Abstract
Here, we propose a drug delivery system for ciprofloxacin (CF) based on cyclodextrin (CD) polymer. We obtained a 3D matrix system with encapsulated drug molecules by crosslinking CF+CD non-covalent complexes with 1.6-hexamethylene isocyanate. The obtained polycarbamide (MAX-system) represents particles (~225 nm in diameter) that demonstrate CF’s sustained release. We investigated how the carrier affects the drug’s interaction with the biological macromolecule human serum albumin (HSA) and CF’s antibacterial properties. Compared to a binary CF–HSA system, CD decreases CF’s binding efficiency to HSA by two times, whereas CF encapsulation in a polymer matrix doubles the Ka value and prevents protein aggregation. The changes in HSA’s secondary structure indicate no alterations in the main mechanism of complex formation between CF and HSA in the presence of both CD-based carriers. CD as well as MAX systems practically do not change CF’s activity against E. coli and B. subtilis, but for MAX systems, prolonged action is realized due to CF’s sustained release. We believe that our findings are important for the further development of new, efficient drug forms.
1. Introduction
According to the World Health Organization, bacterial infections remain one of the leading causes of death in the world [1,2]. The infections are especially threatening among people with weakened immune systems; likewise, secondary bacterial infections might follow surgery or serious illnesses. For example, the COVID-19 pandemic demonstrated the occurrence of severe bacterial pneumonia that required complex and lengthy treatment regimens at high doses of antibiotics and challenged scientists to design new antibacterial drugs [3]. Currently, the development of novel drugs and their clinical trials take a long time and require expensive research. Therefore, the most urgent task of pharmacology is the design of new efficient drug delivery systems to improve the properties of bioactive molecules. Such systems can contribute to increased therapy effectiveness, a controlled release rate, targeted delivery and a reduction in the probability of side effects [1,2,4].
Fluoroquinolone ciprofloxacin (CF) demonstrates a wide spectrum of activity in vitro against Gram-negative and Gram-positive, aerobic and anaerobic bacteria, and even mycobacteria, as well as chemical and biological stability [5]. However, long-term CF therapy and high drug dosages might lead to a number of side effects. Most of the undesirable effects occur in the digestive system: nausea, abdominal discomfort, vomiting and diarrhea. In addition, CF has a negative effect on the central nervous system; patients report dizziness, headaches, nervousness and insomnia. Skin irritations have also been reported; a rash or itching may appear, probably of allergic origin [5,6,7]. Moreover, often patients have weakness, fever and heart palpitations. The CF’s dosage decrease helps reduce the probability of side effects [8,9].
An increase in solubility and, as a result, bioavailability of CF can be achieved using drug delivery systems. The promising approach is to obtain non-covalent CF complexes with oligosaccharides and cyclodextrins. We propose the use of cyclodextrins (CDs) and their derivatives, as they were approved by the FDA and proved to be effective drug carriers [10]. CDs find a wide spectrum of practical applications in various fields; for example, CDs are used in the food and textile industries. CDs are valuable in pharmaceutics as they increase drugs’ bioavailability, solubility and stability and reduce toxicity due to the formation of guest-host complexes [11]. CF complexes with different CD derivatives are well described in the literature [12]. CF’s aromatic fragment penetrates CD’s hydrophobic cavity, and the complex is stabilized by the additional interactions between CD’s substitutes and other CF functional groups.
Recently, particular attention has been drawn to CD polymers synthesized in the presence of another organic molecule (such as a medicinal substance)—a template. The polymerization process leads to the encapsulation of the template into the carrier’s matrix and might also result in a specific network formation with unique properties [13,14,15]. The obtained polymer is characterized by uniform distribution of the drug in the carrier through the particle volume as well as high loading efficiency [16]. For example, dihydroxyphenylalanine (L-DOPA) was incorporated with 90% efficiency into a β-CD polymer using N,N’-carbonyldiimidazole as a linker; the obtained sample demonstrated prolonged release [17].
Here, we design CF’s novel drug delivery: CD polymer based on methyl-β-cyclodextrin (MCD) crosslinked by isocyanate in the presence of CF (a template)—3D polymer matrix with encapsulated CF (MAX). We apply spectroscopic methods to uncover the details of MAX’s physical-chemical properties: the size, structure, release profile and CF’s encapsulation efficacy.
Moreover, we uncover how the drug delivery system might change CF’s biological properties, including its interactions with human serum albumin (HSA) and its antibacterial activity. HSA is the main plasma protein that transports drug molecules in the blood stream. Clearly, the drug’s binding to HSA is a crucial parameter since it significantly affects the drug’s biodistribution, pharmacokinetics, and, therefore, therapeutic efficacy. We investigate the changes in the mechanism and efficacy of CF’s binding to HSA in the drug delivery system. Another important drug characteristic is its antibacterial activity. We are interested to know if the encapsulation of CF in MAX will change the in vitro activity. For this purpose, we conducted experiments on Gram-positive and Gram-negative bacterial strains. We believe that our results will contribute to the development of new, effective drug delivery systems.
2. Materials and Methods
2.1. Materials
Methyl-β-cyclodextrin (MCD; 1.5–2.1-CH3 group per one anhydroglucose), ciprofloxacin (CF), human serum albumin (HSA), 1,6–hexamethylene diisocyanate (HMI) and dimethyl sulfoxide (DMSO) from Sigma-Aldrich (St. Louis, MO, USA). Hydrochloric acid is from Reachim (Moscow, Russia). The tablets for the preparation of phosphate buffered saline (PBS) pH 7.4 were purchased from Pan-Eco (Moscow, Russia). Escherichia coli ATCC 25922 and Bacillus subtilis ATCC 6633 are from the VKPM Kurchatov Institute (Moscow, Russia). These strains are standard for testing the antibacterial properties of substances and susceptibility to antibiotics (All-Russian Collection of Microorganisms, http://www.vkm.ru/, accessed on 7 June 2023).
2.2. Preparation of Ciprofloxacin Complexes with Methyl-β-Cyclodextrin
The MCD powder was dissolved in an aqueous solution (0.03 M, HCl pH 4.0), and then CF solution (0.03 M, pH 4.0 HCl) was added to achieve a 1:1 molar ratio. The resulting mixture was incubated at 37 °C with gentle shaking (100 rpm) for 1 h.
2.3. Synthesis of MAX via Crosslinking CF+MCD Complexes
We chose the volumes of the compounds to achieve the final V = 3 mL (H2O:DMSO = 1:1 vol/vol). A total of 1500 μL of 7.5 mM CF+MCDs complex (1:1 mol:mol) solution (pH 4.0 HCl) were kept at 37 °C for 20–30 min. Then, 1300 μL of dimethyl sulfoxide (DMSO) were added with active stirring. After 2 min, 200 μL of hexamethylene diisocyanate (HDI) solution in DMSO (0.1125 M) were added dropwise, so that the molar ratio of HMI:(CF+MCD) was 1:1. The mixture was incubated for 3 h before being placed in a refrigerator at 4 °C for 12 h. The sample purification was performed by 6 h dialysis (the membrane was MWCO 3 kDa Serva (Heidelberg, Germany)). The resulting solution was freeze-dried at −70 °C for 24 h and then lyophilized at −60 °C for 2 days (Edwards 5 BOC Edwards (West Sussex, UK)).
The reaction yield (measured by MCD) refers to the percentage of MCD tori in the MAX particles compared to the amount used in the synthesis. MCD’s content was determined by FTIR spectroscopy (the intensity of the 1047 cm−1 absorption band corresponded to the oscillation of the C-O-C bond of the CD). The CF’s encapsulation efficiency into the MCD polymer matrix was determined by UV spectroscopy.
2.4. Preparation of Binary (CF–HSA) and Ternary (HSA−(CF+CD Carrier)) Systems
A certain amount (from 10 μL to 800 μL) of 1.5 mM CF (binary system) or 1.5 mM CF+MCD/MAX (ternary system) solution was transferred to the 0.01 M sodium-phosphate buffer (pH 7.4) and 200 μL 0.3 mM. Then HSA solution (pH 7.4) was added. The volume was adjusted to 1 mL. The HSA concentration was maintained at 0.06 mM, while the molar excess of CF (individual, in the complex with MCD, or in MAX) ranged from 0.25 to 20. The protein-drug samples were incubated at 37 °C with gentle shaking (100 rpm). For UV spectroscopy, circular dichroism spectroscopy, and fluorescence spectroscopy analysis, the solutions of HAS (CHSA = 0.02 mM) were used to ensure appropriate signal intensity.
2.5. FTIR Spectroscopy
Tensor 27 spectrometer Bruker (Ettlingen, Germany) with an attenuated total reflection cell and ZnSe single-flection crystal was used to obtain FTIR spectra of the sample’s solutions. The spectrometer is equipped with an MCT detector cooled with liquid N2, a thermo-stat Huber (Offenburg, Germany). FTIR spectra were recorded in the range of 2500–900 cm−1 with a 1 cm−1 resolution 3 times (70 scans each) at 22 °C). Dry nitrogen was used to purge the system with an air compressor, Brezza NiGen HF-1 CLAIND (Tremezzina, Italy). The spectra were analyzed by Opus 7.0.
By analyzing the FTIR spectra in the range of 1700–1600 cm−1 for samples with albumin, the content of the secondary structure was determined. The primary bands were detected by the second derivative of the spectrum, and each spectrum was deconvoluted using the Levenberg–Marquardt algorithm.
2.6. Circular Dichroism
The J-815 spectrometer from Jasco (Tokyo, Japan) was applied to record circular dichroism spectra. The measurements were performed at 25 °C using a quartz cuvette (l = 1 mm) within a wavelength range of 200–260 nm. Spectra were scanned five times at 1 nm steps with an HSA concentration of 0.02 mM. To determine the secondary structure content, the spectra were analyzed with the CDNN program Version 2. The experiment was conducted 3 times, and the results were performed with SD = 3 (n = 3).
2.7. UV Spectroscopy
The Ultrospec 2100 Pro equipment from Amersham Biosciences (San Francisco, CA, USA) was used to record UV spectra. The CF/CF-MCD/MAX absorption spectra were measured in a quartz cell with an optical path length of 1 cm from Hellma Analytics (Jena, Germany) within a wavelength range of 200–450 nm.
2.8. The Determination of MAX’s Size
The determination of MAX’s size was performed by dynamic light scattering (DLS). Zetasizer Nano S (Malvern, England), equipped with a mW He–Ne laser (633 nm), was applied to determine MAX’s hydrodynamic diameter at 25 °C. The sample was tested 3 times, and the data is presented with standard deviations (SD). MAX-particle size was also justified using the AFM microscope NTEGRA II.
2.9. Fluorescence Spectroscopy
The Varian Cary Eclipse spectrophotometer from Agilent Technologies (Santa Clara, CA, USA) was applied to conduct fluorescence measurements. The HSA’s emission spectra were recorded at 37 ± 0.1 °C in a 10 mm quartz cuvette, with an excitation wavelength of 280 nm and a 1 nm step in the range of 290–550 nm. The protein concentration in all samples was maintained at 0.02 mM (phosphate buffer solution with a pH of 7.4).
The Stern–Volmer Equation (1) was employed to analyze the quenching of protein fluorescence by small molecules, considering both statistical and dynamic quenching effects [18].
the equation includes 0 and , which represent the fluorescence intensities in the absence and presence of a drug form (quencher), , which is a Stern–Volmer constant, [Q]—the molar concentration of the quencher, and —the bimolecular constants of quenching rate and the lifetime of HSA’s fluorescence in the absence of the quencher, respectively. The binding constant of HSA to the drug form () and the number of binding sites (n) were determined using Equation (2) at 37 ± 0.1 °C:
2.10. Fluorescence Anisotropy
For studying steady-state fluorescence anisotropy (), another series of solutions was prepared. To the solution of 50 μM CF/CF+MCD (PBS, pH 7.4), the required volume of 5 mM HSA’s (PBS, pH 7.4) solution was added, and the volume was adjusted to 1 mL. The concentration of CF/CF+MCD was kept constant (0.01 mM), while the molar excess of HSA ranged from 0 to 20 (from 0 to 0.2 mM). The complexes were then incubated at 37 °C with gentle shaking (100 rpm).
The Varian Cary Eclipse spectrophotometer (Agilent Technologies, United States) equipped with a polarizer was used for spectra registration. The emission spectra were recorded in four positions of polarizers: HH, VH, HV and VV, where the first letter means horizontal (H) or vertical (V) orientation of the excitation polarizer and the second—horizontal (H) or vertical (V) orientation of the emission polarizer. The recording was conducted at a temperature of 37 ± 0.1 °C in a 10 mm thick cuvette, with an excitation wavelength of 340 nm and a step of 1 nm in the range of 340–550 nm. To calculate the anisotropy () of the fluorophore, Equation (3) was used:
where —intensity of fluorescence emission in different positions, and represents the correction factor for the instrument’s detector sensitivity [19,20,21].
The binding constants (Ka) of CF (or CF+MCD) with HSA were obtained by (4) considering the equilibrium AM + nHSA↔AM×nHSA [18,22]:
We varied the concentration of HSA (from 0 to 0.2 mM) and analyzed the changes in the anisotropy by Hill’s linearization in the n-binding site model (5) [23]:
where θ is a fraction of the bound CF’s molecules calculated as (6):
where ξ0 and ξ are the peak intensities of fluorescence spectra of CF in the absence and presence of HSA, ξ∞ is the intensity of the horizontal asymptote.
2.11. Fluorescence Resonance Energy Transfer (FRET)
In order to determine the average distance (r) between the ligand (CF in drug form) and HSA Trp-214 (absorption at 280 nm) [21,24,25]) Equations (7)–(9) were applied. Formula (7):
was employed to calculate energy transfer efficiency (E). In Equation (7), R0 is the critical radius for 50% energy transfer efficiency that was determined by Equation (8):
where is a factor describing the mutual orientation in the space of the dipole moments of the transitions of the donor and acceptor (2/3 for random orientation as in fluid solution); is the refractive index of the medium (1.33 in the present case); is the quantum yield of the donor in the absence of the acceptor (0.118 for Trp in HSA); and is the overlap integral of the donor fluorescence emission and the acceptor absorption spectra [26]. was calculated according to Equation (9):
where is the fluorescence intensity of the donor at wavelength λ and is the molar coefficient of light absorption of the donor at wavelength λ.
2.12. Release Kinetic Studies
The CF’s release was studied by the dialysis: 1 mL of the sample was placed in the dialysis bag (MWCO of 3.5 kDa, Orange Scientific, Belgium). MWCO was chosen so that only CF could penetrate through the membrane (MCF = 331 Da). We used a small bag and a beaker with a wide bottom, so the sample was fully immersed in the solution. The beaker was covered with foil to avoid evaporation. The system was incubated at 37 °C at 150 rpm, and UV spectra of the external solution were recorded. One hundred percent of released CF is assumed when CCF in the dialysis bag equals CCF in the external solution (50% of the initial CF’s concentration).
2.13. In Vitro Studies
The antimicrobial properties of CF, CF+MCD and MAX were studied by agar-well diffusion tests using Gram-negative Escherichia coli ATCC 25922 and Gram-positive Bacillus subtilis ATCC 6633 strains. The overnight culture (Luria Bertuni medium, pH 7.4) was diluted to match the 0.5 McFarland standard [27,28]. After the distribution of bacteria on Petri dishes, 4 wells were cut using 1 mL of sterile plastic [15,29,30,31]. A total of 50 μL of each sample was put into wells: sterile buffer as a negative control, CF, CF+MCD, MAX, HSA−CF, HSA−(MCD+CF) or HSA−MAX (CCF = 0.2, 0.3 and 0.5 μg/mL for E. coli ATCC 25922 and CCF = 0.4, 0.6 and 0.8 μg/mL for B. subtilis ATCC 6633). The Petri dishes were incubated at 37 °C for 22–24 h. We measured the diameters of the agar areas that demonstrated the absence of bacterial growth. The minimum inhibition concentration (MIC) was estimated as the sample’s concentration at which the inhibition zone area equals the incised agar area:
where c is CF’s concentration (μg/mL), Sinh is the inhibition zone area (mm2) [32,33]. Each sample was tested 3 times, and the data are presented with standard deviations.
3. Results and Discussion
CD oligomers and polymers are of particular interest since they can form non-covalent complexes with guest molecules not only in the pores of the branched polymer but also in the cavities of the CDs [34]. The use of such carriers for the drug’s encapsulation seems to be advantageous compared to common drug-CD complexes. For example, the formation of these systems might lead to prolonged release [35], affect the lipid membrane permeability, and increase the drug’s antibacterial activity [28,30]. Here, we design the CF’s drug delivery system based on the cross-linking of CF’s complexes with methyl-β-cyclodextrin (MCD) to obtain a 3D polymer matrix with encapsulated drug molecules—MAX. Thus, we consider three drug forms: CF, CF+MCD (common non-covalent complex) and MAX (CF imprinted into a polymer matrix) to estimate how different kinds of CD carriers affect CF’s properties.
First, we obtained the MAX system with encapsulated CF molecules by crosslinking CF-MCD complexes with 1.6-hexamethelene diisocyanate (Figure 1). Our previous research [16] demonstrated that MCD+CF is formed by the penetration of CF’s –COOH group and aromatic fragment into the CD cavity. CF’s heterocycle is located outside the CD’s cavity and might react with the linker’s isocyanate group. Nevertheless, we suppose MCD hides CF’s heterocycle, while MCD’s OH groups are oriented in the water–organic medium and are more available for the reaction. Indeed, no CF covalent conjugates were detected in the final sample. This statement will be confirmed further.
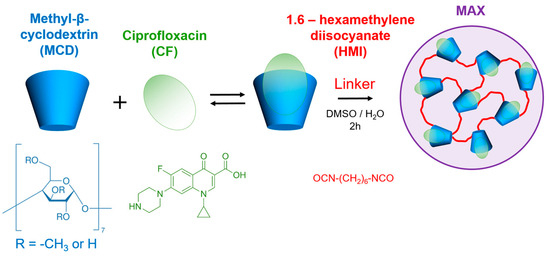
Figure 1.
CF’s and MCD’s chemical structures and the schematic representation of MAX synthesis.
According to dynamic light scattering, MAX forms particles in aqueous media (phosphate buffer, pH 7.4) with an average hydrodynamic diameter of 225 ± 17 nm. The MAX’s structure was investigated by FTIR. In the MAX spectrum, we found the band 1047 cm−1 attributed to stretching vibrations of the C–O–C band assigned to MCD. Additionally, the band’s shoulder appears to indicate crosslinking with MAX formation (Figure S1). These spectral changes are explained by the changes in the microenvironment of the C–O–C bands between glucopyranose residues in β-CD derivatives upon crosslinking. Additionally, we observed the appearance of new low intensity bands: 951 cm−1, 1540–1570 cm−1 and 1610–1680 cm−1 corresponding to C–N, amide I and amide II of disubstituted carbamide, respectively. The CN groups might appear due to the hydrolysis of isocyanate groups. The insignificant intensity of these bands might be explained by the low degree of crosslinking.
Comparing the FTIR spectra of MCD and MAX, we also observed bands in the range 1150–1400 cm−1 in MAX’s spectrum, which indicates the presence of CF. The low intensities might be associated with the complex formation (CF’s incorporation in MCD), which hinders the band of CF. On the other hand, FTIR requires high concentrations to maintain quantitative analysis (0.002–0.02 M for CF), so we believe that the absence of noticeable CF bands indicates the drug’s low content in MAX.
The yield of the reaction by MCD (the proportion of MCD tori present in the MAX particles compared to the amount incorporated during synthesis) was 40%. The CF’s encapsulation efficiency into the MCD polymer matrix, determined by UV spectroscopy, was ~1 μg per 1 mg of MAX. In our previous research [16], we studied a similar system based on another fluoroquinolone, moxifloxacin and sulfobutyl ether-β-cyclodextrin that demonstrated ~0.4 mg per 1 mg. Such a difference might be explained by the low binding constant of CF+MCD (Ka = (1.3 ± 0.3) × 103 M−1 [12]) and/or a higher degree of MCD’s substitution (1.5–2.1 per one glucose residue) compared with sulfobutyl ether-β-cyclodextrin (1.0–1.5 per one glucose residue), so fewer -OH groups are available for reaction with HMI. Nevertheless, the CF’s carrier might significantly affect its interactions with biological macromolecules as it hides the drug.
3.1. Fluorescence Spectroscopy
HSA is the major protein in human blood plasma. Albumin binds and transports biologically active molecules, so the drug-HSA complex formation might significantly affect the bioactivity and bioavailability of small drugs [36]. With the rise of attention to CD polymers, we are interested in how CD carriers would change the CF’s binding with HSA. Fluorescence is a valuable approach for studying the interaction between small molecules and proteins. When tryptophan (Trp) and/or tyrosine (Tyr) are present in the protein, their aromatic fragments can emit intrinsic fluorescence upon absorption of UV light, which changes the protein’s structure and microenvironment of these residues. The changes in the protein’s fluorescence via interaction with other molecules uncover the binding mechanism, mode, constants, intermolecular distances, etc. Commonly, molecular interactions cause fluorescence quenching, including reactions in the excited state, energy transfer, and static and dynamic quenching. Intrinsic fluorescence quenching measurement of proteins is widely used to elucidate the mechanism of their interaction with ligands or drug molecules [37,38,39].
The interaction between CF and HSA is well studied [26,37,40,41] in the literature, while ternary systems such as HSA–(CF+MCD) or HSA–MAX are not reported. At λex = 280 nm, the individual CF and HSA demonstrate emission peaks at 415 and 340 nm, respectively. The spectra of both components are resolved, so it is possible to investigate the state of each component in binary and ternary systems simultaneously (Figure 2a). The complex formation of CF with MCD as well as CF’s encapsulation in MAX do not obviously influence the position of the drug’s emission maximum.
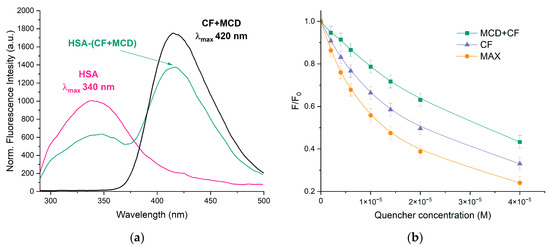
Figure 2.
(a) Normalized emission spectra of HSA (pink), HSA–(CF+MCD) (green), CF+MCD (black), λex = 280 nm, CHSA = 0.02 mM, CCF+MCD = 0.02 mM, 37 °C, PBS, pH 7.4; (b) The intensity of HSA’s peak via the increase of the drug’s concentration, λex = 280 nm, CHSA = 0.02 mM, pH 7.4, 37 °C.
With the increase in CF’s concentration, the HSA’s fluorescence intensity decreased significantly (Figure 2b), and the band shifted at 7 nm towards the higher wavelengths (Figure 2a). The effect suggests that the Trp-214 microenvironment changes to become more hydrophilic via CF’s binding as hydrophilization of the fluorophore environment leads to its red shift [24,42,43]. The data is confirmed by [44,45]. In our previous research, we showed that free MCD does not change the intensity or maximum position of the HSA’s emission spectrum [31,46]. For HSA−(CF+MCD), we observed less pronounced quenching and a more significant red shift (11 nm) compared to HSA−CF. We suggest that more hydrophilic CF+MCD complexes (than free CF) have difficulty binding to the hydrophobic pocket of HSA in subdomain IIA, where CF binds [46,47,48], or that MCD and HSA compete for binding with CF.
Let us consider how MAX affects CF binding with HSA. We expected that encapsulated CF would be less available in MAX for interactions with HSA. However, MAX demonstrates a more significant quenching of HSA’s emissions than free CF. That fact could indicate the different mechanisms of interaction between HSA–CF and HSA–MAX systems. We suppose the interaction between the MAX and HSA is the key parameter of the observed effect. We suppose the protein corona is formed (the interaction between HSA and MAX’s polymer matrix) that brings CF molecules closer to albumin and, consequently, causes the pronounced quenching of HSA’s fluorescence. As MAX may interact only with the surface of HSA, no significant changes in Trp’s microenvironment occur, and the maximum of HSA’s emission does not change.
The decrease in the relative fluorescence intensities was analyzed using the Stern–Volmer Equation (1), and the results are presented in Figure 3a,b and Table 1. KSV values for both binary and ternary systems were calculated at 105 M−1, alluding to the fact that a significant interaction between HSA and CF was responsible for the quenching mechanism. The linear dependence of quenching suggested that only one type of quenching mechanism (either static or dynamic) dominated. To elucidate the most likely mechanism that causes protein quenching, the values of the quenching rate constants kq were determined. Equation (1) was used to calculate kq. KSV was defined as the slope of the Stern–Volmer dependence, and τ0 = 5.71 × 10−9 s is a constant [21]. The observed value of kq is significantly higher than the bimolecular quenching rate constant of 2 × 1010 M−1·s−1, indicating the prevailing contribution of statistical quenching [18,49], i.e., the quenching was not initiated by dynamic collision but via complex formation.
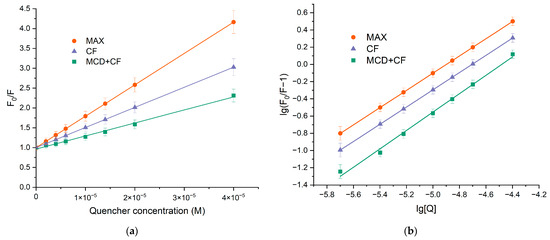
Figure 3.
(a) Stern−Volmer dependences expressing the effect of HSA quenching via binding CF’s forms F0/F, C(HSA) = 0.02 mM, 37 °C, pH 7.4. (b) Plot versus for HSA-drugs, C(HSA) = 0.02 mM, 37 °C, pH 7.4.

Table 1.
Stern–Volmer quenching constants at λex = 280 nm and binding parameters for interaction of drugs with HSA, pH 7.4, CHSA = 0.02 mM.
To determine the binding constants () and stoichiometric coefficient (n), Equation (1) was converted into Equation (2) due to the statistical nature of fluorescence quenching at low CF concentrations. Figure 3b demonstrates the plots of versus . The binding constant for HSA−CF was (1.0 ± 0.3) × 105 M−1 at 37 °C, and there was only one binding site of CF on HSA. The formation of ternary systems led to changes in Ka values while the stoichiometry remained one. For HSA–(CF+MCD), the binding constant was almost two times lower compared to the binary system. The finding might be due to the competition between albumin and MCD for drug binding. Since the binding efficiency of HSA–CF and CF+MCD is close ( M−1 [12]), the drug is distributed between the two ligands.
For MAX, we observed the opposite effect: an increase in Ka value by 1.5 times. That confirms our suggestion about the interaction of HSA with MAX particles, bringing CF and Trp’s residue closer together and hiding the solvent interactions with HSA’s hydrophobic pocket [41,44]. However, this statement is debatable and requires further research.
3.2. Steady-State Fluorescence Anisotropy
Monitoring the modulation of the fluorescence characteristics of CF in the presence of HSA involves excitation of the samples at λex~340 nm. At these wavelengths, n-π* transitions in the absorption spectrum of CF occurred [50], and hence there was no light absorption by HSA. Figure 4 illustrates the impact of added HSA on the steady-state fluorescence anisotropy of CF, providing further evidence of the CF–HSA interaction. The steep increase in anisotropy within the protein indicates significant motional constraints on the drug molecules upon binding to the protein, followed by saturation at higher protein concentrations. This finding is consistent with previous studies [50,51]. In ternary systems, we also observed an increase in anisotropy with increasing HSA concentration. However, in the ternary system, the level of anisotropy was approximately 1.5 times lower than in the binary system HSA+CF, indicating that some CF is still attached to MCD even after reaching a plateau. We suggest that MCD and HSA concurred in binding with CF. The calculated Ka for binary and ternary systems ( M−1 and M−1) agree with the data obtained by the analysis of quenching fluorescence emission of HSA.
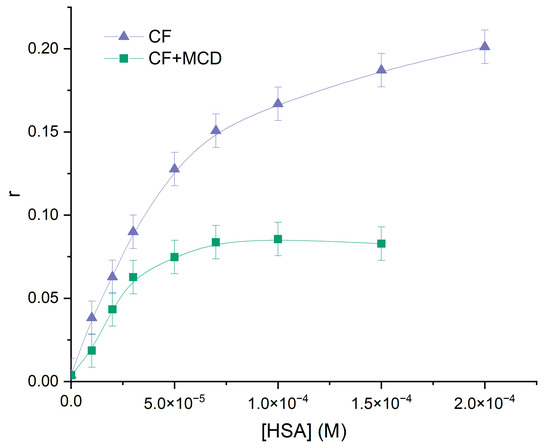
Figure 4.
Anisotropy changes of CF (purple) and (CF+MCD) (green), PBS, 37 °C, CCF = C(CF+MCD) = 0.01 mM.
3.3. Fluorescence Resonance Energy Transfer (FRET)
The use of fluorescence resonance energy transfer (FRET) as a spectroscopic method allows monitoring fluorophore proximity and angular orientation, making it a valuable tool for measuring molecular distances in complex biological systems. Forster’s nonradiative energy transfer theory outlines the conditions necessary for energy transfer to occur, including fluorescent light production by the donor, overlap of the donor’s fluorescence emission spectrum and the acceptor’s UV absorbance spectrum, and a distance of approach between donor and acceptor of less than 7 nm [21,52]. Equations (7)–(9) can be used with FRET to calculate the distance between drugs and HSA.
As the HSA fluorescence spectrum overlaps the CF absorption spectrum, FRET might occur in that system (Figure 5). We calculated the parameters for the binary and ternary systems. The results of the calculations are presented in Table 2. The distance between HSA and CF/CF+MCD/MAX was in the range of 32–35 Å. The parameters are typical for such kinds of systems and correspond to the literature data for HSA−CF and BSA−CF [37,52]. We found that ternary systems have a higher efficiency of energy transfer than binary systems. The probability of energy transfer from HSA to CF is high, as the distance between them is less than 7 nm and between 0.5R0 and 1.5R0. Moreover, this fact might indicate the existence of static quenching due to complex formation [21].
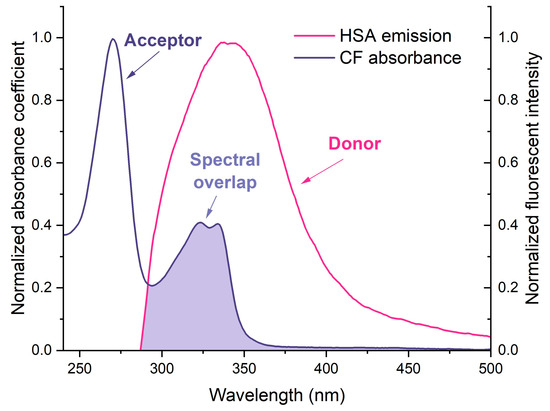
Figure 5.
Normalized overlap of the CF absorption spectra with the fluorescence spectra of HSA at 25 °C, both in PBS pH 7.4.

Table 2.
Energy transfer parameters for the interactions of the drugs with human serum albumin (25 °C, PBS, pH 7.4).
3.4. The Analysis of HSA’s Secondary Structure
Several studies have reported that albumin’s secondary structure undergoes changes upon the formation of the drug-HSA complex [20,45,51,53,54]. Our research involved the use of FTIR [24,55,56] and circular dichroism spectroscopy [45,57,58,59] to investigate the changes in the albumin’s secondary structure content upon the formation of drug-form HSA complexes. We focused on the Amide I and Amide II bands in the FTIR spectra, which are sensitive to changes in the protein’s secondary structure. The Amide I band (1600–1700 cm−1) represents the oscillation of ν(C=O)~80% and ν(C-N)~15%, while the Amide II band (1500–1600 cm−1) corresponds to the δ(N-H)~60%, ν(C-N)~20% and (C-C)~10% (Figure 6a). By deconvolution of the Amide I band, we determined the content of α-helixes, β-structures and random coils, providing important insights into changes in HSA’s secondary structure (Figure 6b).
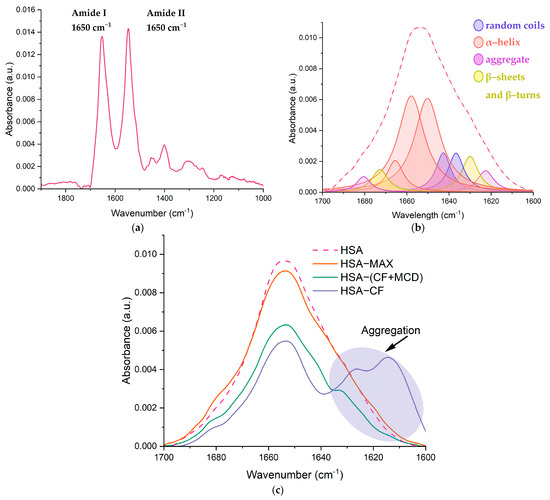
Figure 6.
(a) The FTIR spectra of HSA, pH 7.4; (b) the deconvolution of HSA FTIR spectra, CHSA = 0.06 mM, the simulated spectra demonstrated in the dashed pink line, pH 7.4; (c) changes in the structure of Amide I, CHSA = 0.06 mM, CCF = 1.2 mM, pH 7.4.
Our findings confirmed that native HSA contains approximately 65% α-helix [19,24,44,47,53]. The presence of MCD does not affect HSA’s secondary structure content, while CF causes a noticeable decrease in α-helix by 6–10%, primarily due to an increase in β-structures and random coils that was reported for other drug-HSA systems [24,53,60]. We observed similar changes in HSA’s structure in the case of CF+MCD, although the effect was less pronounced for the ternary system. MCD slightly alters CF–HSA binding, possibly by preventing CF from binding to HSA. The alterations in HSA’s secondary structure for CF+MCD were similar to those in the binary system but less pronounced, so indeed MCD and HSA concur for CF binding. For the HSA–MAX system, the secondary structure of proteins changes insignificantly. Our previous works [31,61] demonstrated that the complex formation between fluoroquinolone drugs and CDs might be accompanied by the increasing role of hydrogen bond formation and the decreasing force of hydrophobic associations [18,45,47]. Therefore, we might suggest that CF+MCD and MAX interact mainly with the surface of the protein due to the formation of hydrogen interactions; therefore, in these systems, changes in the secondary structure are minimal (Table 3).

Table 3.
The content of secondary structures in HSA: CHSA = CCF = 0.06 mM, pH 7.4, SD (n = 3).
Protein aggregation plays a key role in the pathophysiology of many diseases and may have serious consequences [62,63]. In FTIR spectra, characteristic peaks could be observed in regions 1614–1618, 1622–1626 and 1685–1696 cm−1, which correspond to anti-parallel β-layers and are sufficiently distant from the peaks of parallel β-layers and from other elements of the secondary structure [64]. These peaks were not considered in calculating the percentages in Table 3, as aggregation refers to intermolecular contacts. We consider how CF, CF+MCD and MAX would affect the process of aggregation (Figure 6c). When CF was added to HSA, the intensity of the bands at 1626 and 1616 cm−1 assigned to aggregation was expressively increased. We suggested that CF causes the conformational changes in HSA structure that provoke the aggregation. Therefore, it might be concluded that high CF concentrations lead to the aggregation of HSA. It is an undesirable process that should be prevented. Surprisingly, we observe much less expressed aggregation of protein in HSA−(CF+MCD) (Figure 6c). We suppose MCD makes the surface of proteins more hydrophilic, which prevents albumin’s aggregation. In the HSA−MAX system, protein aggregation almost does not occur, even with the large CF’s molar excesses. As MAX could form a more hydrophilic surface on the protein than MCD, the effect of preventing aggregation is more significant. These observations suggest that encapsulating CF into CD-based carriers might significantly decrease adverse events associated with CF.
Circular dichroism (CD) was also used to study the changes in the secondary structure of HSA that occur during the interaction of albumin with CF and their complexes with CD. In the range 200–260 nm, neither CF nor MCD have a CD spectrum, which makes it possible to study complex multicomponent systems. Since α-helices are mainly present in the secondary structure of albumin, as expected, the minimum ellipticity values in the CD spectrum are observed at 208 nm and 220 nm, corresponding to n–π* transitions of the peptide bond in α-helices [48] (Figure 7a). The proportion of α helices in free HSA at pH 7.4 is about 64% (Figure 7b), which is consistent with the literature data and data obtained from FTIR (Table 3). The interaction of proteins with CF’s forms leads to changes in the secondary structure of the protein, and the carrier for CF has a significant impact on these changes. Circular dichroism spectroscopy confirmed the structural changes observed in HSA obtained by FTIR spectroscopy.
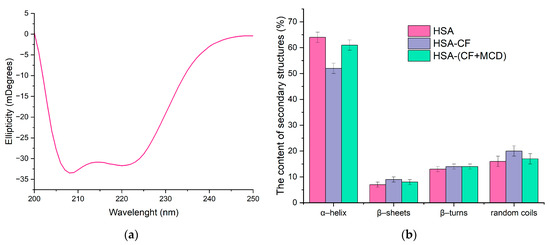
Figure 7.
(a) Circular dichroism spectrum of HSA, CHSA = 0.02 mM, pH 7.4. (b) The content of secondary structures in HSA: CHSA = CCF+MCD = CCF = 0.02 mM, pH 7.4.
3.5. Study of the Kinetics of Ciprofloxacin Release
The method of equilibrium dialysis was used as a model system for studying the effect of complexation on the drug’s kinetic release since the parameter is crucial for discovering the trend toward prolonged drug release. The drug’s complexation with CD might slightly affect its release. For instance, in [12], the formation of CF complexes with β-CD derivatives leads to a slowdown of the drug’s release that correlates with the dissociation constants of the complexes. In this work, we studied not only the CD carrier’s effect on CF’s release but also the role of HSA’s presence (Figure 8 and Table 4). The dialysis membrane’s pore diameter was chosen at 3.5 kDa in such a way that only CF can be released while MCD and MCD-based macromolecules are held inside the dialysis membrane [28,61].
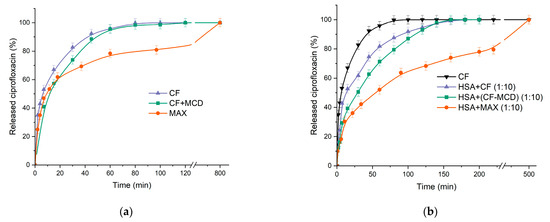
Figure 8.
(a) Free CF and CF complex with MCD and MAX release curves at pH 7.4. (b) Free CF (black); HSA with a tenfold molar excess of free CF (purple); HSA with a tenfold molar excess of CF complex with MCD (in a ratio of 1:1) (green); HSA with a tenfold molar excess of MAX (orange); 37 °C, pH 7.4.

Table 4.
The release rate of CF or CF+CD carrier in the absence and presence of HSA, represented as slopes of the initial sections of the CF release curves (Figure 8), the relative changes in concentration per minute, pH 7.4, 37 °C.
As we expected, the formation of the CF+MCD complex leads to a slowdown in the release of CF by ~15% (according to the tangent of the initial section) compared to free CF (Figure 8a and Table 4). Surprisingly, there was no significant change in the CF’s release profile from MAX within the first 20 min compared to free CF. However, after 40 min, we observed a noticeable decrease in the release rate from MAX compared to the HSA−CF system (~30%). The significant decrease in release rate might be explained by the fast release of CF molecules located near the MAX surface and then the slower release of CFs encapsulated in the inner MAX volume. It is worth noting that 100% of encapsulated CF is released from MAX, which confirms the absence of a covalent link between CF and the polymer matrix.
Thus, MAX demonstrates a longer drug release compared to free CF and CF in complex with MCD.
Let us consider how HSA affects CF’s release profile. We used a tenfold excess of HSA to model in vitro the physiological conditions where the drug is surrounded by many molecules of HSA [65]. For the HSA−CF system, there is a noticeable decrease in the release rate of ~56% in 60 min. One hundred percent of CF releases after 3 h, which is 2.5 times longer than in the case of free CF. A significant decrease in the release rate might be due to the binding of CF in complex with HSA.
In the case of the CF+MCD complex (ratio 1:1) preincubated with HSA, we observed a dramatic decrease in the CF’s release rate (by ~90% in 1 h), whereas the plateau is reached at 220 min (2.75 times longer than free CF) (Figure 8b). The additional decrease in the release rate of ~20% in the presence of MCD is probably due to the competition between MCD and HSA for CF’s binding [12].
The lowest release rate occurs in the HSA−MAX system (Figure 8b). In 3 h, only 70% of CF is released. The data obtained allow us to suggest that CF incapsulated in MCD-based carriers would circulate in the blood flow and be transported by protein for a longer time compared to free CF. Slow-release CF from MAX and the formation of the protein crown may change the antibacterial properties of CF in MAX [66].
3.6. Antibacterial Activity of CF’s Drug Forms in the Presence of HSA
We used Gram-positive Bacillus subtilis and Gram-negative Escherichia coli to investigate the effect of CF’s carrier and HSA’s presence on the drug’s antibacterial properties. We performed a rapid and reliable agar-well diffusion test to determine minimum inhibition concentrations (MIC) [15,29,30,31].
First, we compared the action of free CF, CF+MCD, and MAX: MICCF = MICCF+MCD = 0.02 μg/mL (E. coli) and 0.14 μg/mL (B. subtilis), whereas MICMAX = 0.06 and 0.2 μg/mL for E. coli and B. subtilis, respectively (Table 5). We found that MCD does not change CF’s activity, whereas the encapsulation of CF into MCD polymer slightly increases the MIC value (both strains). We suggested that the increase in MIC might be associated mainly with the particle’s limited agar diffusion.

Table 5.
MIC (µg/mL), pH 7.4 (PBS sterile buffer), agar-well diffusion method, 37 °C.
One can conclude that CD as well as MAX systems practically do not change CF’s activity against both E. coli and B. subtilis, but for MAX systems, prolonged action is realized due to CF’s sustained release.
Pre-incubation of CF/CF+MCD/MAX with HSA does not affect the MIC values. Similar results were obtained earlier in our research group [61]. The data uncovers that CD-based carriers will not change CF’s antimicrobial properties during intravenous injection.
4. Conclusions
For the first time, the MCD-based polymer MAX with encapsulated CF was obtained via crosslinking with isocyanate. MAX represents polycarbamide that forms particles with a hydrodynamic diameter ~220 nm. CF’s structure was not modified during the synthesis. Drug molecules are located inside the polymer matrix.
We compared MAX to the common CF+MCD non-covalent complex in their binding to the biological macromolecule (HAS) that might affect CF’s pharmacokinetic properties. MCD prevents CF’s binding to albumin, probably due to the competition with HSA for drug binding, whereas for MAX, an increase of Ka (~2.5 times) is observed. The latter effect might be associated with the formation of the protein corona (mainly based on albumin) on MAX. Furthermore, the formation of MAX particles and CF+MCD non-covalent complexes to a lesser extent resulted in the prevention of HSA’s extensive aggregation observed upon binding of HAS with non-complexed CF. This aspect is important in terms of the fact that when the structure of albumin changes and aggregation occurs upon its interactions with the particles in the bloodstream, the formed particles acquire the properties of immunogenicity and are subject to absorption by macrophages. Therefore, MAX particles can be considered a system for the drug’s pharmacokinetic regulation.
MAX decreases CF’s release rate significantly (100% is achieved at 8 h), whereas MCD demonstrates only a slight effect on the drug’s release profile (100% at 50 min). In the case of MAX, the carrier might enhance the drug’s lifetime in vivo and prolong the therapeutic effect. MCD as well as MAX systems practically do not change CF’s activity against both E. coli and B. subtilis, but for MAX systems, prolonged action is realized due to CF’s sustained release. Since pre-incubation of CF/CF+MCD/MAX with HSA does not affect the MIC values, one can conclude that CD-based carriers will not change CF’s antimicrobial properties upon intravenous injection. Therefore, MAX is a promising system for drug delivery due to prolonged drug release, the increase of CF’s binding efficiency to HSA, and good in vitro activity.
We have demonstrated that encapsulation of CF in a polymer matrix pronouncedly affects the drug’s properties to a greater extent compared to MCD. MAX is a preferable carrier due to its prolonged release profile and minimal impact on HSA’s secondary structure and aggregation process. This finding is crucial in the development of new drug delivery systems utilizing CD and their polymers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/chemistry5030132/s1, Figure S1: (a) Normalized FTIR spectra of MCD (green) and MAX (orange) pH 7.4, 22 °C. (b) Normalized FTIR spectra of MCD (green) and MAX (orange) range 1240–900 cm−1 in details pH 7.4, 22 °C.
Author Contributions
Conceptualization: A.A.S. and E.V.K.; experimental work: T.Y.K.; methodology of microbiological experiments, N.G.B.; data analysis and interpretation: T.Y.K.; writing—original draft preparation: T.Y.K.; writing—review and editing: A.A.S. and E.V.K.; supervision: A.A.S. and E.V.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Science Foundation grant number 22-24-00604.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information.
Acknowledgments
The work was performed using equipment (FTIR spectrometer Bruker Tensor 27, Jasco J-815 CD Spectrometer and AFM microscope NTEGRA II) from the program for the development of Moscow State University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fanali, G.; di Masi, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human serum albumin: From bench to bedside. Mol. Aspects Med. 2012, 33, 209–290. [Google Scholar] [CrossRef] [PubMed]
- Talebi Bezmin Abadi, A.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. Bionanoscience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- Mahady, G. Medicinal Plants for the Prevention and Treatment of Bacterial Infections. Curr. Pharm. Des. 2005, 11, 2405–2427. [Google Scholar] [CrossRef]
- Chong, W.H.; Saha, B.K.; Ananthakrishnan, R.; Chopra, A. State-of-the-art review of secondary pulmonary infections in patients with COVID-19 pneumonia. Infection 2021, 49, 591–605. [Google Scholar] [CrossRef]
- Tiwari, G.; Tiwari, R.; Bannerjee, S.; Bhati, L.; Pandey, S.; Pandey, P.; Sriwastawa, B. Drug delivery systems: An updated review. Int. J. Pharm. Investig. 2012, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, J.S.; Hooper, D.C. Fluoroquinolone antimicrobial agents. Clin. Microbiol. Rev. 1989, 2, 378–424. [Google Scholar] [CrossRef]
- Bennett, A.C.; Bennett, C.L.; Witherspoon, B.J.; Knopf, K.B. An evaluation of reports of ciprofloxacin, levofloxacin, and moxifloxacin-association neuropsychiatric toxicities, long-term disability, and aortic aneurysms/dissections disseminated by the Food and Drug Administration and the European Medicines Agency. Expert Opin. Drug Saf. 2019, 18, 1055–1063. [Google Scholar] [CrossRef]
- Alhajj, N.; O’Reilly, N.J.; Cathcart, H. Developing ciprofloxacin dry powder for inhalation: A story of challenges and rational design in the treatment of cystic fibrosis lung infection. Int. J. Pharm. 2022, 613, 121388. [Google Scholar] [CrossRef]
- Yayehrad, A.T.; Wondie, G.B.; Marew, T. Different Nanotechnology Approaches for Ciprofloxacin Delivery Against Multidrug-Resistant Microbes. Infect. Drug Resist. 2022, 15, 413–426. [Google Scholar] [CrossRef]
- Schacht, P.; Arcieri, G.; Hullmann, R. Safety of oral ciprofloxacin. Am. J. Med. 1989, 87, S98–S102. [Google Scholar] [CrossRef]
- Loftsson, T.; Jarho, P.; Másson, M.; Järvinen, T. Cyclodextrins in drug delivery. Expert Opin. Drug Deliv. 2005, 2, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Brewster, M.E. Cyclodextrin-based pharmaceutics: Past, present and future. Nat. Rev. Drug Discov. 2004, 3, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Skuredina, A.A.; Kopnova, T.Y.; Le-deygen, I.M.; Kudryashova, E.V. Physical and Chemical Properties of the Guest—Host Inclusion Complexes of Cyprofloxacin with β -Cyclodextrin Derivatives. Moscow Univ. Chem. Bull. 2020, 75, 218–224. [Google Scholar] [CrossRef]
- Chen, G.; Jiang, M. Cyclodextrin-based inclusion complexation bridging supramolecular chemistry and macromolecular self-assembly. Chem. Soc. Rev. 2011, 40, 2254. [Google Scholar] [CrossRef]
- Hishiya, T.; Asanuma, H.; Komiyama, M. Spectroscopic Anatomy of Molecular-Imprinting of Cyclodextrin. Evidence for Preferential Formation of Ordered Cyclodextrin Assemblies. J. Am. Chem. Soc. 2002, 124, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Skuredina, A.A.; Tychinina, A.S.; Le-Deygen, I.M.; Golyshev, S.A.; Belogurova, N.G.; Kudryashova, E.V. The formation of quasi-regular polymeric network of cross-linked sulfobutyl ether derivative of β-cyclodextrin synthesized with moxifloxacin as a template. React. Funct. Polym. 2021, 159, 104811. [Google Scholar] [CrossRef]
- Egawa, Y.; Shimura, Y.; Nowatari, Y.; Aiba, D.; Juni, K. Preparation of molecularly imprinted cyclodextrin microspheres. Int. J. Pharm. 2005, 293, 165–170. [Google Scholar] [CrossRef]
- Trotta, F.; Caldera, F.; Cavalli, R.; Soster, M.; Riedo, C.; Biasizzo, M.; Uccello Barretta, G.; Balzano, F.; Brunella, V. Molecularly imprinted cyclodextrin nanosponges for the controlled delivery of L-DOPA: Perspectives for the treatment of Parkinson’s disease. Expert Opin. Drug Deliv. 2016, 13, 1671–1680. [Google Scholar] [CrossRef]
- Zhang, L.-W.; Wang, K.; Zhang, X.-X. Study of the interactions between fluoroquinolones and human serum albumin by affinity capillary electrophoresis and fluorescence method. Anal. Chim. Acta 2007, 603, 101–110. [Google Scholar] [CrossRef]
- McCarty, T.A.; Page, P.M.; Baker, G.A.; Bright, F.V. Behavior of Acrylodan-Labeled Human Serum Albumin Dissolved in Ionic Liquids. Ind. Eng. Chem. Res. 2008, 47, 560–569. [Google Scholar] [CrossRef]
- Vlasova, I.M.; Bukharova, E.M.; Kuleshova, A.A.; Saletsky, A.M. Spectroscopic investigations of interaction of fluorescent nanomarkers of fluorescein family with human serum albumin at different values of pH. Curr. Appl. Phys. 2011, 11, 1126–1132. [Google Scholar] [CrossRef]
- Rehman, M.T.; Shamsi, H.; Khan, A.U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharm. 2014, 11, 1785–1797. [Google Scholar] [CrossRef] [PubMed]
- Astray, G.; Mejuto, J.C.; Morales, J.; Rial-Otero, R.; Simal-Gándara, J. Factors controlling flavors binding constants to cyclodextrins and their applications in foods. Food Res. Int. 2010, 43, 1212–1218. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Belogurova, N.G.; Krylov, S.S.; Semenova, M.N.; Semenov, V.V.; Kudryashova, E.V. Plant Alkylbenzenes and Terpenoids in the Form of Cyclodextrin Inclusion Complexes as Antibacterial Agents and Levofloxacin Synergists. Pharmaceuticals 2022, 15, 861. [Google Scholar] [CrossRef]
- Poureshghi, F.; Ghandforoushan, P.; Safarnejad, A.; Soltani, S. Interaction of an antiepileptic drug, lamotrigine with human serum albumin (HSA): Application of spectroscopic techniques and molecular modeling methods. J. Photochem. Photobiol. B Biol. 2017, 166, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Datta, A. Steady state and time-resolved fluorescence investigation of the specific binding of two chlorin derivatives with human serum albumin. J. Phys. Chem. B 2007, 111, 10557–10562. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Y.; Wang, Q.; Shi, Z.-H.; Xia, X.-H.; Sun, H.-W. Interaction Characteristic Studies of Ciprofloxacin and/or Sulphadiazine with Bovine Serum Albumin by Spectroscopic Technique. Asian J. Chem. 2015, 27, 818–826. [Google Scholar] [CrossRef]
- Missoun, F.; de los Ríos, A.P.; Ortiz-Martínez, V.; Salar-García, M.J.; Hernández-Fernández, J.; Hernández-Fernández, F.J. Discovering Low Toxicity Ionic Liquids for Saccharomyces cerevisiae by Using the Agar Well Diffusion Test. Processes 2020, 8, 1163. [Google Scholar] [CrossRef]
- Skuredina, A.A.; Kopnova, T.Y.; Tychinina, A.S.; Golyshev, S.A.; Le-Deygen, I.M.; Belogurova, N.G.; Kudryashova, E.V. The New Strategy for Studying Drug-Delivery Systems with Prolonged Release: Seven-Day In Vitro Antibacterial Action. Molecules 2022, 27, 8026. [Google Scholar] [CrossRef]
- Le-Deygen, I.M.; Skuredina, A.A.; Safronova, A.S.; Yakimov, I.D.; Kolmogorov, I.M.; Deygen, D.M.; Burova, T.V.; Grinberg, N.V.; Grinberg, V.Y.; Kudryashova, E.V. Moxifloxacin interacts with lipid bilayer, causing dramatic changes in its structure and phase transitions. Chem. Phys. Lipids 2020, 228, 104891. [Google Scholar] [CrossRef]
- Skuredina, A.A.; Tychinina, A.S.; Le-Deygen, I.M.; Golyshev, S.A.; Kopnova, T.Y.; Le, N.T.; Belogurova, N.G.; Kudryashova, E.V. Cyclodextrins and Their Polymers Affect the Lipid Membrane Permeability and Increase Levofloxacin’s Antibacterial Activity In Vitro. Polymers 2022, 14, 4476. [Google Scholar] [CrossRef]
- Yakupova, L.R.; Kopnova, T.Y.; Skuredina, A.A.; Le-Deygen, I.M.; Shustrov, P.N.; Novoselov, A.M.; Kudryashova, E.V. The Formation of β-Cyclodextrin Complexes with Levofloxacin and Ceftriaxone as an Approach to the Regulation of Drugs’ Pharmacokinetic. Colloid J. 2023, 85, 114–127. [Google Scholar] [CrossRef]
- Bhargav, H.S.; Shastri, S.D.; Poornav, S.P.; Darshan, K.M.; Nayak, M.M. Measurement of the Zone of Inhibition of an Antibiotic. In Proceedings of the 2016 IEEE 6th International Conference on Advanced Computing (IACC), Bhimavaram, India, 27–28 February 2016; pp. 409–414. [Google Scholar]
- Cooper, K.E. The Theory of Antibiotic Inhibition Zones. In Analytical Microbiology; Elsevier: Amsterdam, The Netherlands, 1963; pp. 1–86. [Google Scholar]
- Sainz-Rozas, P.R.; Isasi, J.R.; González-Gaitano, G. Binding of dibenzofuran and its derivatives to water-soluble β-cyclodextrin polymers. J. Photochem. Photobiol. A Chem. 2005, 173, 248–257. [Google Scholar] [CrossRef]
- Argenziano, M.; Haimhoffer, A.; Bastiancich, C.; Jicsinszky, L.; Caldera, F.; Trotta, F.; Scutera, S.; Alotto, D.; Fumagalli, M.; Musso, T.; et al. In Vitro Enhanced Skin Permeation and Retention of Imiquimod Loaded in β-Cyclodextrin Nanosponge Hydrogel. Pharmaceutics 2019, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Iranfar, H.; Rajabi, O.; Salari, R.; Chamani, J. Probing the Interaction of Human Serum Albumin with Ciprofloxacin in the Presence of Silver Nanoparticles of Three Sizes: Multispectroscopic and ζ Potential Investigation. J. Phys. Chem. B 2012, 116, 1951–1964. [Google Scholar] [CrossRef]
- Zhang, H.-M.; Wang, Y.-Q.; Jiang, M.-L. A fluorimetric study of the interaction of C.I. Solvent Red 24 with haemoglobin. Dye. Pigment. 2009, 82, 156–163. [Google Scholar] [CrossRef]
- Wei, Y.; Li, J.; Dong, C.; Shuang, S.; Liu, D.; Huie, C.W. Investigation of the association behaviors between biliverdin and bovine serum albumin by fluorescence spectroscopy. Talanta 2006, 70, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.V.; Jain, N.K. Suppression of agglomeration of ciprofloxacin-loaded human serum albumin nanoparticles. AAPS PharmSciTech 2007, 8, E118–E123. [Google Scholar] [CrossRef]
- Fick, A.C.; Reinscheid, U.M. Characterization of the binding epitope of ciprofloxacin bound to human serum albumin. J. Pharm. Biomed. Anal. 2006, 41, 1025–1028. [Google Scholar] [CrossRef]
- Valeur, B. Molecular Fluorescence; John Wiley & Sons: Hoboken, NJ, USA, 2009; ISBN 9783527407736. [Google Scholar]
- Gunasekaran, S.; Rajalakshmi, K.; Kumaresan, S. Vibrational analysis, electronic structure and nonlinear optical properties of Levofloxacin by density functional theory. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 112, 351–363. [Google Scholar] [CrossRef]
- Ahmad, B.; Parveen, S.; Khan, R.H. Effect of Albumin Conformation on the Binding of Ciprofloxacin to Human Serum Albumin: A Novel Approach Directly Assigning Binding Site. Biomacromolecules 2006, 7, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Seedher, N.; Agarwal, P. Complexation of fluoroquinolone antibiotics with human serum albumin: A fluorescence quenching study. J. Lumin. 2010, 130, 1841–1848. [Google Scholar] [CrossRef]
- Yakupova, L.R.; Kopnova, T.Y.; Skuredina, A.A.; Kudryashova, E.V. Effect of Methyl-β-Cyclodextrin on the Interaction of Fluoroquinolones with Human Serum Albumin. Russ. J. Bioorganic Chem. 2022, 48, 163–172. [Google Scholar] [CrossRef]
- Seedher, N.; Agarwal, P. Competitive Binding of Fluoroquinolone Antibiotics and Some Other Drugs to Human Serum Albumin: A Luminescence Spectroscopic Study. Luminescence 2013, 28, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Varshney, A.; Ansari, Y.; Zaidi, N.; Ahmad, E.; Badr, G.; Alam, P.; Khan, R.H. Analysis of Binding Interaction Between Antibacterial Ciprofloxacin and Human Serum Albumin by Spectroscopic Techniques. Cell Biochem. Biophys. 2014, 70, 93–101. [Google Scholar] [CrossRef]
- Kaur, A.; Khan, I.A.; Banipal, P.K.; Banipal, T.S. Deciphering the complexation process of a fluoroquinolone antibiotic, levofloxacin, with bovine serum albumin in the presence of additives. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 191, 259–270. [Google Scholar] [CrossRef]
- Paul, B.K.; Guchhait, N.; Bhattacharya, S.C. Binding of ciprofloxacin to bovine serum albumin: Photophysical and thermodynamic aspects. J. Photochem. Photobiol. B Biol. 2017, 172, 11–19. [Google Scholar] [CrossRef]
- Paul, B.K.; Ghosh, N.; Mukherjee, S. Interplay of Multiple Interaction Forces: Binding of Norfloxacin to Human Serum Albumin. J. Phys. Chem. B 2015, 119, 13093–13102. [Google Scholar] [CrossRef]
- Hu, Y.J.; Ou-Yang, Y.; Zhang, Y.; Liu, Y. Affinity and specificity of ciprofloxacin-bovine serum albumin interactions: Spectroscopic approach. Protein J. 2010, 29, 234–241. [Google Scholar] [CrossRef]
- Abu, T.M.M.; Ghithan, J.; Abu-Taha, M.I.; Darwish, S.M.; Abu-hadid, M.M. Spectroscopic approach of the interaction study of ceftriaxone and human serum albumin. J. Biophys. Struct. Biol. 2014, 6, 1–12. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Hirano, A.; Arakawa, T.; Shiraki, K. Effects of alcohol on the solubility and structure of native and disulfide-modified bovine serum albumin. Int. J. Biol. Macromol. 2012, 50, 1286–1291. [Google Scholar] [CrossRef]
- Tatulian, S.A. Structural Characterization of Membrane Proteins and Peptides by FTIR and ATR-FTIR Spectroscopy. In Lipid-protein Interactions: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2013; pp. 177–218. [Google Scholar]
- Tretiakova, D.; Le-Deigen, I.; Onishchenko, N.; Kuntsche, J.; Kudryashova, E.; Vodovozova, E. Phosphatidylinositol Stabilizes Fluid-Phase Liposomes Loaded with a Melphalan Lipophilic Prodrug. Pharmaceutics 2021, 13, 473. [Google Scholar] [CrossRef]
- Yan, J.; Wu, D.; Ma, X.; Wang, L.; Xu, K.; Li, H. Spectral and molecular modeling studies on the influence of β-cyclodextrin and its derivatives on aripiprazole-human serum albumin binding. Carbohydr. Polym. 2015, 131, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.K.; Guchhait, N. A spectral deciphering of the binding interaction of an intramolecular charge transfer fluorescence probe with a cationic protein: Thermodynamic analysis of the binding phenomenon combined with blind docking study. Photochem. Photobiol. Sci. 2011, 10, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Dockal, M.; Carter, D.C.; Rüker, F. Conformational Transitions of the Three Recombinant Domains of Human Serum Albumin Depending on pH. J. Biol. Chem. 2000, 275, 3042–3050. [Google Scholar] [CrossRef] [PubMed]
- Trynda-Lemiesz, L. Paclitaxel-HSA interaction. Binding sites on HSA molecule. Bioorganic Med. Chem. 2004, 12, 3269–3275. [Google Scholar] [CrossRef]
- Skuredina, A.A.; Yakupova, L.R.; Kopnova, T.Y.; Le-Deygen, I.M.; Belogurova, N.G.; Kudryashova, E.V. Cyclodextrins and Their Polymers Affect Human Serum Albumin’s Interaction with Drugs Used in the Treatment of Pulmonary Infections. Pharmaceutics 2023, 15, 1598. [Google Scholar] [CrossRef] [PubMed]
- Rostagno, A.; Ghiso, J.A. Misfolding, Aggregation, and Amyloid Formation: The Dark Side of Proteins. In Protein Folding Disorders of the Central Nervous System; World Scientific: Singapore, 2017; pp. 1–31. [Google Scholar]
- Akter, R.; Cao, P.; Noor, H.; Ridgway, Z.; Tu, L.-H.; Wang, H.; Wong, A.G.; Zhang, X.; Abedini, A.; Schmidt, A.M.; et al. Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology. J. Diabetes Res. 2016, 2016, 2798269. [Google Scholar] [CrossRef] [PubMed]
- Kudryashova, E.V. Reversible self-association of ovalbumin at air-water interfaces and the consequences for the exerted surface pressure. Protein Sci. 2005, 14, 483–493. [Google Scholar] [CrossRef]
- Davis, R.; Markham, A.; Balfour, J.A. Ciprofloxacin. Drugs 1996, 51, 1019–1074. [Google Scholar] [CrossRef]
- Kianfar, E. Protein nanoparticles in drug delivery: Animal protein, plant proteins and protein cages, albumin nanoparticles. J. Nanobiotechnol. 2021, 19, 159. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).