes in bacterial and fungal communities across compost recipes, preparation methods, and composting times. PLoS ONE 2013, 8, e79512. [Google Scholar] [CrossRef]
- Jurjevic, Z.; Peterson, S.W.; Solfrizzo, M.; Peraica, M. Sterigmatocystin production by nine newly described Aspergillus species in section Versicolores grown on two different media. Mycotoxin Res. 2013, 29, 141–145. [Google Scholar] [CrossRef]
- Stein, R.A.; Bulboacӑ, A.E. Mycotoxins. In Foodborne Diseases; Dodd, C.E.R., Aldsworth, T., Stein, R.A., Cliver, D.O., Riemann, H.P., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 407–446. [Google Scholar]
- Frisvad, J.C.; Larsen, T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Front. Microbiol. 2015, 6, 1485. [Google Scholar] [CrossRef]
- Nayak, A.P.; Green, B.J.; Beezhold, D.H. Fungal hemolysins. Med. Mycol. 2013, 51, 1–16. [Google Scholar] [CrossRef]
- Wan, L.; Luo, G.; Lu, H.; Xuan, D.; Cao, H.; Zhang, J. Changes in the hemolytic activity of Candida species by common electrolytes. BMC Microbiol. 2015, 15, 171. [Google Scholar] [CrossRef][Green Version]
- Andrews, T.; Neher, D.A.; Weicht, T.R.; Barlow, J.W. Mammary microbiome of lactating organic dairy cows varies by time, tissue site, and infection status. PLoS ONE 2019, 14, e0225001. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microb. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B.; Keady, P.B.; Brewer, T.E.; Clements, N.; Morgan, E.E.; Awerbuch, J.; Miller, S.L.; Fierer, N. Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 Colorado Front Range flood. Environ. Sci. Technol. 2015, 49, 2675–2684. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M. Towards a Unified Paradigm for Sequence-Based Identification of Fungi; Wiley Online Library, 2013; Available online: https://onlinelibrary.wiley.com/doi/10.1111/mec.12481 (accessed on 2 August 2022).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package: Ordination Methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists. Ver. 2.5-7. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 2 August 2022).
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Samson, R.A.; Pitt, J.I. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification; Harwood Academic: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium—A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–173. [Google Scholar]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.-B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2005, 53, 53–62. [Google Scholar] [CrossRef]
- Hecht, K.A.; O’Donnell, A.F.; Brodsky, J.L. The proteolytic landscape of the yeast vacuole. Cell Logist. 2014, 4, e28023. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome. Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Ter Braak, C.J.F.; Šmilauer, P. Canoco Reference Manual and User’s Guide. Software for Ordination, Version 5; Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Metzner, M.; Sauter-Louis, C.; Seemueller, A.; Petzl, W.; Klee, W. Infrared thermography of the udder surface of dairy cattle: Characteristics, methods, and correlation with rectal temperature. Vet. J. 2014, 199, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yin, S.; Xu, Y.; Xiang, L.; Wang, H.; Li, Z.; Fan, K.; Pan, G. The richness and diversity of catalases in bacteria. Front. Microbiol. 2021, 12, 645477. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Cruz, M.; Briones-Martin-del-Campo, M.; Canas-Villamar, I.; Montalvo-Arredondo, J.; Riego-Ruiz, L.; Castano, I.; De Las Penas, A. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot. Cell 2008, 7, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, L.M.; Sumner, S.S. Antibacterial activity of the lactoperoxidase system: A review. J. Food Prot. 1993, 56, 887–892. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Sakamoto, M. The family Porphyromonadaceae. In The Prokaryotes: Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Júnior, J.C.R.; De Oliveira, A.M.; de Silva, F.G.; Tamanini, R.; de Oliveira, A.L.M.; Beloti, V. The main spoilage-related psychrotrophic bacteria in refrigerated raw milk. J. Dairy Sci. 2018, 101, 75–83. [Google Scholar] [CrossRef]
- Trujillo, M.E.; Dedysh, S.; DeVos, P.; Hedlund, B.; Kämpfer, P.; Rainey, F.A.; Whitman, W.B. Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut Bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Y.; Li, Y.; Zhang, Y.; Liu, T.; Wang, Y.; Sharpton, T.J.; Zhu, W. Progressive colonization of bacteria and degradation of rice straw in the rumen by Illumina sequencing. Front. Microbiol. 2017, 8, 2165. [Google Scholar] [CrossRef]
- Sun, L.; Han, X.; Li, J.; Zhao, Z.; Liu, Y.; Xi, Q.; Guo, X.; Gun, S. Microbial community and its association with physicochemical factors during compost bedding for dairy cows. Front. Microbiol. 2020, 11, 254. [Google Scholar] [CrossRef]
- Neher, D.A.; Limoges, M.A.; Weicht, T.R.; Sharma, M.; Millner, P.D.; Donnelly, C. Bacterial community dynamics distinguish poultry compost from dairy compost and non-amended soils planted with spinach. Microorganisms 2020, 8, 1601. [Google Scholar] [CrossRef]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 2017, 93, fix006. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.L.; Yu, X.B.; Zhang, C.; Wang, G.X. Growth inhibition and microcystin degradation effects of Acinetobacter guillouiae A2 on Microcystis Aeruginosa. Res. Microbiol. 2015, 166, 93–101. [Google Scholar] [CrossRef]
- Majumder, S.; Gangadhar, G.; Raghuvanshi, S.; Gupta, S. A comprehensive study on the behavior of a novel bacterial strain Acinetobacter guillouiae for bioremediation of divalent copper. Bioprocess. Biosyst. Eng. 2015, 38, 1749–1760. [Google Scholar] [CrossRef]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. The causes of Pseudomonas diversity. Microbiology (Reading) 2000, 146, 2345–2350. [Google Scholar] [CrossRef]
- Narancic, T.; Salvador, M.; Hughes, G.M.; Beagan, N.; Abdulmutalib, U.; Kenny, S.T.; Wu, H.H.; Saccomanno, M.; Um, J.; O’Connor, K.E.; et al. Genome analysis of the metabolically versatile Pseudomonas umsongensis GO16: The genetic basis for PET monomer upcycling into polyhydroxyalkanoates. Microb. Biotechnol. 2021, 14, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Attia, M.A.; Nelson, C.E.; Offen, W.A.; Jain, N.; Davies, G.J.; Gardner, J.G.; Brumer, H. In vitro and in vivo characterization of three Cellvibrio japonicus glycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions. Biotechnol. Biofuels 2018, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Young, C.C.; Kampfer, P.; Chen, W.M.; Yen, W.S.; Arun, A.B.; Lai, W.A.; Shen, F.T.; Rekha, P.D.; Lin, K.Y.; Chou, J.H. Luteimonas composti sp. Nov., a moderately thermophilic bacterium isolated from food waste. Int. J. Syst. Evol. Microbiol. 2007, 57, 741–744. [Google Scholar] [CrossRef]
- Wong, K.; Shaw, T.I.; Oladeinde, A.; Glenn, T.C.; Oakley, B.; Molina, M. Rapid microbiome changes in freshly deposited cow feces under field conditions. Front. Microbiol. 2016, 7, 500. [Google Scholar] [CrossRef]
- Bonner, R.D.; Fergus, C.L. The fungus flora of cattle feeds. Mycologia 1959, 51, 855–863. [Google Scholar] [CrossRef]
- Morton, A.G. A study of nitrate reduction in mould fungi. J. Exp. Bot. 1956, 7, 97–112. [Google Scholar] [CrossRef]
- Xu, J.; Song, Y.C.; Guo, Y.; Mei, Y.N.; Tan, R.X. Fumigaclavines D-H, new ergot alkaloids from endophytic Aspergillus fumigatus. Planta. Med. 2014, 80, 1131–1137. [Google Scholar] [CrossRef]
- Heo, Y.M.; Lee, H.; Kim, K.; Kwon, S.L.; Park, M.Y.; Kang, J.E.; Kim, G.H.; Kim, B.S.; Kim, J.J. Fungal diversity in intertidal mudflats and abandoned solar salterns as a source for biological resources. Mar. Drugs 2019, 17, 601. [Google Scholar] [CrossRef]
- Fernandes, T.; Carvalho, B.F.; Mantovani, H.C.; Schwan, R.F.; Avila, C.L.S. Identification and characterization of yeasts from bovine rumen for potential use as probiotics. J. Appl. Microbiol. 2019, 127, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Losada, L.; Ajayi, O.; Frisvad, J.C.; Yu, J.; Nierman, W.C. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med. Mycol. 2009, 47 (Suppl. 1), S88–S96. [Google Scholar] [CrossRef]
- Magan, N. Fungal colonization and decomposition of cereal straw. Int. Biodeterior. 1988, 24, 435–443. [Google Scholar] [CrossRef]
- Karapetsa, M.; Tsolaki, V.; Arabatzis, M.; Petinaki, E.; Velegraki, A.; Zakynthinos, E. Septic shock due to Candida famata (Debaryomyces hansenii) candidemia in an ICU immunocompetent trauma-patient. J. Infect. Public Health 2019, 12, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L. Etiological agents of bovine mastitis. Vet. Microbiol. 1988, 16, 41–66. [Google Scholar] [CrossRef]
- Banjara, N.; Nickerson, K.W.; Suhr, M.J.; Hallen-Adams, H.E. Killer toxin from several food-derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. Int. J. Food Microbiol. 2016, 222, 23–29. [Google Scholar] [CrossRef]
- Breuer, U.; Harms, H. Debaryomyces hansenii—an extremophilic yeast with biotechnological potential. Yeast 2006, 23, 415–437. [Google Scholar] [CrossRef]
- Dworecka-Kaszak, B.; Krutkiewicz, A.; Szopa, D.; Kleczkowski, M.; Bieganska, M. High prevalence of Candida yeast in milk samples from cows suffering from mastitis in Poland. Sci. World J. 2012, 2012, 196347. [Google Scholar] [CrossRef]
- Moravkova, M.; Huvarova, V.; Vlkova, H.; Kostovova, I.; Bacova, R. Raw bovine milk as a reservoir of yeast with virulence factors and decreased susceptibility to antifungal agents. Med. Mycol. 2021, 59, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Andrews, T.D. Ecology of Composted Bedded Pack and Its Impact on the Udder Microbiome with an Emphasis on Mastitis Epidemiology. Master’s Thesis, University of Vermont, Burlington, VT, USA, 2018. [Google Scholar]
- Al-Qaysi, S.A.S.; Al-Haideri, H.; Thabit, Z.A.; Al-Kubaisy, W.H.A.A.; Ibrahim, J.A.A. Production, characterization, and antimicrobial activity of mycocin produced by Debaryomyces hansenii DSMZ70238. Int. J. Microbiol. 2017, 2017, 2605382. [Google Scholar] [CrossRef] [PubMed]
- Padovan, A.C.; Melo, A.S.; Colombo, A.L. Systematic review and new insights into the molecular characterization of the Candida rugosa species complex. Fungal Genet. Biol 2013, 61, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Sugita, T.; Hata, E.; Katsuda, K.; Zhang, E.; Kiku, Y.; Sugawara, K.; Ozawa, T.; Matsubara, T.; Ando, T.; et al. Molecular-based identification of yeasts isolated from bovine clinical mastitis in Japan. J. Vet. Med. Sci. 2013, 75, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Pongcharoen, P. The ability of Pichia kudriavzevii to tolerate multiple stresses makes it promising for developing improved bioethanol production processes. Lett. Appl. Microbiol. 2022, 75, 36–44. [Google Scholar] [CrossRef]
- Bajaj, B.K.; Raina, S.; Singh, S. Killer toxin from a novel killer yeast Pichia kudriavzevii RY55 with idiosyncratic antibacterial activity. J. Basic Microbiol. 2013, 53, 645–656. [Google Scholar] [CrossRef]
- Antunes, J.; Aguiar, C. Search for killer phenotypes with potential for biological control. Ann. Microbiol. 2012, 62, 427–433. [Google Scholar] [CrossRef]
- Gomes, S.I.F.; van Bodegom, P.M.; van Agtmaal, M.; Soudzilovskaia, N.A.; Bestman, M.; Duijm, E.; Speksnijder, A.; van Eekeren, N. Microbiota in dung and milk differ between organic and conventional dairy farms. Front. Microbiol. 2020, 11, 1746. [Google Scholar] [CrossRef]
- Mbareche, H.; Veillette, M.; Bilodeau, G.J.; Duchaine, C. Fungal aerosols at dairy farms using molecular and culture techniques. Sci. Total Environ. 2019, 653, 253–263. [Google Scholar] [CrossRef]
- Fourie, R.; Kuloyo, O.O.; Mochochoko, B.M.; Albertyn, J.; Pohl, C.H. Iron at the centre of Candida albicans interactions. Front. Cell Infect. Microbiol. 2018, 8, 185. [Google Scholar] [CrossRef]
- Luo, G.; Samaranayake, L.P.; Yau, J.Y. Candida species exhibit differential in vitro hemolytic activities. J. Clin. Microbiol. 2001, 39, 2971–2974. [Google Scholar] [CrossRef] [PubMed]
- Schlander, M.; Distler, U.; Tenzer, S.; Thines, E.; Claus, H. Purification and properties of yeast proteases secreted by Wickerhamomyces anomalus 227 and Metschnikovia pulcherrima 446 during growth in a white grape juice. Fermentation 2016, 3, 2. [Google Scholar] [CrossRef]
- Montoya, A.M.; Luna-Rodriguez, C.E.; Gracia-Robles, G.; Rojas, O.C.; Trevino-Rangel, R.J.; Gonzalez, G.M. In vitro virulence determinants, comparative pathogenicity of Diutina (Candida) mesorugosa clinical isolates and literature review of the D. rugosa complex. Mycologia 2019, 111, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Cadez, N.; Raspor, P.; Turchetti, B.; Cardinali, G.; Ciafardini, G.; Veneziani, G.; Peter, G. Candida adriatica sp. nov. and Candida molendinolei sp. nov., two yeast species isolated from olive oil and its by-products. Int. J. Syst. Evol. Microbiol. 2012, 62, 2296–2302. [Google Scholar] [CrossRef]
- Lagace, L.S.; Bisson, L.F. Survey of yeast acid proteases for effectiveness of wine haze reduction. Am. J. Enol. Viticult. 1990, 41, 147–155. [Google Scholar]
- Bolumar, T.; Sanz, Y.; Aristoy, M.C.; Toldra, F. Purification and characterisation of proteases A and D from Debaryomyces hansenii. Int. J. Food Microbiol. 2008, 124, 135–141. [Google Scholar] [CrossRef]
- Nascimento, B.L.; Delabeneta, M.F.; Rosseto, L.R.B.; Junges, D.S.B.; Paris, A.P.; Persel, C.; Gandra, R.F. Yeast mycocins: A great potential for application in health. FEMS Yeast Res. 2020, 20, foaa016. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Swiderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkolazka, A.; Paszczynski, A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Vissers, M.M.; Driehuis, F.; Te Giffel, M.C.; De Jong, P.; Lankveld, J.M. Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J. Dairy Sci. 2007, 90, 928–936. [Google Scholar] [CrossRef]
- Vissers, M.M.; Driehuis, F.; Te Giffel, M.C.; De Jong, P.; Lankveld, J.M. Short communication: Quantification of the transmission of microorganisms to milk via dirt attached to the exterior of teats. J. Dairy Sci. 2007, 90, 3579–3582. [Google Scholar] [CrossRef] [PubMed]
- Quintana, A.R.; Perea, J.M.; Garcia-Bejar, B.; Jimenez, L.; Garzon, A.; Arias, R. Dominant yeast community in raw sheep’s milk and potential transfers of yeast species in relation to farming practices. Animal 2020, 10, 906. [Google Scholar] [CrossRef] [PubMed]
- Stergiadis, S.; Cabeza-Luna, I.; Mora-Ortiz, M.; Stewart, R.D.; Dewhurst, R.J.; Humphries, D.J.; Watson, M.; Roehe, R.; Auffret, M.D. Unravelling the role of rumen microbial communities, genes, and activities on milk fatty acid profile using a combination of omics approaches. Front. Microbiol. 2021, 11, 590441. [Google Scholar] [CrossRef] [PubMed]
- Porcellato, D.; Smistad, M.; Bombelli, A.; Abdelghani, A.; Jorgensen, H.J.; Skeie, S.B. Longitudinal study of the bulk tank milk microbiota reveals major temporal shifts in composition. Front. Microbiol. 2021, 12, 616429. [Google Scholar] [CrossRef]
- Zhang, L.L.; Ma, H.X.; Zhang, H.Q.; Xun, L.Y.; Chen, G.J.; Wang, L.S. Thermomyces lanuginosus is the dominant fungus in maize straw composts. Bioresour. Technol. 2015, 197, 266–275. [Google Scholar] [CrossRef]
- Yan, T.; Frost, J.P.; Agnew, R.E.; Binnie, R.C.; Mayne, C.S. Relationships among manure nitrogen output and dietary and animal factors in lactating dairy cows. J. Dairy Sci. 2006, 89, 3981–3991. [Google Scholar] [CrossRef]
- Lawther, J.M.; Sun, R.C.; Banks, W.B. Extraction, fractionation, and characterization of structural polysaccharides from wheat-straw. J. Agr Food Chem 1995, 43, 667–675. [Google Scholar] [CrossRef]
- Tullus, A.; Mandre, M.; Soo, T.; Tullus, H. Relationships between cellulose, lignin and nutrients in the stemwood of hybrid aspen in Estonian plantations. Cell Chem.Technol. 2010, 44, 101–109. [Google Scholar]
- Roussel, S.; Reboux, G.; Dalphin, J.C.; Bardonnet, K.; Millon, L.; Piarroux, R. Microbiological evolution of hay and relapse in patients with farmer’s lung. Occup Environ. Med. 2004, 61, e3. [Google Scholar]


| Management Practice | Farm Name | ||||
|---|---|---|---|---|---|
| C | B | D | L | S | |
| Bedding Style | DBP | DBP | DBP | CBP | DBP |
| Primary Bedding | hay | hay | hay | sawdust | straw |
| Secondary Bedding | wood chips | wood chips, straw | wood chips | hay | wood shavings |
| Breed | Holstein—Jersey cross | Jersey | Jersey | Holstein—Jersey cross | Jersey |
| Stocking Density (m2/cow) | 8.18 | 7.43 | 8.23 | 9.3 | 12.01 |
| Bedding (kg/cow/day) a | 4.14 | 2.45 | 12.47 | 5.33 | 5.02 |
| Median Temperature °C | 30.5 (n = 12) | 41 (n = 4) | 36 (n = 3) | 33.5 (n = 4) | 13 (n = 2) |
| Median Oxygen (%) | 8.66 (n = 9) | 1.23 (n = 4) | 2.3 (n = 3) | 2.10 (n = 4) | 7.8 (n = 1) |
| Isolate Contig | Sample Source a | Isolation Media b | Culture Temperature (°C) | Species | % Match |
|---|---|---|---|---|---|
| 1 | BP | CZA | 22 | Aspergillus niger | 99.67 |
| 5 | BP | CZA | 22 | Penicillium solitum strain 20-01 | 99.83 |
| 6 | BP | CZA | 22 | Penicillium solitum strain 20-01 | 99.83 |
| 9 | TS | CYA | 35 | Aspergillus fumigatus strain cy018 | 99.65 |
| 11 | TS | CYA | 35 | Uncultured fungus CMH603 | 99.78 |
| 14 | BP | Unknown | 22 | Penicillium solitum strain 20-01 | 99.82 |
| 16 | TS | CYA | 22 | Penicillium solitum strain 20-01 | 99.82 |
| 17 | TS | CYA | 22 | Penicillium solitum strain 20-01 | 99.82 |
| 19 | TS | CZA | 22 | Penicillium solitum strain 20-01 | 99.83 |
| 21 | BP | DG18 | 22 | Penicillium solitum strain 20-01 | 99.83 |
| 27 | BP | YE20S | 22 | Penicillium chrysogenum strain ZJ-T2 | 99.65 |
| 32 | BP | YE20S | 22 | Aspergillus fumigatus strain cy018 | 99.65 |
| 33 | TS | DG18 | 22 | Penicillium janthinellum series | 99.65 |
| 37 | BP | DG18 | 22 | Penicillium solitum strain 20-01 | 95.65 |
| 39 | BP | Humidity Chamber | 22 | Penicillium solitum strain 20-01 | 99.82 |
| 41 | TS | CZA | 22 | Hypocreales uncultured | 99.26 |
| 47 | TS | CYA | 22 | Talaromyces radicus (Penicillium radicum) | 96.91 |
| 95 | BP | CMA | 22 | Aspergillus luchuensis | 100 |
| 129 | BP | Humidity Chamber | 34 | Talaromyces verruculosus (Penicillium verruculosum) | 99.13 |
| CG48 | BP | YES | 34 | Aspergillus fumigatus strain cy018 | 99.82 |
| CH4 | TS | DG18 | 35 | Aspergillus fumigatus strain cy018 | 99.83 |
| Isolate Contig | Farm | Sample Source a | Isolation Media b | Species | % Match | Protease | Hemolytic | 35 °C | Catalase | Peroxidase |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | BTM | RBC | Diutina rugosa | 95.7 | 1 | 0 | 1 | 1 | 0 |
| 2 | M | BTM | RBC | Diutina rugosa | 95.7 | 1 | 0 | 1 | 1 | 0 |
| 3 | M | BTM | RBC | Diutina rugosa | 95.7 | 1 | 0 | 1 | 1 | 0 |
| 5 | M | BTM | RBC | Uncultured Tremellomycetes | 98.9 | 1 | 0 | 0 | 0 | 0 |
| 6 | C | High SCC | CAF | Uncultured Tremellomycetes | 99.8 | 1 | 0 | 0 | 0 | 0 |
| 7 | C | BTM | CAF | Pichia holstii | 97.2 | 1 | 0 | 1 | 0 | 0 |
| 8 | C | BTM | AWPY | Pichia kudriavzevii | 99.8 | 1 | 0 | 1 | 0 | 0 |
| 9 | C | BTM | AWPY | Wickerhamomyces anomalus | 99.8 | 0 | 0 | 1 | 0 | 0 |
| 10 | C | BTM | CYA | Wickerhamomyces anomalus | 82.4 | 1 | 0 | 1 | 0 | 0 |
| 11 | C | BTM | CYA | Wickerhamomyces anomalus | 99.7 | 1 | 0 | 1 | 0 | 0 |
| 12 | C | QM | Blood Agar | Diutina rugosa | 96.0 | 1 | 0 | 1 | 0 | 0 |
| 13 | C | BP | BHI | Diutina catenulata | 100.0 | 1 | 1 | 1 | 0 | 0 |
| 14 | C | BP | BHI | Wickerhamomyces anomalus | 99.7 | 1 | 0 | 1 | 0 | 0 |
| 15 | C | BP | RUM | Pichia fermentans | 99.8 | 1 | 1 | 1 | 0 | 0 |
| 16 | C | BP | RUM | Pichia fermentans | 99.6 | 1 | 0 | 0 | 1 | 0 |
| 17 | C | BP | CAF | Pichia fermentans | 99.6 | 1 | 0 | 1 | 0 | 0 |
| 22 | C | BP | YLA | (Candida) glaebosa | 99.3 | 1 | 0 | 0 | 0 | 0 |
| 23 | C | BP | BHI | Wickerhamomyces anomalus | 100.0 | 1 | 0 | 0 | 0 | 0 |
| 25 | C | BP | RUM | (Candida) glaebosa | 99.0 | 1 | 0 | 0 | 0 | 0 |
| 26 | C | BP | CAF | Hyphopichia pseudoburtonii | 99.6 | 1 | 0 | 0 | 0 | 0 |
| 27 | C | TS | SDA | (Candida) glaebosa | 98.7 | 1 | 0 | 0 | 0 | 0 |
| 28 | C | TS | SDA | (Candida) glaebosa | 98.4 | 1 | 0 | 1 | 0 | 0 |
| 29 | C | TS | SDA | Debaryomyces hansenii | 100.0 | 1 | 0 | 0 | 0 | 0 |
| 30 | C | TS | RUM | Debaryomyces hansenii | 99.8 | 1 | 0 | 0 | 1 | 0 |
| 31 | C | TS | RUM | Debaryomyces hansenii | 100.0 | 1 | 0 | 0 | 1 | 0 |
| 32 | C | TS | RUM | Hyphopichia burtonii | 99.8 | 1 | 0 | 1 | 0 | 0 |
| 33 | C | TS | CAF | Diutina catenulata | 99.4 | 1 | 0 | 1 | 1 | 0 |
| 34 | C | TS | CAF | Debaryomyces hansenii | 100.0 | 1 | 0 | 0 | 1 | 0 |
| 35 | C | TS | BHI | Debaryomyces hansenii | 100.0 | 1 | 0 | 0 | 0 | 0 |
| 36 | C | TS | BHI | Debaryomyces hansenii | 99.8 | 1 | 0 | 0 | 0 | 0 |
| 37 | C | TS | BHI | Diutina catenulata | 100.0 | 1 | 0 | 1 | 0 | 0 |
| 38 | C | TS | BHI | Debaryomyces hansenii | 99.7 | 1 | 0 | 1 | 0 | 0 |
| 39 | C | TS | YLA | Wickerhamomyces anomalus | 99.7 | 1 | 0 | 1 | 0 | 0 |
| 40 | C | TS | YNG | Hyphopichia burtonii | 99.8 | 1 | 0 | 1 | 0 | 0 |
| 41 | C | BP | SDA | Debaryomyces hansenii | 99.7 | 1 | 0 | 1 | 0 | 0 |
| 42 | C | BP | CAF | Wickerhamomyces anomalus | 100.0 | 1 | 0 | 1 | 0 | 0 |
| 43 | C | BP | YLA | Debaryomyces coudertii | 99.5 | 0 | 0 | 1 | 0 | 0 |
| 44 | C | BP | BHI | Diutina catenulata | 100.0 | 1 | 0 | 1 | 0 | 0 |
| 45 | C | BP | YLA | Wickerhamomyces anomalus | 99.7 | 1 | 0 | 1 | 0 | 0 |
| 46 | C | BP | SDA | Diutina catenulata | 100.0 | 1 | 0 | 1 | 0 | 0 |
| Pichia kudriavzevii | Pichia holstii | Wickerhamomyces anomalus | |
|---|---|---|---|
| Samples in which isolate is present | 90% | 8% | 3% |
| Median relative abundance among samples in which isolate is present | 12.2% | 0.1% | 0.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neher, D.A.; Andrews, T.D.; Weicht, T.R.; Hurd, A.; Barlow, J.W. Organic Farm Bedded Pack System Microbiomes: A Case Study with Comparisons to Similar and Different Bedded Packs. Dairy 2022, 3, 587-607. https://doi.org/10.3390/dairy3030042
Neher DA, Andrews TD, Weicht TR, Hurd A, Barlow JW. Organic Farm Bedded Pack System Microbiomes: A Case Study with Comparisons to Similar and Different Bedded Packs. Dairy. 2022; 3(3):587-607. https://doi.org/10.3390/dairy3030042
Chicago/Turabian StyleNeher, Deborah A., Tucker D. Andrews, Thomas R. Weicht, Asa Hurd, and John W. Barlow. 2022. "Organic Farm Bedded Pack System Microbiomes: A Case Study with Comparisons to Similar and Different Bedded Packs" Dairy 3, no. 3: 587-607. https://doi.org/10.3390/dairy3030042
APA StyleNeher, D. A., Andrews, T. D., Weicht, T. R., Hurd, A., & Barlow, J. W. (2022). Organic Farm Bedded Pack System Microbiomes: A Case Study with Comparisons to Similar and Different Bedded Packs. Dairy, 3(3), 587-607. https://doi.org/10.3390/dairy3030042





