A Ratiometric Fluorescent Probe for pH Measurement over a Wide Range Composed of Three Types of Fluorophores Assembled on a DNA Scaffold
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of OG-Modified DNA (OG-ODN3) and OG-AE
2.3. Preparation of Fluorophore-Assembled DNA Origami Scaffold (rec-Ori)
2.4. Agarose Gel Electrophoresis Analysis
2.5. AFM Imaging
2.6. Fluorescence Spectra Measurement and pH Titration
2.7. Measurement of UV-vis Absorption Spectra
3. Results
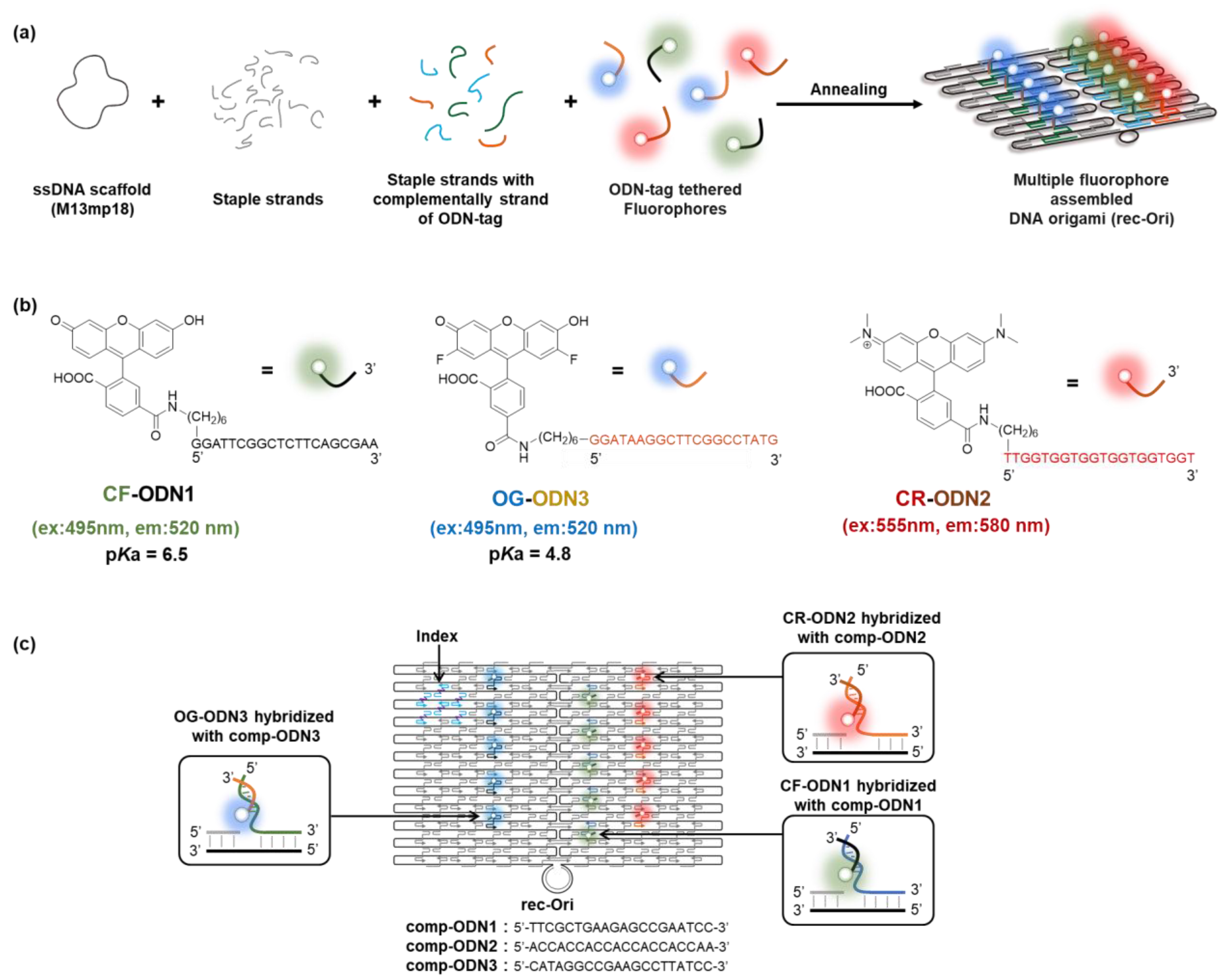
3.1. Design of Fluorophore-Assembled DNA Scaffolds
| Combination of ODN-tag and Comp-ODN | ||
|---|---|---|
| Fluorophore | DNA Sequence | |
| CF-ODN1 | CF at 5′ of ODN1 | 5′-GGATTCGGCTCTTCAGCGAA-3′ |
| comp-ODN1 | 3′-CCTAAGCCGAGAAGTCGCTT-5′ | |
| OG-ODN3 | OG at 5′ of ODN3 | 5′-GGATAAGGCTTCGGCCTATG-3′ |
| comp-ODN3 | 3′-CCTATTCCGAAGCCGGATAC-5′ | |
| CR-ODN2 | CR at 5′ of ODN2 | 5′-TTGGTGGTGGTGGTGGTGGT-3′ |
| comp-ODN2 | 3′-AACCACCACCACCACCACCA-5′ | |
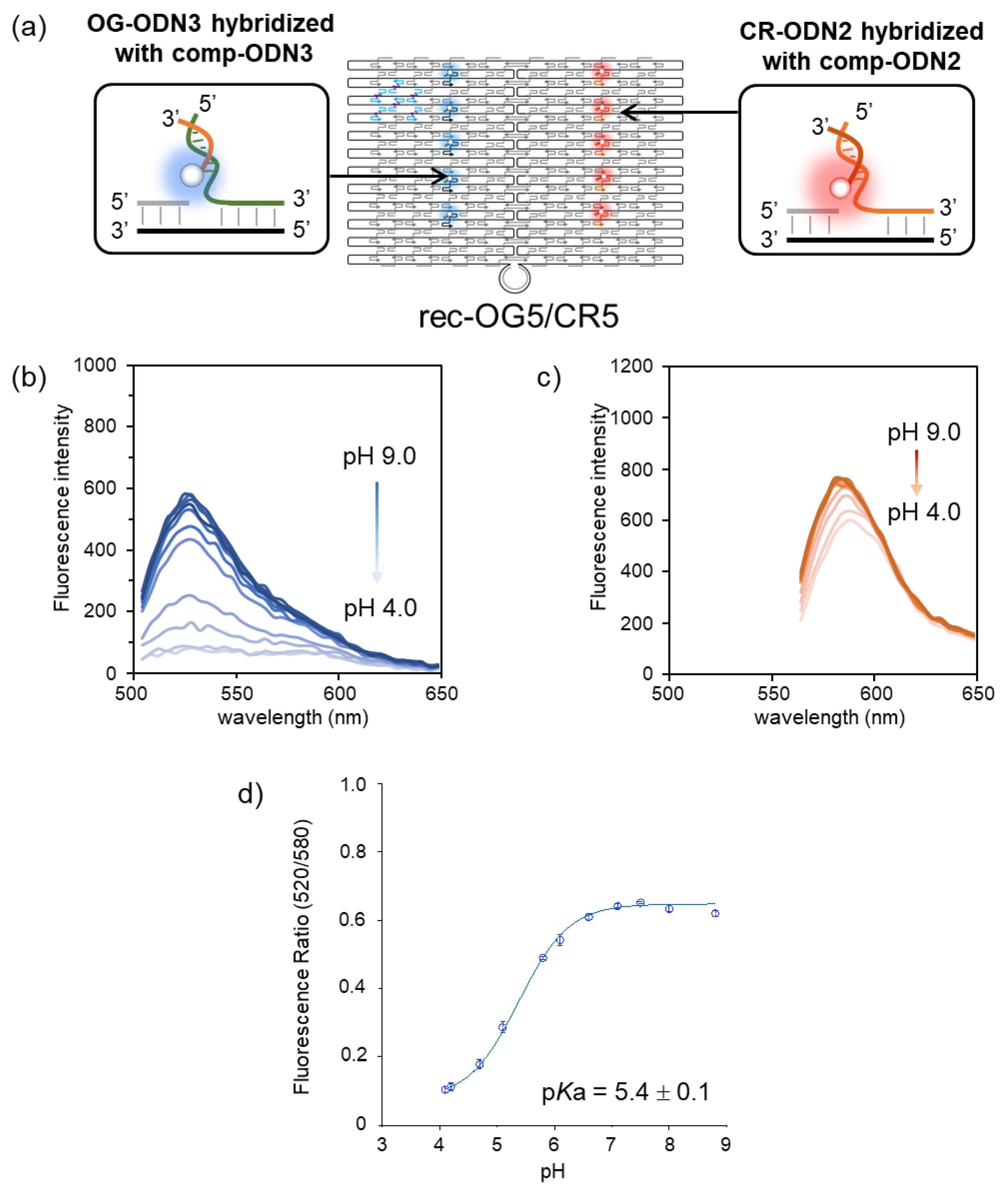
3.2. Ratiometric Detection of pH Changes by Rec-Ori Assembled with OG and CR
3.3. Estimation of the Ratio of Fluorescence Emission Intensities of CF, OG, and CR Assembled on Rec-Ori with Different Numbers of CF and OG
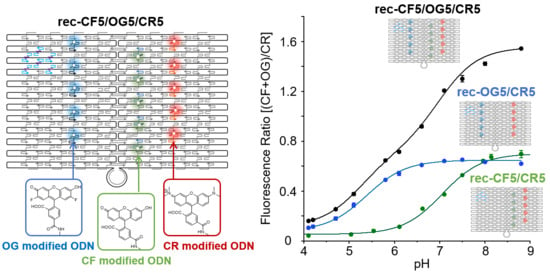
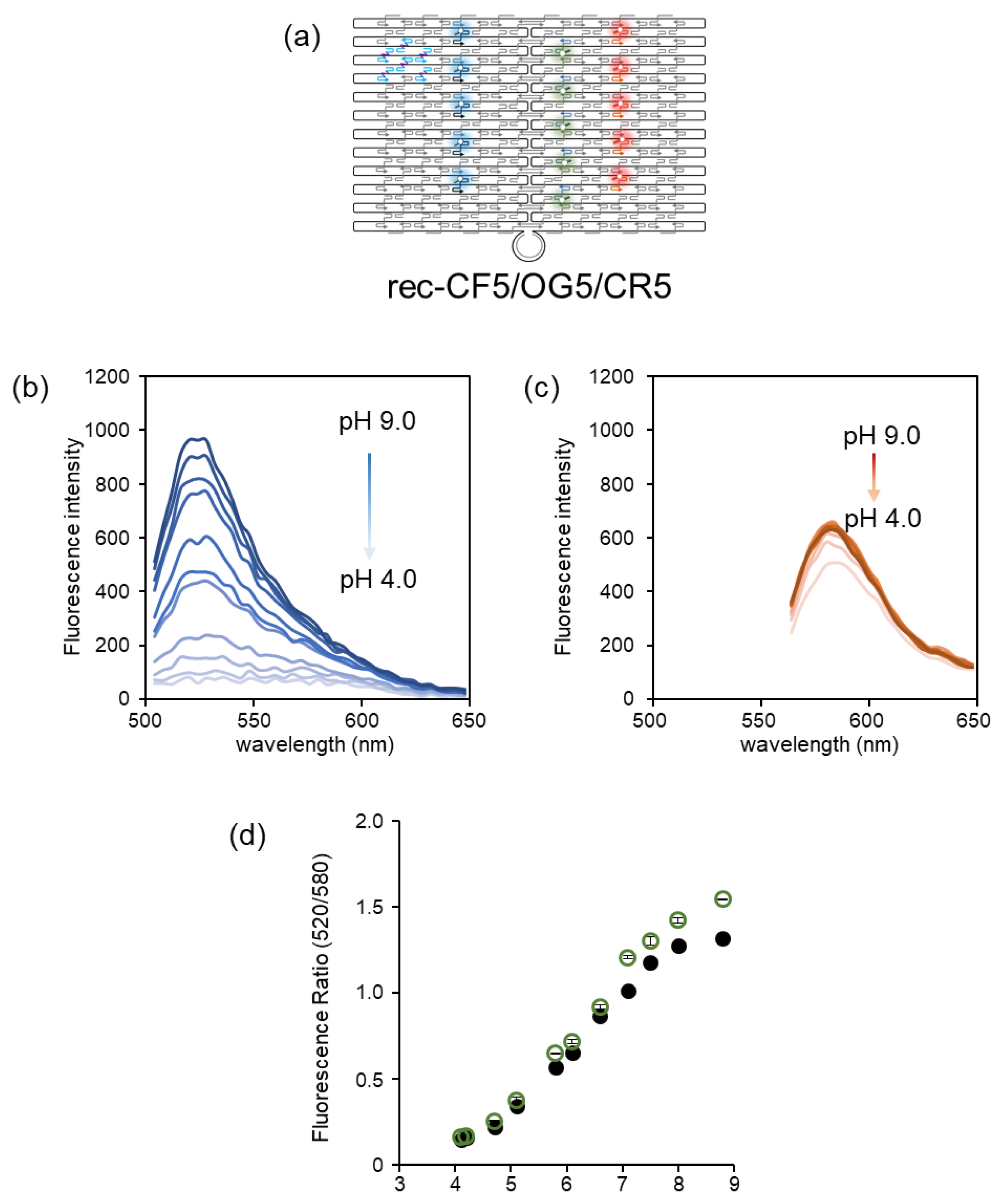
3.4. Ratiometric Detection of pH Changes by Rec-Ori Assembled with CF, OG, and CR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roos, A.; Boron, W.F. Intracellular pH. Physiol. Rev. 1981, 61, 296–434. [Google Scholar] [CrossRef]
- Srivastava, J.; Barber, D.L.; Jacobson, M.P. Intracellular pH sensors: Design principles and functional significance. Physiology 2007, 22, 30–39. [Google Scholar] [CrossRef]
- Willmann, J.K.; Bruggen, N.V.; Dinkelbuorg, L.M.; Gambhir, S.S. Molecular imaging in drug development. Nat. Rev. Drug Disc. 2008, 7, 591–607. [Google Scholar] [CrossRef]
- Hargreaves, R.J. The role of molecular imaging in drug discovery and development. Clinical pharmacology and therapeutics. Clin. Pharmacol. Ther. 2008, 83, 349–353. [Google Scholar] [CrossRef]
- Han, J.; Burgess, K. Fluorescent indicators for intracellular pH. Chem. Rev. 2010, 110, 2709–2728. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, A.; Wolfbeis, O.S.; Borisov, S.M. Optical sensing and imaging of pH values: Spectroscopies, materials, and applications. Chem. Rev. 2020, 120, 12357–12489. [Google Scholar] [CrossRef]
- Urano, Y.; Asanuma, D.; Hama, Y.; Koyama, Y.; Barrett, T.; Kamiya, M.; Nagano, T.; Watanabe, T.; Hasegawa, A.; Choyke, P.L.; et al. Selective molecular imaging of viable cancer cells with pH-activatable fluorescence probes. Nat. Med. 2009, 15, 104–109. [Google Scholar] [CrossRef]
- Han, J.; Loudet, A.; Barhoumi, R.; Burghardt, R.C.; Burgess, K. A Ratiometric pH Reporter for Imaging Protein-dye Conjugates In Living Cells. J. Am. Chem. Soc. 2009, 131, 1642–1643. [Google Scholar] [CrossRef]
- Tang, B.; Yu, F.; Li, P.; Tong, L.; Duan, X.; Xie, T.; Wang, X. A Near-Infrared Neutral pH Fluorescent Probe for Monitoring Minor pH Changes: Imaging in Living HepG2 and HL-7702 Cells. J. Am. Chem. Soc. 2009, 131, 3016–3023. [Google Scholar] [CrossRef]
- Nakata, E.; Yukimachi, Y.; Nazumi, Y.; Uto, Y.; Maezawa, H.; Hashimoto, T.; Okamoto, Y.; Hori, H. A newly designed cell-permeable SNARF derivative as an effective intracellular pH indicator. Chem. Commun. 2010, 46, 3526–3528. [Google Scholar] [CrossRef]
- Wann, J.G.; Hsu, Y.; Yang, C.; Lin, C.; Tai, D.W.; Chen, J.; Hsiao, C.; Chen, C. Neutrophils in acidotic haemodialysed patients have lower intracellular pH and inflamed state. Nephrol. Dial. Transpl. 2007, 22, 2613–2622. [Google Scholar] [CrossRef]
- Nilsson, C.; Kågedal, K.; Johansson, U.; Ollinger, K. Analysis of cytosolic and lysosomal pH in apoptotic cells by flow cytometry. Methods Cell Sci. 2004, 25, 185–194. [Google Scholar] [CrossRef]
- Sennoune, S.R.; Bakunts, K.; Martinez, G.M.; Chua-Tuan, J.L.; Kebir, Y.; Attaya, M.N.; Martinez-Zaguilan, R. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: Distribution and functional activity. Am. J. Physiol. Cell Physiol. 2004, 286, C1443–C1452. [Google Scholar] [CrossRef]
- Nilsson, C.; Johansson, U.; Johansson, A.; Ollinger, K.K. Cytosolic acidification and lysosomal alkalinization during TNF-α induced apoptosis in U937 cells. Apoptosis 2006, 11, 1149–1159. [Google Scholar] [CrossRef]
- Huang, X.; Song, J.; Yung, B.C.; Huang, X.; Xiong, Y.; Chen, X. Ratiometric optical nanoprobes enable accurate molecular detection and imaging. Chem. Soc. Rev. 2018, 47, 2873–2920. [Google Scholar] [CrossRef]
- Méndez-Ardoy, A.; Reina, J.; Montenegro, J. Synthesis and supramolecular functional assemblies of ratiometric pH probes. Chem. Eur. J. 2020, 26, 7516–7536. [Google Scholar] [CrossRef]
- Di Costanzo, L.; Panunzi, B. Visual pH Sensors: From a Chemical Perspective to New Bioengineered Materials. Molecules 2021, 26, 2952. [Google Scholar] [CrossRef]
- Chen, Y. Recent advances in fluorescent probes for extracellular pH detection and imaging. Anal. Biochem. 2021, 612, 113900. [Google Scholar] [CrossRef]
- Nakata, E.; Gerelbaatar, K.; Komatsubara, F.; Morii, T. Stimuli-Responsible SNARF Derivatives as a Latent Ratiometric Fluorescent Probe. Molecules 2022, 27, 7181. [Google Scholar] [CrossRef]
- Hou, J.T.; Ren, W.X.; Li, K.; Seo, J.; Sharma, A.; Yu, X.Q.; Kim, J.S. Fluorescent bioimaging of pH: From design to applications. Chem. Soc. Rev. 2017, 46, 2076–2090. [Google Scholar] [CrossRef]
- Jaworska, A.; Malek, K.; Kudelski, A. Intracellular pH–Advantages and pitfalls of surface-enhanced Raman scattering and fluorescence microscopy–A review. Spectrochim. Acta Part A 2021, 251, 119410. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.; Grinstein, S.; Orlowski, J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Brahimi-Horn, M.C.; Pouysségur, J. Oxygen, a source of life and stress. FEBS Lett. 2007, 581, 3582–3591. [Google Scholar] [CrossRef] [PubMed]
- Nakata, E.; Hirose, H.; Gerelbaatar, K.; Arafiles, J.V.V.; Zhang, Z.; Futaki, S.; Morii, T. A facile combinatorial approach to construct a ratiometric fluorescent sensor: Application for the real-time sensing of cellular pH changes. Chem. Sci. 2021, 12, 8231–8240. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Sugiyama, H. Single-molecule analysis using DNA origami. Angew. Chem. Int. Ed. 2012, 51, 874–890. [Google Scholar] [CrossRef]
- Linko, V.; Nummelin, S.; Aarnos, L.; Tapio, K.; Toppari, J.J.; Kostiainen, M.A. DNA-Based Enzyme Reactors and Systems. Nanomaterials 2016, 6, 139. [Google Scholar] [CrossRef]
- Rajendran, A.; Nakata, E.; Nakano, S.; Morii, T. Nucleic-Acid-Templated Enzyme Cascades. ChemBioChem 2017, 18, 696–716. [Google Scholar] [CrossRef]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef]
- Ngo, T.A.; Dinh, H.; Nguyen, T.M.; Liew, F.F.; Nakata, E.; Morii, T. Protein adaptors assemble functional proteins on DNA scaffolds. Chem. Commun. 2019, 55, 12428–12446. [Google Scholar] [CrossRef]
- Zhan, P.; Andreas, P.; Qiao, J.; Dongfang, W.; Shikufa, M.; Qiancheng, X.; Qi, S.; Yingxu, S.; Baoquan, D.; Chenxiang, L.; et al. Recent Advances in DNA Origami-Engineered Nanomaterials and Applications. Chem. Rev. 2023, 123, 3976–4050. [Google Scholar] [CrossRef]
- Lin, C.; Jungmann, R.; Leifer, A.M.; Li, C.; Levner, D.; Church, G.M.; Shih, W.M.; Yin, P. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 2012, 4, 832–839. [Google Scholar] [CrossRef]
- Woehrstein, J.B.; Strauss, M.T.; Ong, L.L.; Wei, B.; Zhang, D.Y.; Jungmann, R.; Yin, P. Sub–100-nm metafluorophores with digitally tunable optical properties self-assembled from DNA. Sci. Adv. 2017, 3, 18–26. [Google Scholar] [CrossRef]
- Arafiles, J.V.V.; Hirose, H.; Akishiba, M.; Tsuji, S.; Imanishi, M.; Futaki, S. Stimulating Macropinocytosis for Intracellular Nucleic Acid and Protein Delivery: A Combined Strategy with Membrane-Lytic Peptides to Facilitate Endosomal Escape. Bioconjugate Chem. 2020, 31, 547–553. [Google Scholar] [CrossRef]
- Sun, H.; Almdal, K.; Andresen, T.L. Expanding the dynamic measurement range for polymeric nanoparticle pH sensors. Chem. Commun. 2011, 47, 5268–5270. [Google Scholar] [CrossRef] [PubMed]
- Benjaminsen, R.V.; Sun, H.; Henriksen, J.R.; Christensen, N.M.; Almdal, K.; Andresen, T.L. Evaluating nanoparticle sensor design for intracellular pH measurements. ACS Nano 2011, 5, 5864–5873. [Google Scholar] [CrossRef]
- Chauhan, V.M.; Burnett, G.R.; Aylott, J.W. Dual-fluorophore ratiometric pH nanosensor with tuneable pKa and extended dynamic range. Analyst 2011, 136, 1799–1801. [Google Scholar] [CrossRef]
- Chauhan, V.M.; Orsi, G.; Brown, A.; Pritchard, D.I.; Aylott, J.W. Mapping the pharyngeal and intestinal pH of Caenorhabditis elegans and real-time luminal pH oscillations using extended dynamic range pH-sensitive nanosensors. ACS Nano 2013, 7, 5577–5587. [Google Scholar] [CrossRef]
- Desai, A.S.; Chauhan, V.M.; Johnston, A.P.; Esler, T.; Aylott, J.W. Fluorescent nanosensors for intracellular measurements: Synthesis, characterization, calibration, and measurement. Front. Physiol. 2014, 4, 401. [Google Scholar] [CrossRef]
- Doussineau, T.; Schulz, A.; Lapresta-Fernandez, A.; Moro, A.; Körsten, S.; Trupp, S.; Mohr, G.J. On the design of fluorescent ratiometric nanosensors. Chem. Eur. J. 2010, 16, 10290–10299. [Google Scholar] [CrossRef]
- Johnson, I.; Spence, M.T.Z. The Molecular Probes Handbook, 11th ed.; Invitrogen Corp.: Carlsbad, CA, USA, 2010. [Google Scholar]
- Ogawa, M.; Kosaka, N.; Choyke, P.L.; Kobayashi, H. H-type dimer formation of fluorophores: A mechanism for activatable, in vivo optical molecular imaging. ACS Chem. Biol. 2009, 4, 535. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Aguilera, O.; Neckers, D.C. Aggregation phenomena in xanthene dyes. Acc. Chem. Res. 1989, 22, 171. [Google Scholar] [CrossRef]
- Selwyn, J.E.; Steinfeld, J.I. Aggregation of equilibriums of xanthene dyes. J. Phys. Chem. 1972, 76, 762. [Google Scholar] [CrossRef]
- Zhang, Z.; Nakata, E.; Shibano, Y.; Morii, T. FRET-Based Cathepsin Probes for Simultaneous Detection of Cathepsin B and D Activities. ChemBioChem 2022, 23, e202200319. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakata, E.; Gerelbaatar, K.; Asif, M.; Konishi, H.; Shibano, Y.; Lin, P.; Morii, T. A Ratiometric Fluorescent Probe for pH Measurement over a Wide Range Composed of Three Types of Fluorophores Assembled on a DNA Scaffold. Chemistry 2023, 5, 1832-1842. https://doi.org/10.3390/chemistry5030125
Nakata E, Gerelbaatar K, Asif M, Konishi H, Shibano Y, Lin P, Morii T. A Ratiometric Fluorescent Probe for pH Measurement over a Wide Range Composed of Three Types of Fluorophores Assembled on a DNA Scaffold. Chemistry. 2023; 5(3):1832-1842. https://doi.org/10.3390/chemistry5030125
Chicago/Turabian StyleNakata, Eiji, Khongorzul Gerelbaatar, Mashal Asif, Hiroaki Konishi, Yuya Shibano, Peng Lin, and Takashi Morii. 2023. "A Ratiometric Fluorescent Probe for pH Measurement over a Wide Range Composed of Three Types of Fluorophores Assembled on a DNA Scaffold" Chemistry 5, no. 3: 1832-1842. https://doi.org/10.3390/chemistry5030125
APA StyleNakata, E., Gerelbaatar, K., Asif, M., Konishi, H., Shibano, Y., Lin, P., & Morii, T. (2023). A Ratiometric Fluorescent Probe for pH Measurement over a Wide Range Composed of Three Types of Fluorophores Assembled on a DNA Scaffold. Chemistry, 5(3), 1832-1842. https://doi.org/10.3390/chemistry5030125