From SARS to MERS and SARS-CoV-2: Comparative Spike Protein Remodeling and Ligand-Binding Hot-Spots Revealed by Multiscale Simulations
Abstract
1. Introduction
- -
- XBB.1.16: emerged mid-2023, widely disseminated, and outcompeted other Omicron subvariants, showing comparable ACE2 affinity [13].
- -
- BA.2.75: detected in Singapore and India, shows mutations likely responsible for immune escape and high transmissibility [14].
- -
- EG.5 (“Eris”): derivative of XBB with spike protein alterations, contributed significantly to global case increases [15].
- -
- CH.1.1: identified in Southeast Asia, shares mutations with Delta and BA subvariants, notably L452R, enhancing transmissibility and RBD interaction [16].
2. Materials and Methods
2.1. In Silico Mutagenesis and System Preparation
2.2. Binding Site Identification and Molecular Docking Studies
2.3. Molecular Dynamic Simulations
2.4. Quantum Mechanical Studies
2.5. Cross-Validation Across Computational Tiers
3. Results
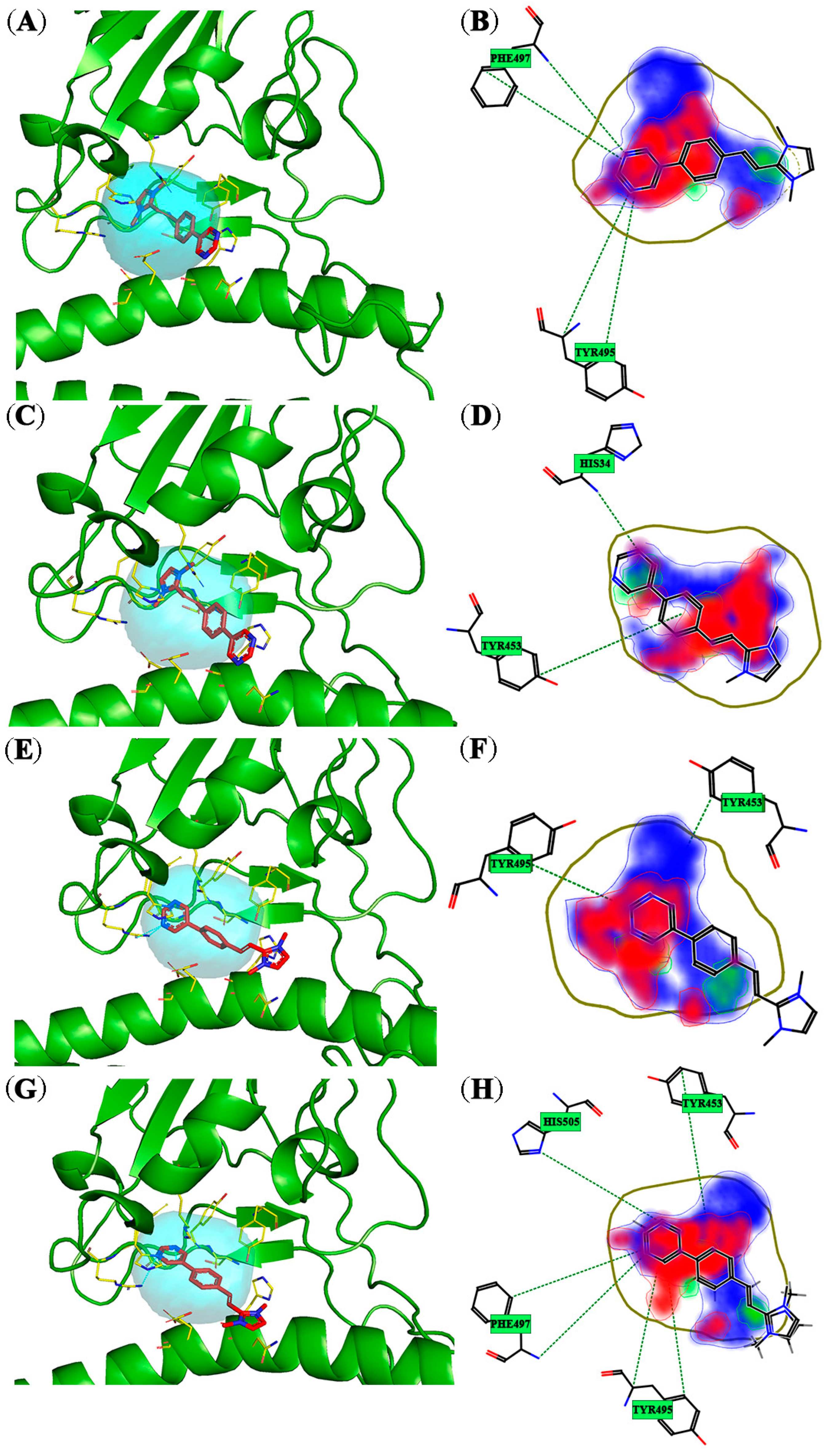
3.1. Molecular Docking Studies
3.2. Molecular Dynamics Simulations
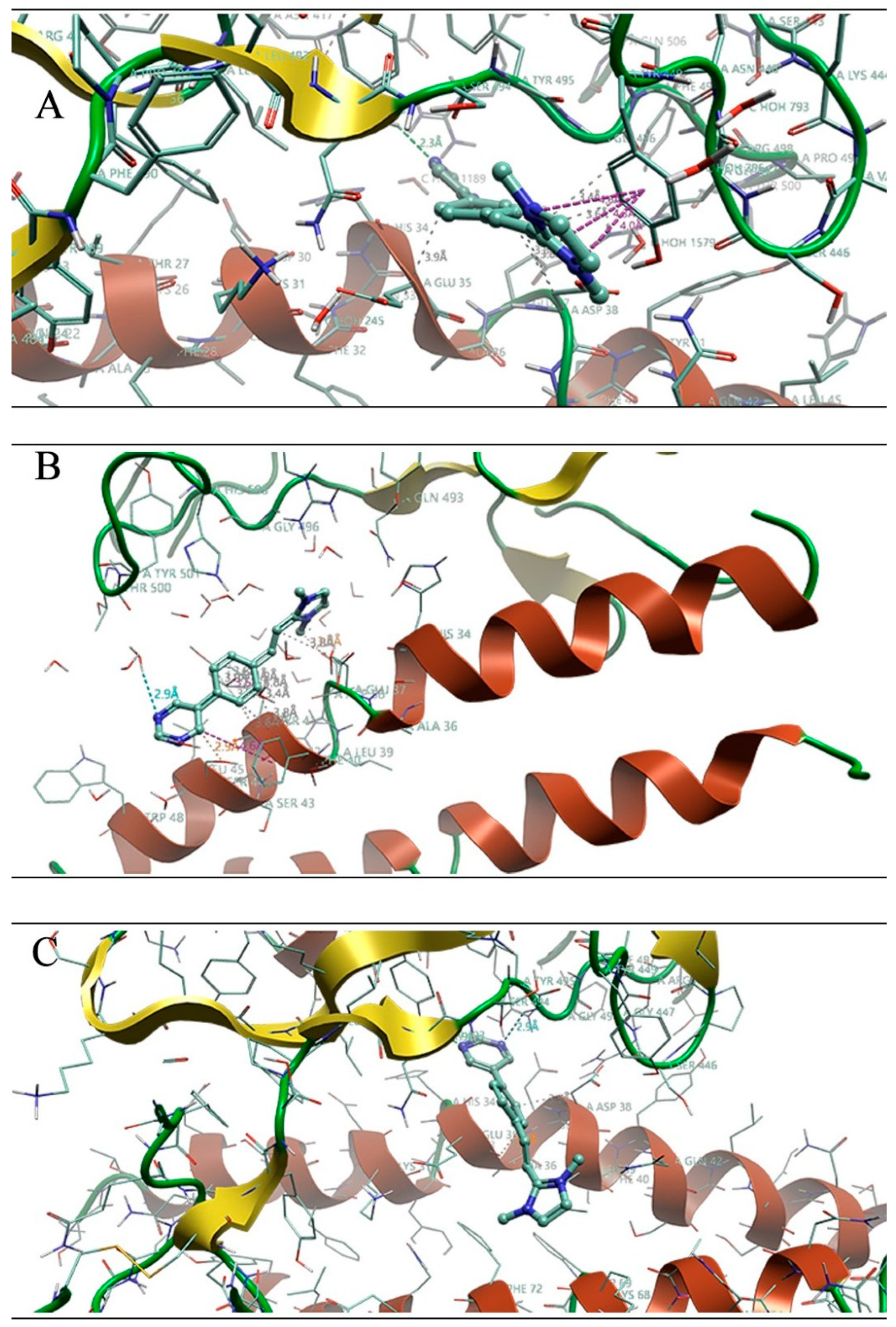
3.3. Quantum Mechanical Studies
| Target | ΔE (kcal/mol) | Interacting Residues |
|---|---|---|
| BA | −16.41 | Asn33, His34, Tyr449, Tyr453, Tyr495, Arg498 |
| CH | −25.33 | Asn33, His34, Glu37, Asp38, Tyr449, Tyr453, Tyr495, Arg498 |
| EG | −20.54 | His34, Glu37, Tyr453, Tyr495, Phe497, Arg498, His505 |
| XBB | −19.72 | Asn33, His34, Asn405, Tyr449, Tyr453, Gln493, Tyr495, |
| Mers | −9.28 | Asn33, Asp38, Phe40, Tyr41 |
| SARS-CoV-1 | −12.60 | Asn33, Asp38, Tyr41, Tyr436, Tyr440 |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| SARS | severe acute respiratory syndrome |
| MERS | Middle East respiratory syndrome |
| COVID-19 | coronavirus disease 2019 |
| WHO | World Health Organization |
| RBD | receptor-binding domain |
| SARS-CoV | Severe Acute Respiratory Syndrome Coronavirus |
| MERS-CoV | Middle East Respiratory Syndrome Coronavirus |
| BCC1 | (E)-1-methyl-2-4-(pyrimidin-5-yl) styryl) pyridin-1-ium |
| BCC2 | (E)-1-methyl-2-4-(pyrimidin-5-yl) styryl) quinolin-1-ium |
| BCC3 | (E)-1,3-dimethyl-2-4-(pyrimidin-5-yl)styryl)-1-h-imidazol-3-ium |
References
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-CoV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Vijayanand, P.; Wilkins, E.; Woodhead, M. Severe acute respiratory syndrome (SARS): A review. Clin. Med. 2004, 4, 152–160. [Google Scholar] [CrossRef]
- Azhar, E.I.; Hui, D.S.C.; Memish, Z.A.; Drosten, C.; Zumla, A. The Middle East Respiratory Syndrome (MERS). Infect. Dis. Clin. N. Am. 2019, 33, 891–905. [Google Scholar] [CrossRef]
- Santos-López, G.; Cortés-Hernández, P.; Vallejo-Ruiz, V.; Reyes-Leyva, J. SARS-CoV-2: Basic concepts, origin and treatment advances. Gac. Med. Mex. 2021, 157, 84–89. [Google Scholar] [CrossRef]
- Sankaran, N.; Weiss, R.A. Viruses: Impact on Science and Society. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 671–680. [Google Scholar] [CrossRef]
- Lundstrom, K.; Seyran, M.; Pizzol, D.; Adadi, P.; Mohamed Abd El-Aziz, T.; Hassan, S.S.; Soares, A.; Kandimalla, R.; Tambuwala, M.M.; Aljabali, A.A.A.; et al. The Importance of Research on the Origin of SARS-CoV-2. Viruses 2020, 12, 1203. [Google Scholar] [CrossRef]
- Weng, Y.L.; Naik, S.R.; Dingelstad, N.; Lugo, M.R.; Kalyaanamoorthy, S.; Ganesan, A. Molecular dynamics and in silico mutagenesis on the reversible inhibitor-bound SARS-CoV-2 main protease complexes reveal the role of lateral pocket in enhancing the ligand affinity. Sci. Rep. 2021, 11, 7429. [Google Scholar] [CrossRef]
- Widagdo, W.; Okba, N.M.A.; Raj, V.S.; Haagmans, B.L. MERS-coronavirus: From discovery to intervention. One Health 2017, 3, 11–16. [Google Scholar] [CrossRef]
- Sipala, F.; Cavallaro, G.; Forte, G.; Satriano, C.; Giuffrida, A.; Fraix, A.; Spadaro, A.; Petralia, S.; Bonaccorso, C.; Fortuna, C.G.; et al. Different In Silico Approaches Using Heterocyclic Derivatives against the Binding between Different Lineages of SARS-CoV-2 and ACE2. Molecules 2023, 28, 3908. [Google Scholar] [CrossRef]
- Brüssow, H. COVID-19: Omicron—The latest, the least virulent, but probably not the last variant of concern of SARS-CoV-2. Microb. Biotechnol. 2022, 15, 1927–1939. [Google Scholar] [CrossRef]
- Yamasoba, D.; Uriu, K.; Plianchaisuk, A.; Kosugi, Y.; Pan, L.; Zahradnik, J.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 omicron XBB.1.16 variant. Lancet Infect. Dis. 2023, 23, 655–656. [Google Scholar] [CrossRef]
- Planas, D.; Bruel, T.; Staropoli, I.; Guivel-Benhassine, F.; Porrot, F.; Maes, P.; Grzelak, L.; Prot, M.; Mougari, S.; Planchais, C.; et al. Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat. Commun. 2023, 14, 824. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: A Rapid Global Increase in COVID-19 is Due to the Emergence of the EG.5 (Eris) Subvariant of Omicron SARS-CoV-2. Med. Sci. Monit. 2023, 29, e942244. [Google Scholar] [CrossRef]
- Bazzani, L.; Imperia, E.; Scarpa, F.; Sanna, D.; Casu, M.; Borsetti, A.; Pascarella, S.; Petrosillo, N.; Cella, E.; Giovanetti, M.; et al. SARS-CoV CH.1.1 Variant: Genomic and Structural Insight. Infect. Dis. Rep. 2023, 15, 292–298. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Wrapp, D.; De Vlieger, D.; Corbett, K.S.; Torres, G.M.; Wang, N.; Van Breedam, W.; Roose, K.; van Schie, L.; Hoffmann, M.; Pöhlmann, S.; et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181, 1004–1015.e15. [Google Scholar] [CrossRef]
- Kuhn, M.; Firth-Clark, S.; Tosco, P.; Mey, A.S.J.S.; Mackey, M.; Michel, J. Assessment of binding affinity via alchemical free-energy calculations. J. Chem. Inf. Model. 2020, 60, 3120–3130. [Google Scholar] [CrossRef]
- Bauer, M.R.; Mackey, M.D. Electrostatic complementarity as a fast and effective tool to optimize binding and selectivity of protein–ligand complexes. J. Med. Chem. 2019, 62, 3036–3050. [Google Scholar] [CrossRef]
- Cheeseright, T.; Mackey, M.; Rose, S.; Vinter, A. Molecular field extrema as descriptors of biological activity: Definition and validation. J. Chem. Inf. Model. 2006, 46, 665–676. [Google Scholar] [CrossRef]
- Baroni, M.; Cruciani, G.; Sciabola, S.; Perruccio, F.; Mason, J.S. A Common Reference Framework for Analyzing/Comparing Proteins and Ligands. Fingerprints for Ligands and Proteins (FLAP): Theory and Application. J. Chem. Inf. Model. 2007, 47, 279–294. [Google Scholar] [CrossRef]
- Carosati, E.; Sciabola, S.; Cruciani, G. Hydrogen Bonding Interactions of Covalently Bonded Fluorine Atoms: From Crystallographic Data to a New Angular Function in the GRID Force Field. J. Med. Chem. 2004, 47, 5114–5125. [Google Scholar] [CrossRef] [PubMed]
- Goodford, P.J. A Computational Procedure for Determining Energetically Favorable Binding Sites on Biologically Important Macromolecules. J. Med. Chem. 1985, 28, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Bojadzic, D.; Alcazar, O.; Chen, J.; Chuang, S.T.; Condor Capcha, J.M.; Shehadeh, L.A.; Buchwald, P. Small-Molecule Inhibitors of the Coronavirus Spike: ACE2 Protein–Protein Interaction as Blockers of Viral Attachment and Entry for SARS-CoV-2. ACS Infect. Dis. 2021, 7, 1519–1534. [Google Scholar] [CrossRef]
- Case, D.A.; Babin, V.; Berryman, J.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E.; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. AMBER 14; University of California: San Francisco, CA, USA, 2014; pp. 1–826. [Google Scholar]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. THEOCHEM 1999, 461–462, 1–21. [Google Scholar] [CrossRef]
- Chung, L.W.; Sameera, W.M.C.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.; Liu, F.; et al. The ONIOM Method and Its Applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef] [PubMed]
- Dauber-Osguthorpe, P.; Roberts, V.A.; Osguthorpe, D.J.; Wolff, J.; Genest, M.; Hagler, A.T. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins 1988, 4, 31–47. [Google Scholar] [CrossRef]
- Forte, G.; Grassi, A.; Marletta, G. Molecular modeling of oligopeptide adsorption onto functionalized quartz surfaces. J. Phys. Chem. B 2007, 111, 11237–11243. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Xiong, Q.; Cao, L.; Ma, C.; Tortorici, M.A.; Liu, C.; Si, J.; Liu, P.; Gu, M.; Walls, A.C.; Wang, C.; et al. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature 2022, 612, 748–757. [Google Scholar] [CrossRef]
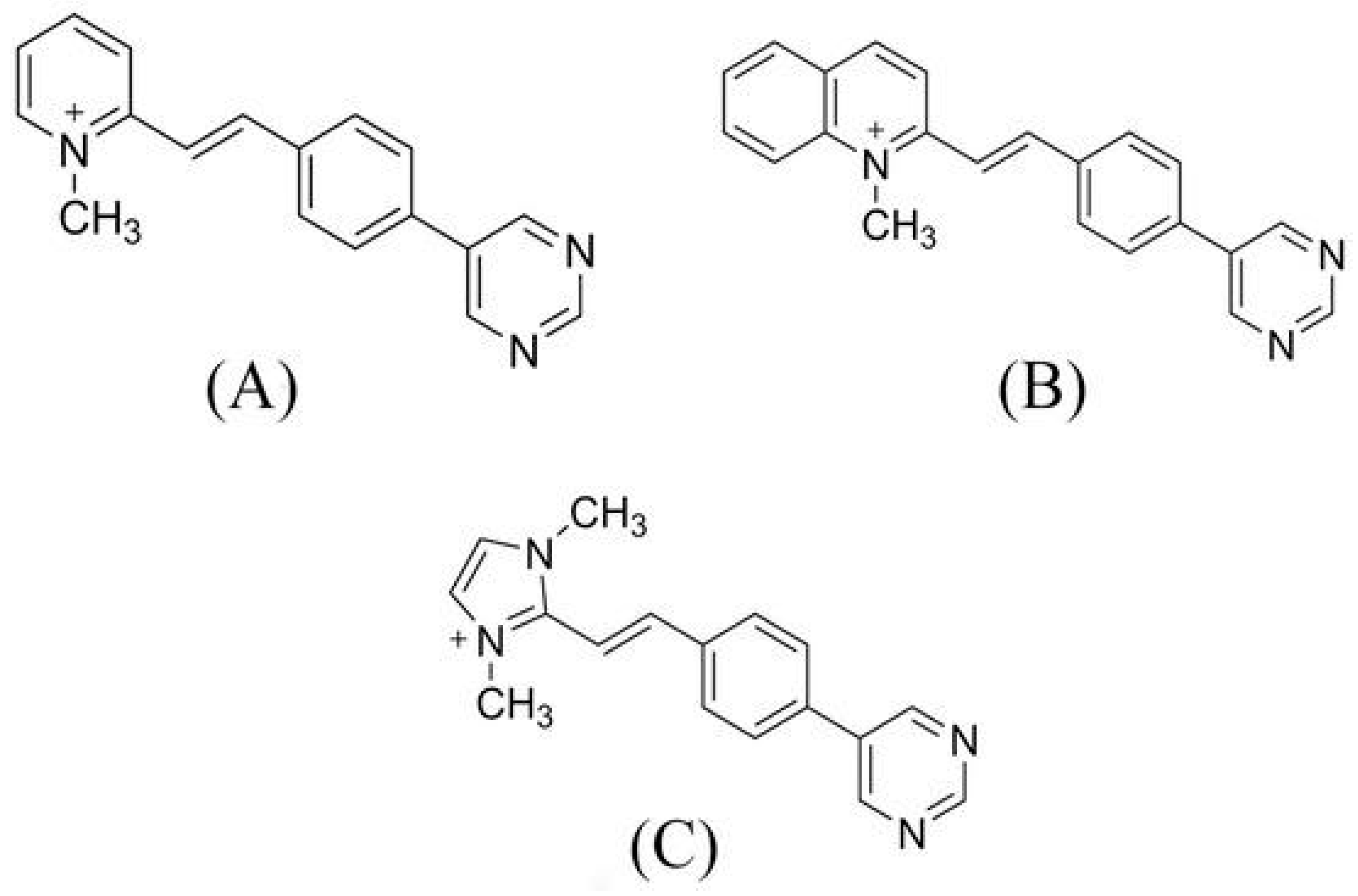


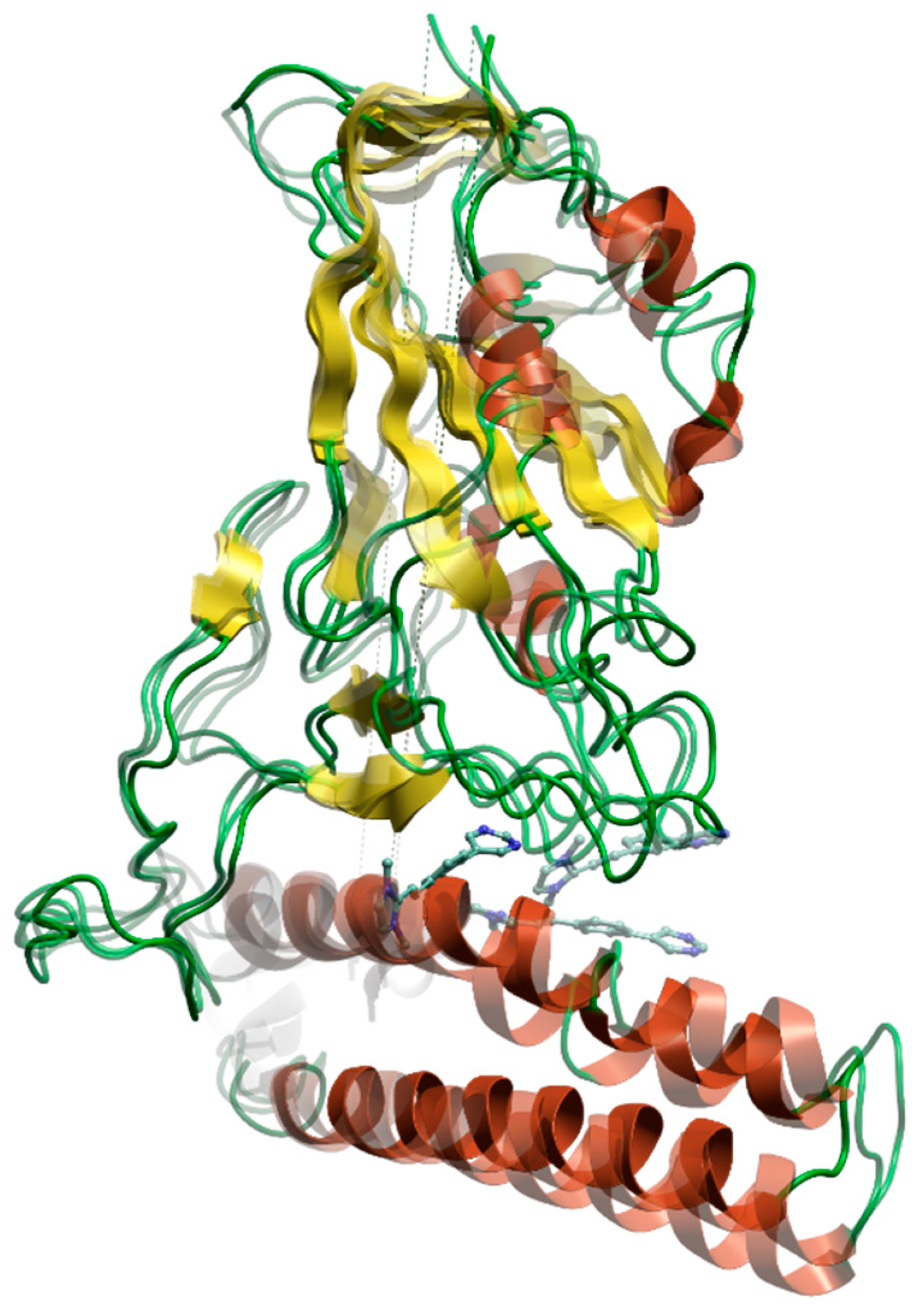
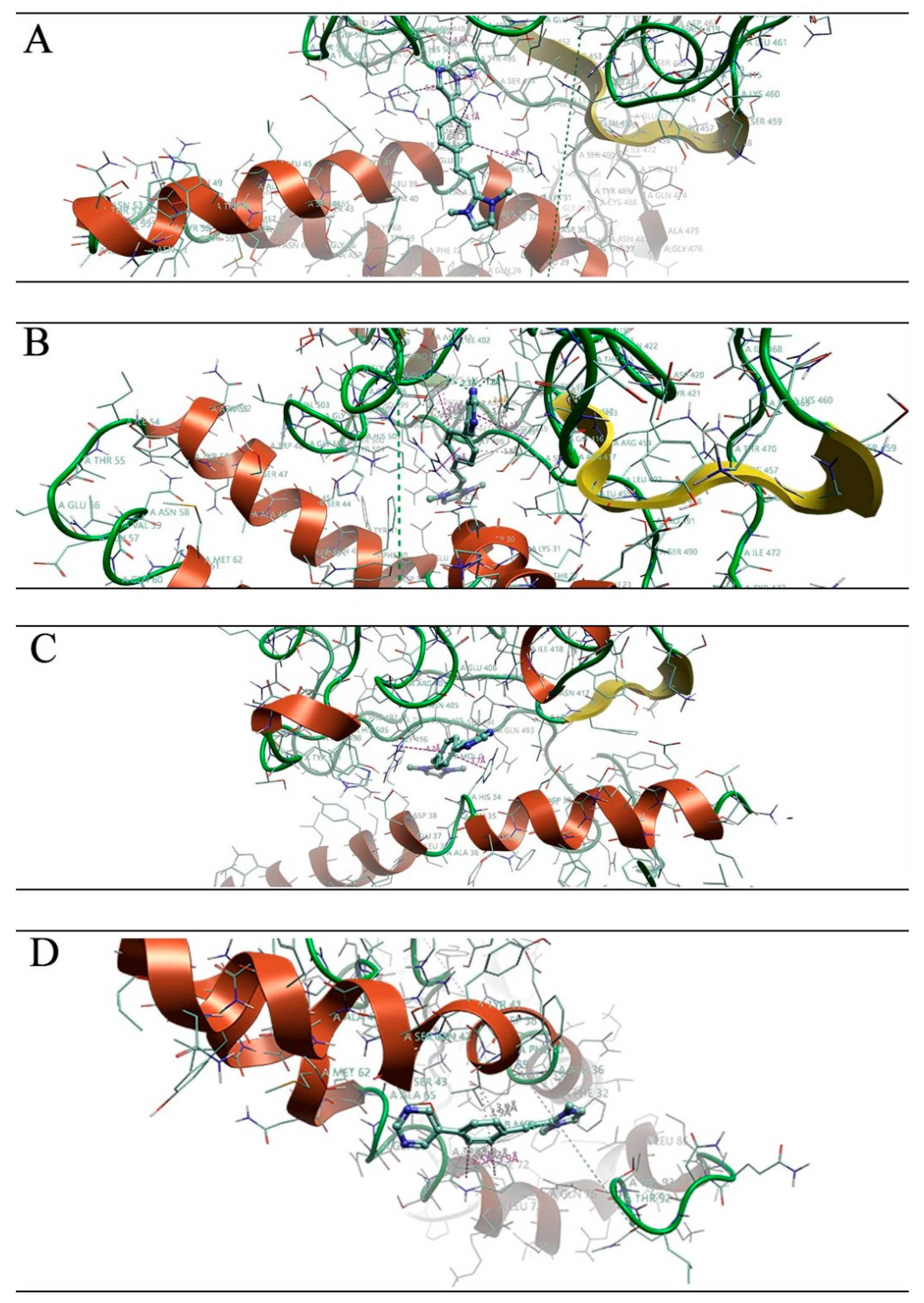
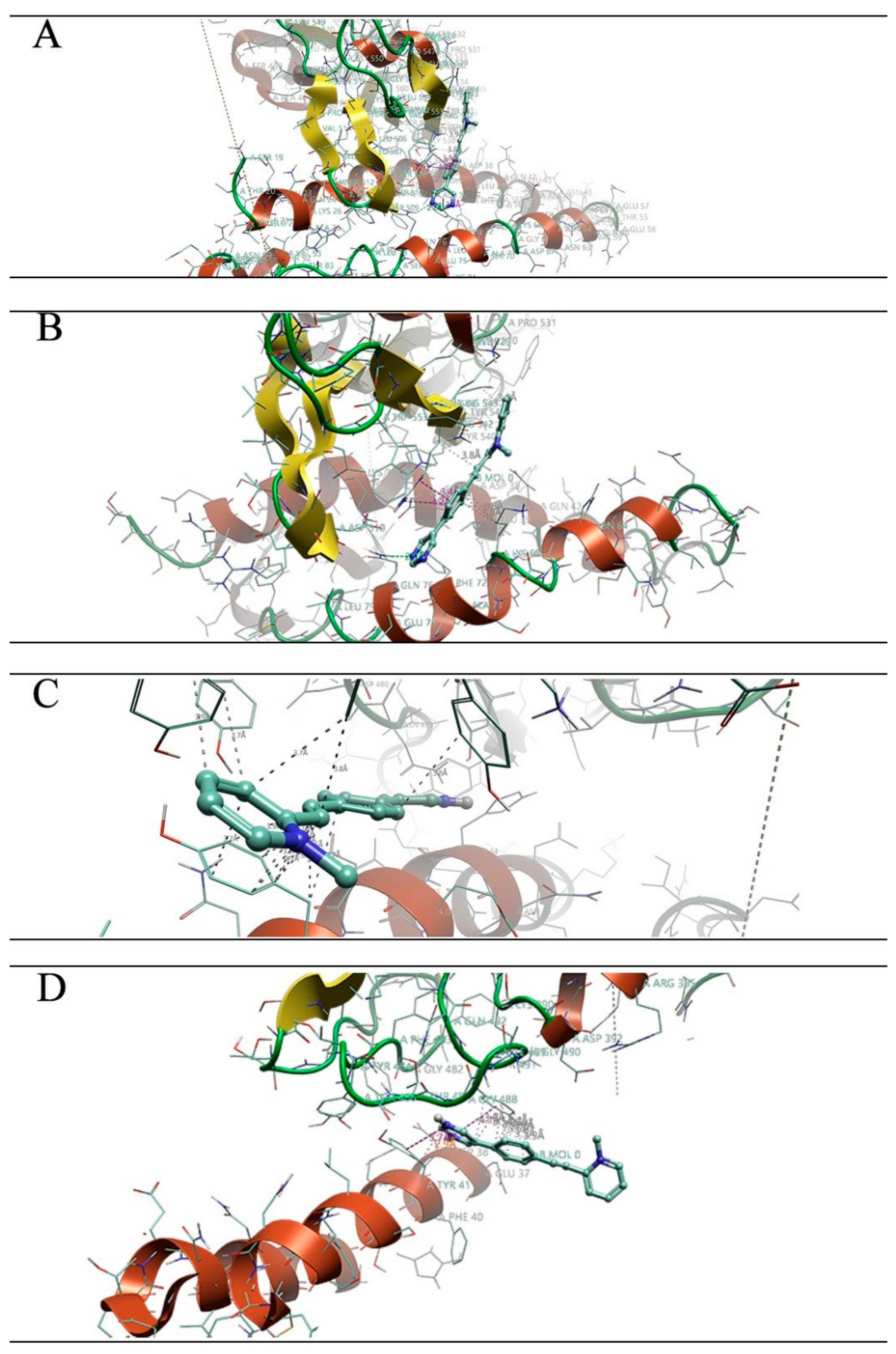
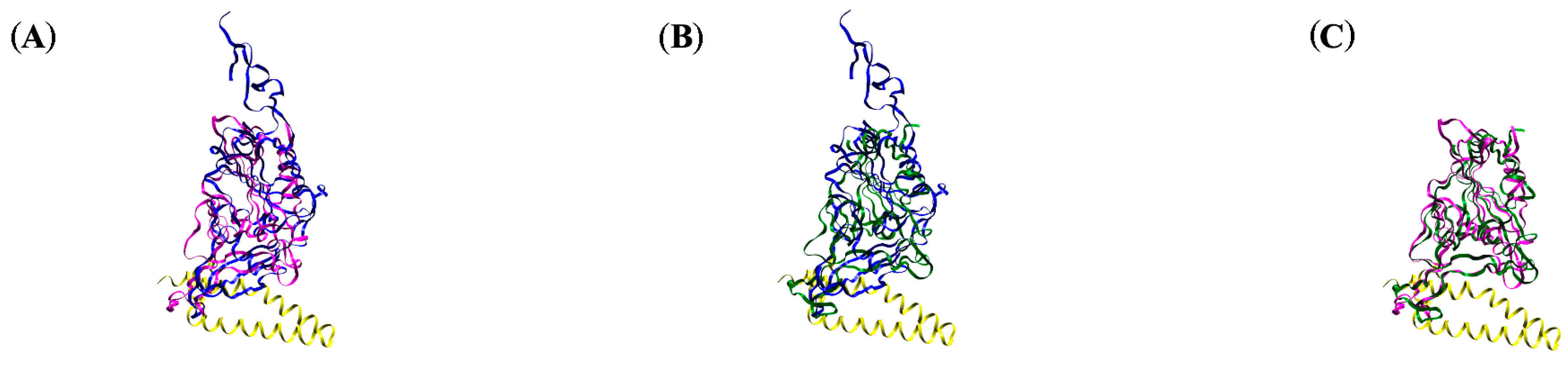

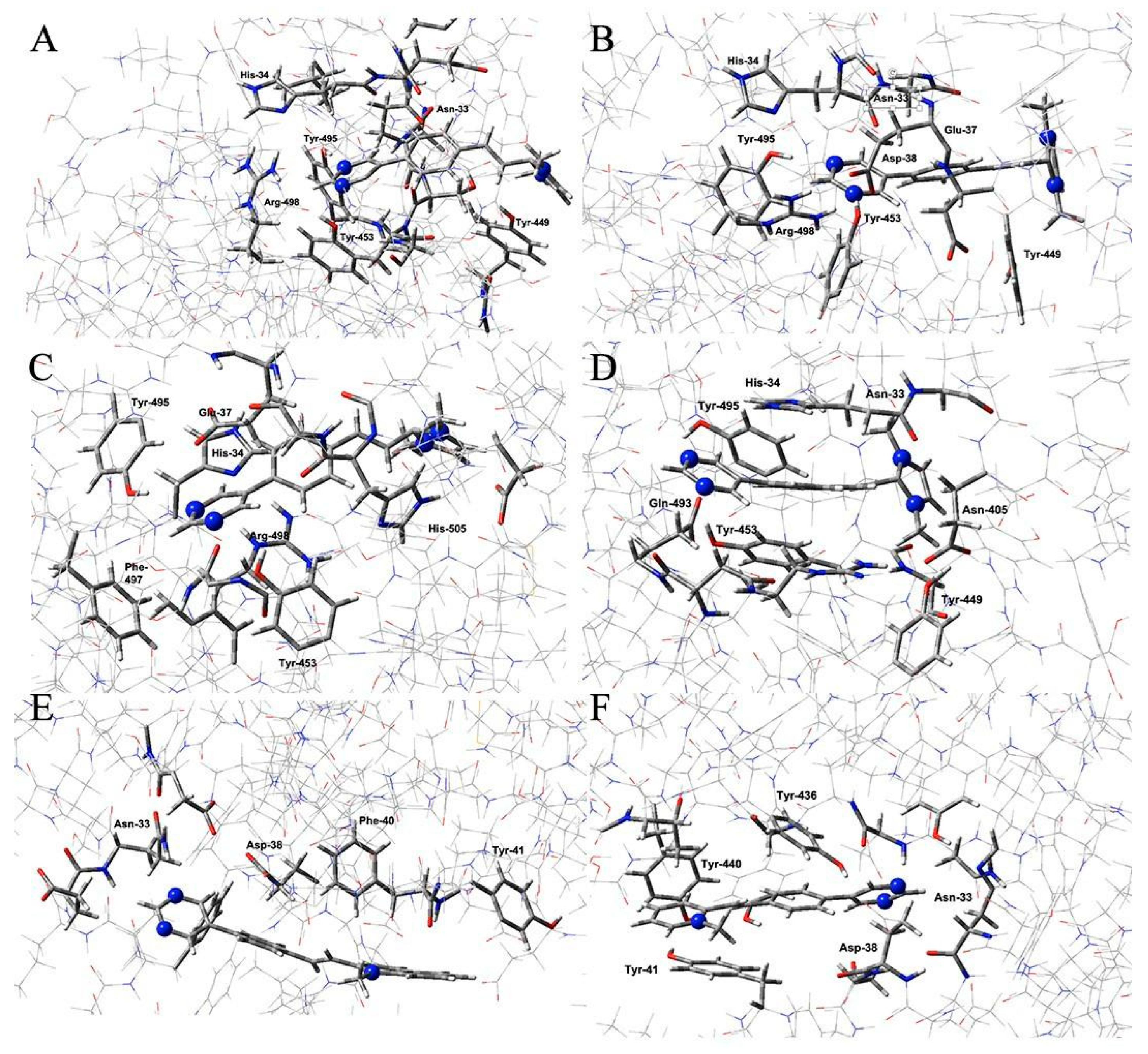
| Omicron XBB | Omicron EG | Omicron CH | Omicron BA |
|---|---|---|---|
| D405N | D405N | D405N | D405N |
| K417N | K417N | K417N | K417N |
| Q498R | Q498R | Q498R | Q498R |
| N501Y | N501Y | N501Y | N501Y |
| Y505H | Y505H | Y505H | Y505H |
| ////////// | ////////// | K444T | K444T |
| V445P | V445P | ////////// | ////////// |
| ////////// | ////////// | L452R | L452R |
| ////////// | F456L | ////////// | ////////// |
| F486P | F486P | ////////// | ////////// |
| ////////// | ////////// | F486S | ////////// |
| F490S | F490S | ////////// | ////////// |
| Variants | Amino Acids Residues | ||
|---|---|---|---|
| Omicron Variants | XBB | Pocket 2 | Asn33, His34, Glu37, Asp38, Arg403, Tyr453, Ser494, Tyr495, Gly496, Phe497, Arg498, His505 |
| BA | |||
| CH | |||
| EG | |||
| Other Virus Strains | SARS-CoV-1 | Pocket 2 | Asn33, His34, Glu37, Asp38, Tyr41, Lys390, Tyr436, Tyr440, Asn479, Asp480, Tyr481, Gly482, Phe483, Tyr484, Thr487, Tyr491 |
| MERS | RBD Pocket 1 | Asn33, His34, Glu35, Ala36, Glu37, Asp38, Leu39, Phe40, Tyr41, Gln42, Ser43, Leu45, Asn64, Ala65, Lys68, Trp69, Phe72, Lys496, Trp535, Glu536, Asp537, Gly538, Asp539, Tyr540, Tyr541, Ser537, Gly558, Ser559, Thr560 | |
| RBD Pocket 2 | Ser19, Thr20, Glu23, Gln24, Lys26, Thr27, Cys503, Ser504, Arg505, Leu506, Leu507, Ser508, Asp509, Asp510, Arg511, Thr512, Glu513, Val514, Pro515, Gln516, Pro525, Leu545, Ser546, Pro547, Leu548, Glu549, Gly550, Gly551, Gly552, Trp553, Leu554 | ||
| Target | Candidate | Glob-Sum | Distance | Glob-Prod | H | DRY | O | N1 |
|---|---|---|---|---|---|---|---|---|
| XBB (Pocket 2) | BCC-3 | 1.252 | 12.859 | 0.530 | 0.704 | 0.561 | 0.212 | 0.000 |
| BCC-1 | 1.183 | 13.113 | 0.481 | 0.676 | 0.513 | 0.244 | 0.000 | |
| BCC-2 | 1.165 | 13.668 | 0.482 | 0.500 | 0.437 | 0.232 | 0.000 | |
| DRI-C23041 | 1.060 | 12.374 | 0.384 | 0.482 | 0.492 | 0.443 | 0.207 | |
| BA (Pocket 2) | BCC-3 | 1.252 | 12.859 | 0.530 | 0.704 | 0.561 | 0.212 | 0.000 |
| DRI-C23041 | 1.221 | 12.279 | 0.417 | 0.482 | 0.492 | 0.443 | 0.211 | |
| BCC-1 | 1.186 | 13.000 | 0.509 | 0.513 | 0.513 | 0.244 | 0.000 | |
| BCC-2 | 1.173 | 13.422 | 0.506 | 0.437 | 0.437 | 0.232 | 0.000 | |
| CH (Pocket 2) | BCC-3 | 1.280 | 12.862 | 0.527 | 0.757 | 0.561 | 0.192 | 0.000 |
| BCC-1 | 1.185 | 13.152 | 0.500 | 0.676 | 0.513 | 0.196 | 0.000 | |
| DRI-C23041 | 1.096 | 12.385 | 0.403 | 0.482 | 0.478 | 0.432 | 0.211 | |
| BCC-2 | 1.025 | 13.729 | 0.443 | 0.602 | 0.342 | 0.266 | 0.000 | |
| EG (Pocket 2) | BCC-3 | 1.323 | 12.695 | 0.555 | 0.778 | 0.561 | 0.212 | 0.000 |
| BCC-1 | 1.186 | 13.000 | 0.509 | 0.710 | 0.513 | 0.244 | 0.000 | |
| BCC-2 | 1.173 | 13.422 | 0.506 | 0.597 | 0.437 | 0.233 | 0.000 | |
| DRI-C23041 | 1.148 | 12.427 | 0.389 | 0.482 | 0.492 | 0.440 | 0.174 |
| Target | Candidate | Glob-Sum | Distance | Glob-Prod | H | DRY | O | N1 |
|---|---|---|---|---|---|---|---|---|
| SARS-CoV-1 (Pocket 2) | BCC-1 | 2.063 | 10.252 | 0.713 | 0.947 | 1.201 | 0.272 | 0.000 |
| BCC-3 | 1.914 | 10.454 | 0.724 | 0.942 | 0.938 | 0.314 | 0.000 | |
| BCC-2 | 1.980 | 10.499 | 0.703 | 0.877 | 1.233 | 0.263 | 0.000 | |
| DRI-C23041 | 1.755 | 9.904 | 0.487 | 0.935 | 1.161 | 0.410 | 0.220 | |
| MERS (RBD Pocket) | BCC-2 | 2.376 | 10.268 | 0.646 | 0.983 | 1.596 | 0.116 | 0.000 |
| BCC-1 | 2.266 | 10.191 | 0.669 | 0.964 | 1.465 | 0.170 | 0.000 | |
| BCC-3 | 2.203 | 10.730 | 0.595 | 0.968 | 1.286 | 0.172 | 0.000 | |
| DRI-C23041 | 2.138 | 8.805 | 0.514 | 0.855 | 1.487 | 0.240 | 0.222 | |
| MERS (RBD Pocket 2) | BCC-2 | 1.824 | 11.268 | 0.678 | 0.960 | 0.802 | 0.185 | 0.000 |
| DRI-C23041 | 1.756 | 9.328 | 0.546 | 0.931 | 0.831 | 0.297 | 0.235 | |
| BCC-3 | 1.654 | 11.705 | 0.654 | 0.980 | 0.676 | 0.162 | 0.000 | |
| BCC-1 | 1.597 | 11.570 | 0.627 | 0.988 | 0.736 | 0.190 | 0.000 |
| Variant | Ligand | ΔG (kcal/mol) | |
|---|---|---|---|
| BA.2.75 | 2 | BCC-3 | −22.47 |
| 3 | BCC-1 | −19.79 | |
| CH.1.1 | 2 | BCC-3 | −19.46 |
| 3 | BCC-1 | −17.28 | |
| EG.5 | 2 | BCC-3 | −24.96 |
| 3 | BCC-1 | −22.65 | |
| XBB.1.16 | 2 | BCC-3 | −19.36 |
| 3 | BCC-1 | −17.07 | |
| SARS-CoV-1 | 2 | BCC-1 | −19.91 |
| 3 | BCC-3 | −10.01 | |
| 4 | BCC-2 | −14.63 | |
| MERS | 1 | BCC-2 | −22.31 |
| 2 | BCC-2 | −13.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavallaro, G.; Forte, G.; Bonaccorso, C.; Nicolosi, M.; Sipala, F.; Varrica, G.; Fortuna, C.G.; Ronsisvalle, S. From SARS to MERS and SARS-CoV-2: Comparative Spike Protein Remodeling and Ligand-Binding Hot-Spots Revealed by Multiscale Simulations. Chemistry 2025, 7, 132. https://doi.org/10.3390/chemistry7040132
Cavallaro G, Forte G, Bonaccorso C, Nicolosi M, Sipala F, Varrica G, Fortuna CG, Ronsisvalle S. From SARS to MERS and SARS-CoV-2: Comparative Spike Protein Remodeling and Ligand-Binding Hot-Spots Revealed by Multiscale Simulations. Chemistry. 2025; 7(4):132. https://doi.org/10.3390/chemistry7040132
Chicago/Turabian StyleCavallaro, Gianfranco, Giuseppe Forte, Carmela Bonaccorso, Milena Nicolosi, Federica Sipala, Giulia Varrica, Cosimo Gianluca Fortuna, and Simone Ronsisvalle. 2025. "From SARS to MERS and SARS-CoV-2: Comparative Spike Protein Remodeling and Ligand-Binding Hot-Spots Revealed by Multiscale Simulations" Chemistry 7, no. 4: 132. https://doi.org/10.3390/chemistry7040132
APA StyleCavallaro, G., Forte, G., Bonaccorso, C., Nicolosi, M., Sipala, F., Varrica, G., Fortuna, C. G., & Ronsisvalle, S. (2025). From SARS to MERS and SARS-CoV-2: Comparative Spike Protein Remodeling and Ligand-Binding Hot-Spots Revealed by Multiscale Simulations. Chemistry, 7(4), 132. https://doi.org/10.3390/chemistry7040132






