Abstract
The 70% ethanol extract of Artemisia campestris was screened to find PPARγ ligands using the PPARγ ligand-responsive chimera luciferase reporter system. Capillartemisin B was identified as a PPARγ ligand that stimulated lipid accumulation in 3T3-L1 cells. By further purification of PPARγ ligands from a large-scale preparation of the methanol extract of Artemisia campestris, we isolated and identified eupatilin and santaflavone as PPARγ ligands. Weak PPARγ ligand activity of eupatilin or santaflavone in reporter assay was enhanced by a PPARγ antagonist, GW9662, suggesting that santaflavone or eupatilin and GW9662 bound simultaneously to the multiple sub-pockets of the PPARγ ligand-binding domain (LBD) and cooperatively activated PPARγ. Docking simulation suggested that eupatilin binds to the Ω-pocket but not to the AF-2 pocket of Y-shaped PPARγ LBD where artepillin C that differs from capillartemisin B at the C-5′ position without hydroxy group binds. Eupatilin or santaflavone with or without GW9662 did not stimulate lipid accumulation in differentiated 3T3-L1 cells, suggesting that binding of each compound alone or with GW9662 to the Ω-pocket which stimulated the PPARγ-responsive reporter expression was not enough to stimulate lipid accumulation. The PPARγ ligands found in this study have a potential to design the fragment-based drug design of a novel PPARγ ligand that cover the Y-shaped PPARγ LBD.
1. Introduction
Artemisia campestris is a perennial herb belonging to the Asteraceae family, native to a wide region of Eurasia and North America, and is known for its medicinal and culinary properties [1]. In Tunisia, Artemisia campestris, locally named “T’gouft”, has been used in folk medicine for its anti-venom, anti-inflammatory, anti-rheumatic and anti-microbial activities [2]. The leaf extract of Artemisia campestris has been reported to alleviate early diabetic nephropathy in rats by inhibiting protein oxidation and nitric oxide production [3]. Meanwhile, thiazolidinediones, which are synthetic ligands of peroxisome proliferator-activated receptor γ (PPARγ), such as pioglitazone and rosiglitazone, are utilized for the treatment of diabetes [4].
After random screening of PPARγ ligands in various plant extracts, PPARγ ligands in the extract of Artemisia campestris were purified using a reporter-guided system in this study. PPARγ is a kind of nuclear receptor superfamily and plays an important role in adipogenesis and glucose metabolism. PPARγ forms a heterodimer with retinoid X receptor (RXR), recruiting the coactivators in a ligand-dependent manner, and binds to PPAR response elements (PPREs) in the regulatory region of target genes [5]. Thiazolidinediones (TZDs), such as rosiglitazone and pioglitazone, have been clinically utilized as PPARγ agonists for the therapy of type 2 diabetes by improving insulin resistance, but have been associated with serious side effects, including increased fluid retention, body weight gain and cardiovascular risk [6]. Progress in the development of selective PPARγ modulators (SPPARγMs) as safer alternatives to full PPARγ agonists has been achieved. SPPARγMs bind differently to the receptor compared to the full agonist and alter the receptor conformation differently, resulting in differential coactivator recruitment and minimizing the side effects of the TZDs [6,7]. The ligand-binding domain of PPARγ forms a Y-shaped ligand-binding pocket that consists of the “AF-2 pocket” near helix H12 and the “Ω-pocket” near helix 3, the Ω-loop and the β-sheet, and different ligands of PPARγ bind to the different sub-pockets [8]. A hydrogen bond with Tyr473 on helix H12 in the AF-2 pocket with a PPARγ full agonist, such as BRL49653, contributes to the activation of PPARγ [9], while a partial agonist, such as the fatty acid ligand 15-oxoETE, binds to the “Ω-pocket” near helix 3, the Ω-loop and the β-sheet [8]. The ligand-bound AF-2 domain with helix H12 forms the binding interfaces with the steroid receptor coactivator (SRC) family members with an LXXLL motif [10].
In our previous study, ethyl-p-methoxycinnamate identified in the extract of Kaempferia galanga bound to the Ω-pocket cooperatively activated PPARγ with PPARγ antagonist GW9662 that covalently bound to Cys285 [11], and we created a potent PPARγ agonist by linking both ligands [12].
In this study, two types of PPARγ ligands with or without activity to stimulate lipid accumulation in differentiated 3T3-L1 cells were identified from the extract of Artemisia campestris and the latter type of ligand cooperatively activated PPARγ-dependent reporter expression with PPARγ antagonist GW9662. Cooperative activation by the ligand was analyzed by docking simulation.
2. Materials and Methods
2.1. Chemical Reagents and Instruments
Organic solvents for fractionation were purchased from Nacalai Tesque, Kyoto, Japan. 1H NMR spectra were measured and recorded on an Avance I 400 (reference TMS, Burker, Germany).
2.2. Preparation of Plant Extracts and Purification of PPARγ Ligands
Dried aerial parts of Artemisia campestris (103.3 g or 557.4 g) were extracted with 70% ethanol (EtOH, 1.5 L) or 100% methanol (MeOH, 4 L), respectively, for 2 weeks at room temperature. After filtration, the filtrate was evaporated to dryness in vacuo at 40 °C to afford the 70% EtOH extract (20.8 g) or MeOH extract (72.3 g). Purification steps of the extracts are described in Supplemental Figure S1a,b. The 70% EtOH extract or the methanol extract were partitioned between ethyl acetate (EtOAc), butanol (BuOH) and H2O. The EtOAc-soluble portions (8.1 g or 52.92 g) of each extract were subjected to further purification. For the further purification, silica gel column chromatography (Wako gel C-200, Fujifilm Wako Pure Chemical Corporation, Osaka, Japan; YFLC AI-580, Yamazen Corporation, Osaka, Japan) or HPLC (Chromaster, Hitachi High-Tech Science Corporation, Tokyo, Japan) using an InertSep Slim C18-B (GL Sciences, Osaka Japan) or ODS-3 column (GL Sciences) was employed as described in Supplemental Figure S1a,b.
2.3. Cell Culture
HepG2 cells or 3T3-L1 cells (JCRB, Osaka, Japan) were grown in Dulbecco’s modified Eagle medium (Fujifilm Wako) supplemented with 10% fetal bovine serum (Biosera, Nuaille, France), 100 U/mL of penicillin and 100 μg/mL streptomycin (GIBCO BRL, Palo Alto, CA, USA) at 37 °C in a humidified 5% CO2 atmosphere.
2.4. PPARγ Reporter Assay
pGal4-PPARγLBD and pUAS-tk-luc were cotransfected with pact-βgal into HepG2 cells using Hilymax (Dojindo, Kumamoto, Japan) and PPARγ ligand activity was determined by luciferase activity using the Luciferase Assay System according to the manufacturer’s protocol (Promega, Madison, WI) normalized by β-galactosidase activity using chlorophenol red β-d-galactopyranoside (Roche Diagnostics GmbH, Germany) as a substrate, as described previously [13]. A schematic reporter construct employed in this study is shown in Supplemental Figure S1c.
2.5. Analysis of Lipid Accumulation in the Differentiated 3T3-L1 Cells
Adipose differentiation was induced by treating confluent 3T3-L1 cells for 2 days with a hormonal cocktail containing insulin (Sigma, St. Louis, MO) (5 μg/mL), isobutylmethylxanthine (Sigma) (0.5 mM) and dexamethasone (Fujifilm Wako) (0.25 μM) and then for another 2 days with insulin (5 μg/mL) alone. After the induction of differentiation, the basal medium with or without test samples was replenished every other day for 8 days [14]. Cells were stained with Oil Red-O as described previously [13]. Briefly, cells were fixed with 10% neutral formalin (Fujifilm Wako) followed by staining with 0.3% Oil Red-O solution.
2.6. Docking Study of Eupatilin to PPARγ LBD
A docking study between eupatilin and the PPARγ LBD was performed as described previously [12]. Briefly, Molegro Virtual Docker ver. 6.0.1 (Molegro Aps, Denmark) was used for the prediction of the binding mode of eupatilin in human PPARγ LBD. The ligand structure of eupatilin was drawn using Marvin Sketch 5.11.5. The data of the protein structure and compound to be docked were imported into the docking program according to the instructions provided. Potential ligand-binding sites of proteins were calculated using the Molegro cavity detection algorithm. The simulation of the binding site of eupatilin in the presence of GW9662 was performed by using the crystal structure between human PPARγ LBD and GW9662 (PDB code 3B0R), in which configuration of GW9662 was anchored to the Cys285 residue in the helix H3. Technically, after conversion of GW9662 to a cofactor, eupatilin was docked as a ligand to the complex between human PPARγ LBD and GW9662.
2.7. Statistics
Data are expressed as means ± standard error of the mean (SEM). Differences were considered statistically significant at p < 0.05, assessed using a Student’s t-test.
3. Results
3.1. Identification of Capillartemisin B as a PPARγ Agonist in the Methanol Extract of Artemisia Campestris and Its Biological Activity
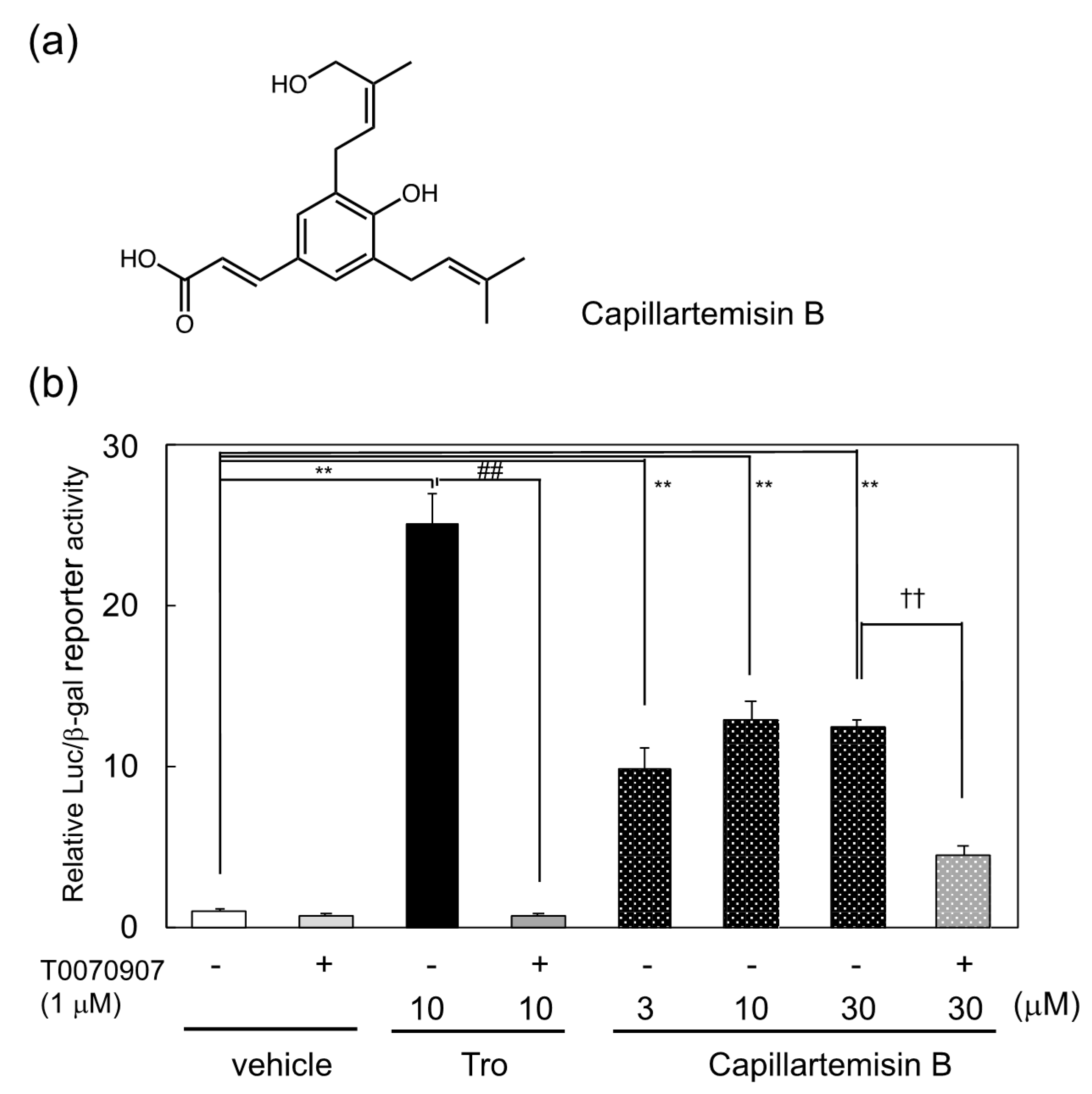
By the use of reporter guided-screening of a PPARγ agonist, capillartemisin B, a kind of phenylpropanoid, was identified as a PPARγ agonist in the 70% ethanol extract of Artemisia campestris. pGAL4-PPARγLBD, a plasmid that expresses a GAL4 DNA-binding domain/PPARγ ligand-binding domain chimera protein, and pUAS-tk-Luc, which contains the target sequence of GAL4, were employed as a PPARγ reporter system as described previously [13]. The structure and reporter activity of capillartemisin B and its purification steps are described in Figure 1a,b and Supplemental Figure S1a. The structure of capillartemisin B purified from the extract was confirmed by NMR spectra to be identical to a previous report [15]. PPARγ ligand activity of capillartemisin B in the reporter assay was suppressed by PPARγ antagonist T0070907, suggesting that capillartemisin B functioned as a PPARγ agonist (Figure 1b).
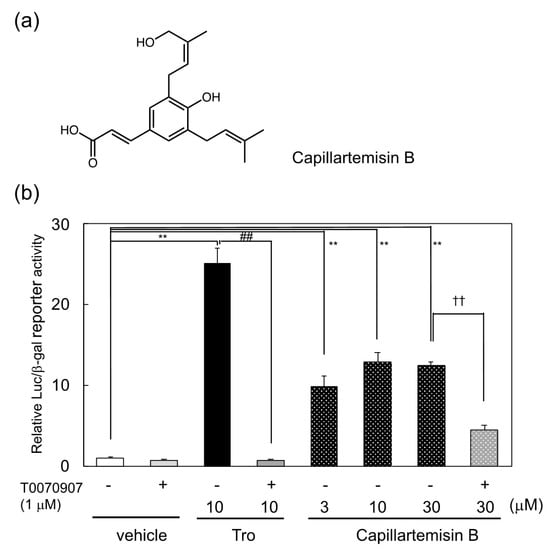
Figure 1.
(a) Chemical structure of capillaetemisin B and (b) induction of PPARγ-dependent reporter activation by capillartemisin B isolated from the 70% ethanol extract of Artemisia campestris. The luciferase reporter assay was performed in HepG2 cells transiently cotransfected with pGal4-PPARγLBD, pUAS-tk-Luc reporter and pact-βgal plasmids. Relative luciferase activities were normalized by β-galactosidase activity. Results are presented as mean ± SD (n = 3). ** p < 0.01 vs. vehicle (DMSO) control. ## p < 0.01 vs. troglitazone treatment. †† p < 0.01 vs. 30 μM carillartemisin B only.
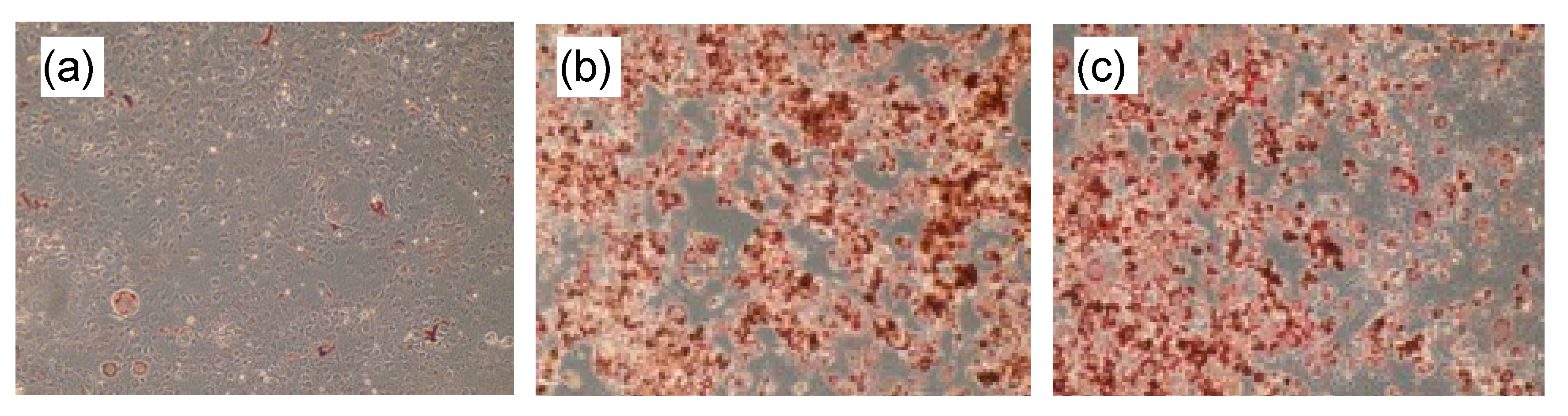
In the differentiated 3T3-L1 preadipocytes to adipocytes, capillartemisin B stimulated lipid accumulation similar to a synthetic PPARγ agonist, troglitazone (Figure 2).
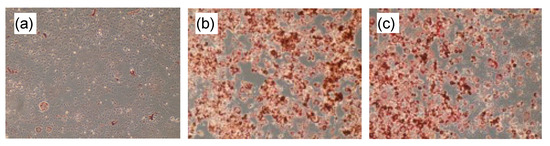
Figure 2.
Lipid accumulation in adipose-differentiated 3T3-L1 cells. After incubation with (a) vehicle (DMSO), (b) 10 μM troglitazone or (c) 30 μM capillartemisin B, cells were stained by Oil Red-O.
3.2. Identification of Santaflavone and Eupatilin as PPARγ Ligands in the Methanol Extracts of Artemisia Campestris That Function as the Cooperating PPARγ Ligands with GW9662
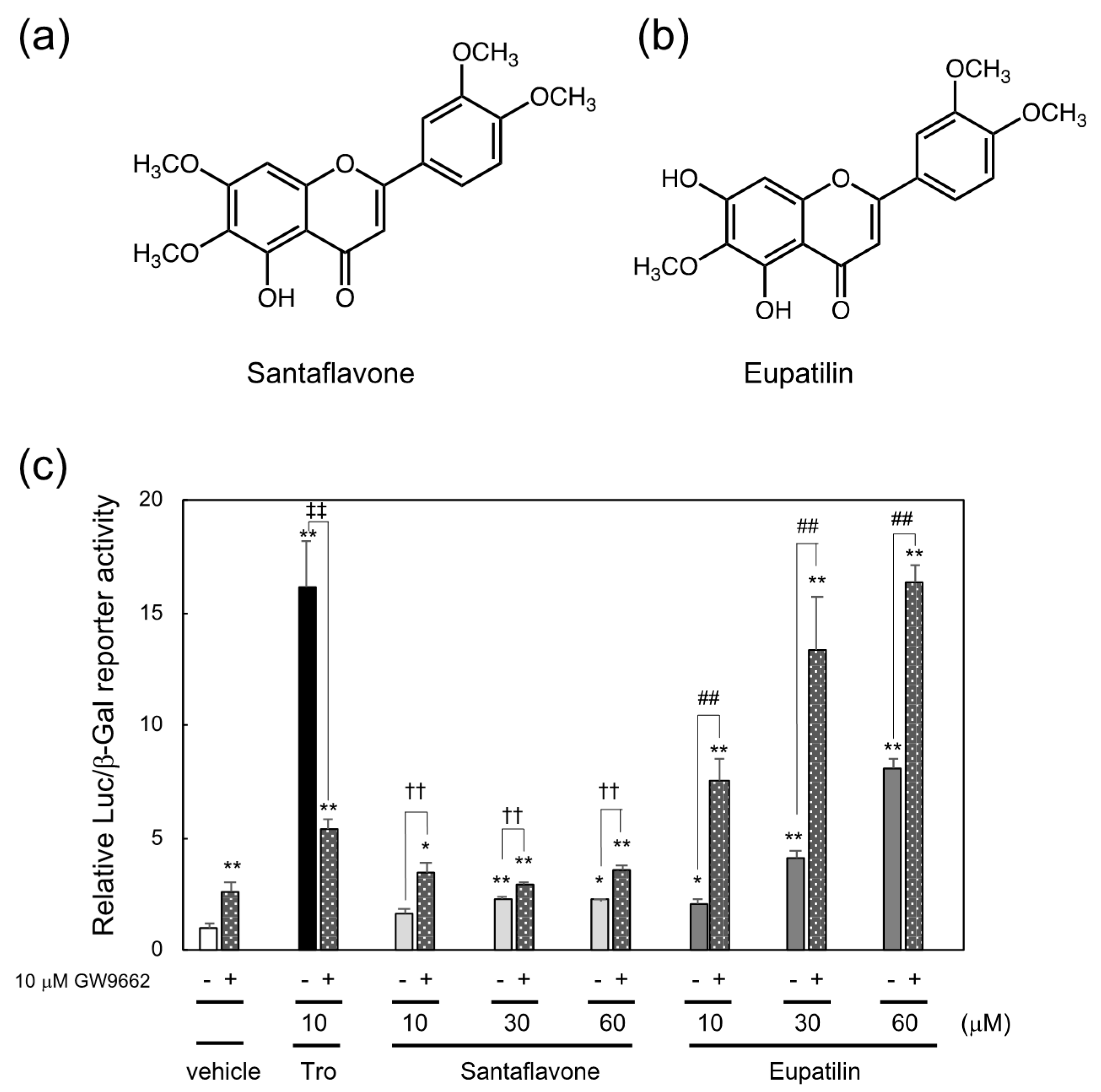
In the second reporter-guided screening of PPARγ ligands in the methanol extract of Artemisia campestris, santaflavone and eupatilin, whose structures were identical to those in previous reports [16,17], were identified as PPARγ ligands (Supplemental Figure S1b, Figure 3c). The structures of santaflavone and eupatilin are shown in Figure 3a,b, respectively. Since santaflavone and eupatilin are structurally related to luteolin, which cooperatively activated PPARγ with PPARγ antagonist GW9662 like ethyl-p-methoxycinnamate as described in the previous study [12], in this study, we examined whether each of these compounds functions as a cooperating ligand for PPARγ and found that santaflavone and eupatilin cooperatively increase PPARγ-dependent reporter expression in the presence of GW9662 (Figure 3c).
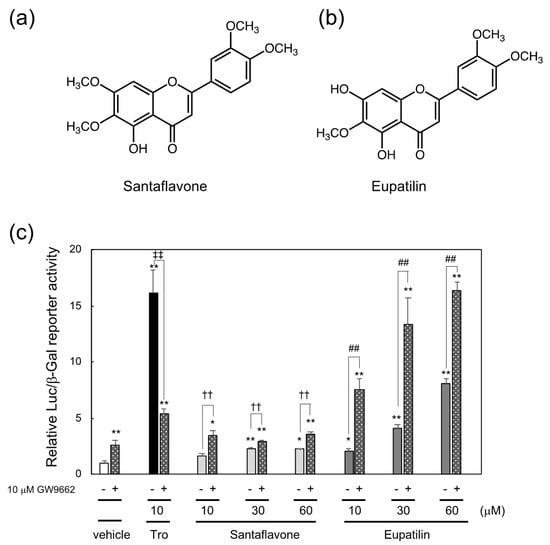
Figure 3.
Chemical structure of (a) santaflavone and (b) eupatilin and (c) induction of PPARγ-dependent reporter activation by santaflavone and eupatilin isolated from the methanol extract of Artemisia campestris and enhancement of their reporter activities by PPARγ antagonist GW9662. * p < 0.05, ** p < 0.01 vs. vehicle control. ‡‡ p < 0.01 vs. 10 μM troglitazone only. †† p < 0.01 vs. santaflavone only, ## p < 0.01 vs. eupatilin only.
3.3. Lipid Accumulation in the 3T3-L1 cells
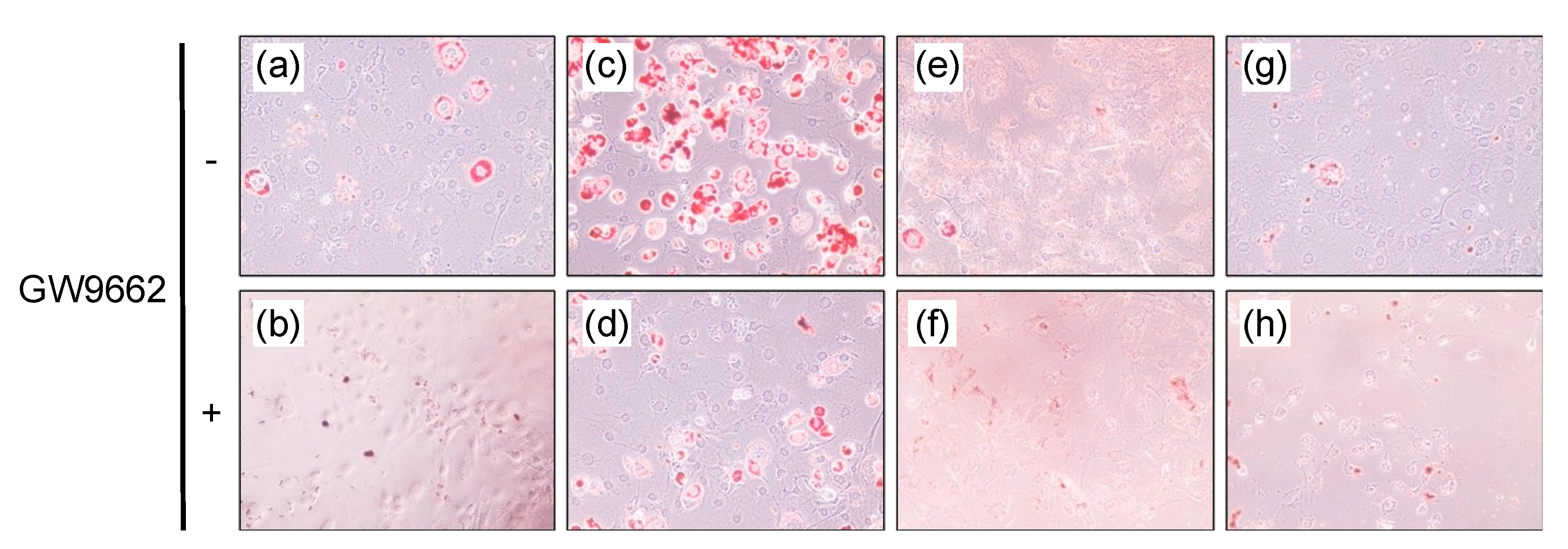
Since a PPARγ agonist, such as troglitazone, stimulates lipid accumulation in adipocytes, differentiated 3T3-L1 cells were employed to examine the effect of santaflavone and eupatilin on lipid accumulation, but santaflavone and eupatilin with or without GW9662 did not stimulate lipid accumulation in 3T3-L1 cells (Figure 4e–h).
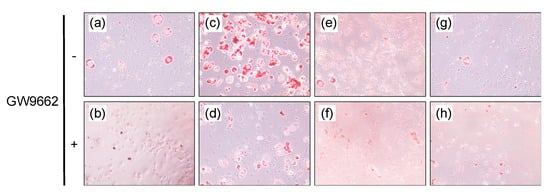
Figure 4.
Lipid accumulation in adipose-differentiated 3T3-L1 cells. After incubation with vehicle (DMSO) (a,b), 10 μM troglitazone (c,d), 60 μM santaflavone (e,f) or 60 μM eupatilin (g,h) without (a,c,e,g) or with (b,d,f,h) 1 μM GW9662, the cells were stained by Oil Red-O.
3.4. Analysis of the Putative Binding Mode of Eupatilin in the GW9662-Bound PPARγ LBD
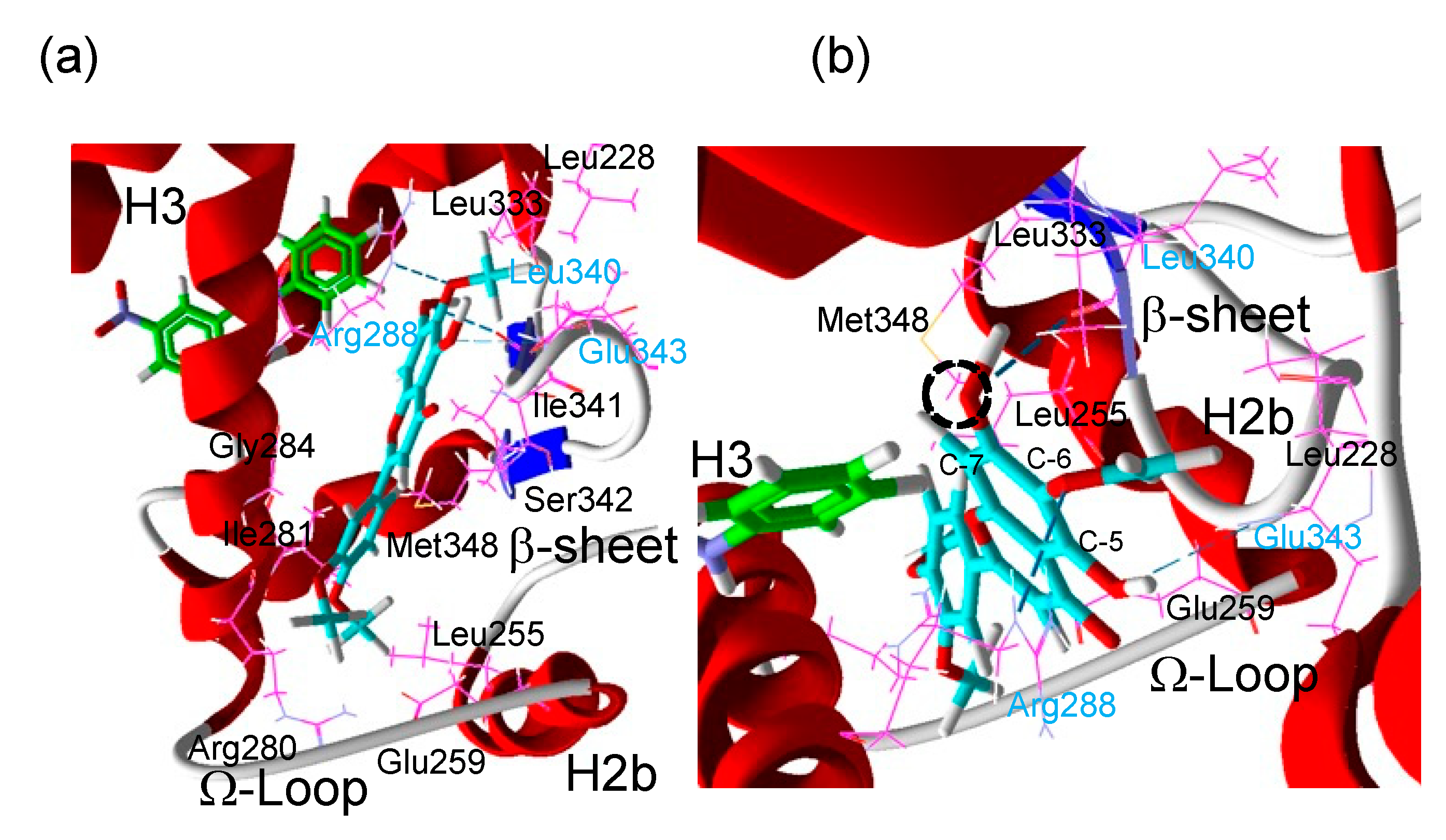
A docking simulation was performed to obtain insight into the putative binding mode of eupatilin. The crystal structure of human PPARγ LBD with GW9662 binding was retrieved from the RCSB Protein Data Bank (PDB code 3B0R), and then after conversion of GW9662 to a cofactor, eupatilin was docked as a ligand to the complex between human PPARγ LBD and GW9662 (Figure 5a,b). The docking predicted that eupatilin occupies the Ω-pocket [8], which is one of the Y-shaped sub-pockets of the PPARγ LBD near helix 3, the Ω-loop and the β-sheet in the presence of GW9662 (Figure 5a,b), in a manner similar to luteolin that cooperatively activated PPARγ with GW9662 [12,18]. Eupatilin was predicted to make three hydrogen bonds with the side chain of Arg288 (on helix 3 (H3)), the carbonyl in the backbone of Leu340 (on β-sheet 3 (β3)) and the amine in the backbone of Glu343 (on β4) and hydrophobic interactions with Leu255 (H2b), Glu259 (Ω-loop), Arg280 (H3), Ile281 (H3), Gly284 (H3), Ile341 (β3), Ser342 (between β3 and β4) and Met348 in the GW9662-bound PPARγ LBD (Figure 6b). From this docking simulation, eupatilin with a hydroxy group at the C-7 position of the flavone (Figure 3b) induced stronger reporter expression (Figure 3c) than santaflavone with a methoxy group at the C-7 position (Figure 3a). This seemed to be reasonable judging from the fitting structure of compounds for this cavity; that is, a small hydroxy group of eupatilin at the C-7 position may be a better fit for this cavity than a large methoxy group of santaflavone at the C-7 position.
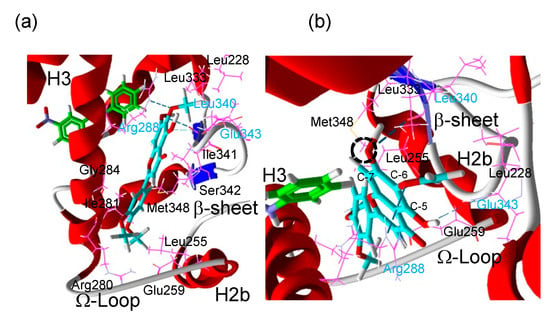
Figure 5.
(a) Putative binding mode of eupatilin (cyan) in the complex between human PPARγ LBD and GW9662 (green). The crystal structure of human PPARγ LBD and GW9662 was retrieved from the RCSB Protein Data Bank (PBD code 3B0R). After conversion of GW9662 to a cofactor, eupatilin was docked as a ligand to the complex of human PPARγ LBD and GW9662. Residues in cyan are predicted to form hydrogen bonds to eupatilin and residues in black are predicted to form hydrophilic interactions with eupatilin. (b) A magnified picture of (a) to show the hydrogen bonds (dashed cyan line). The dashed circle shows an oxygen bound to C-7 in eupatilin where a hydroxy residue (eupatilin) or a methoxy residue (santaflavone) is formed.
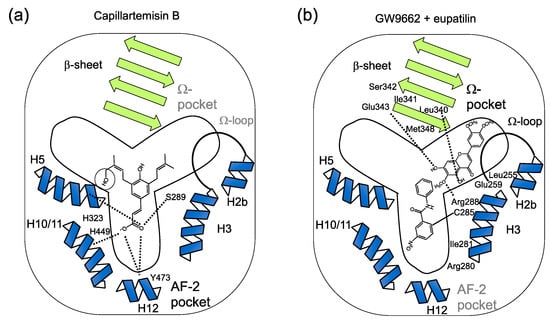
Figure 6.
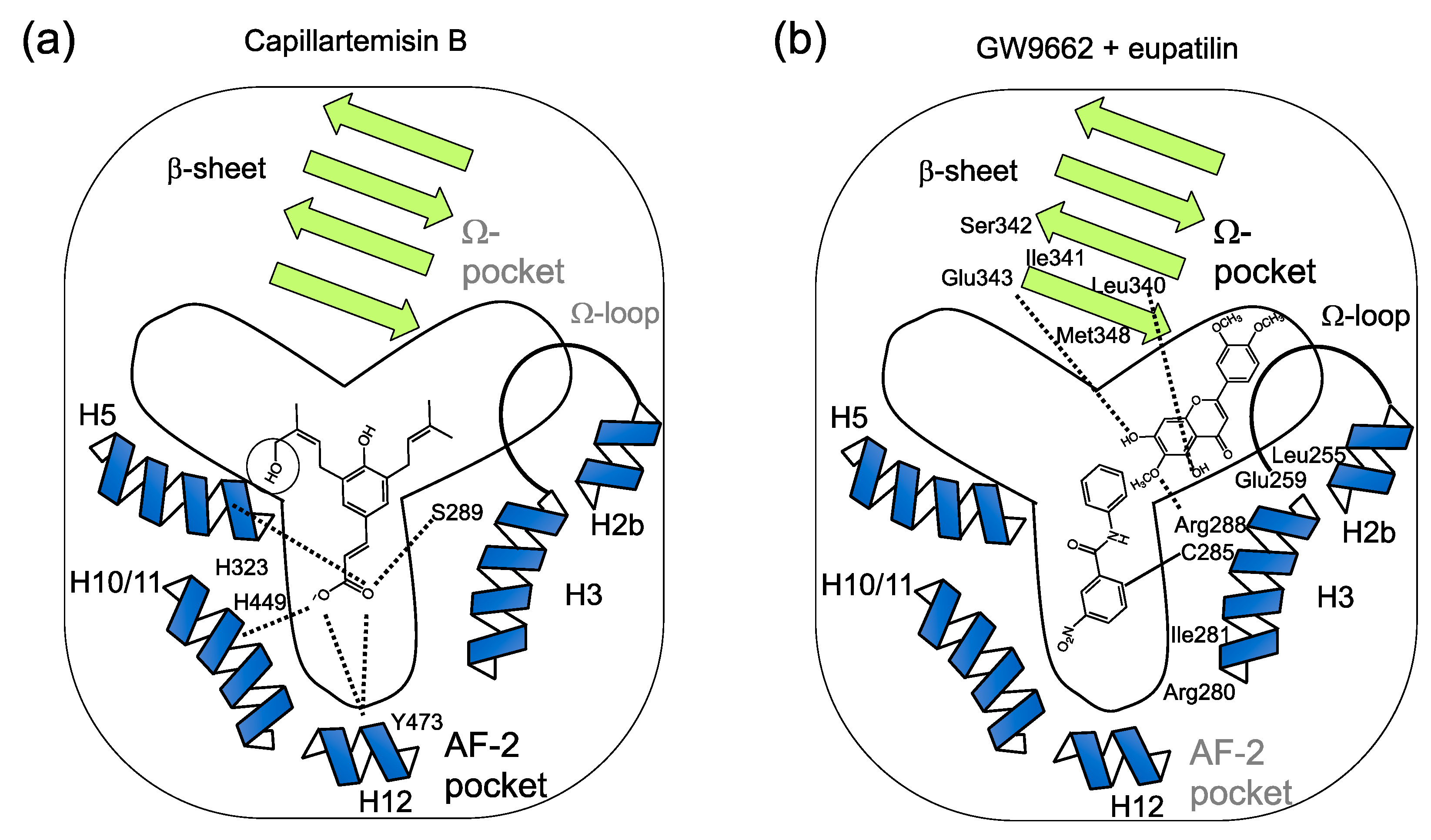
(a) Speculated binding of two different PPARγ ligands to the sub-pockets of a Y-shaped PPARγ ligand-binding pocket. Speculated binding of capillartenisin B to the AF-2 sub-pocket deduced from the binding of artepillin C [19] that differs from capillartemisin B at the C-5’ position without a hydroxy group (circled). Two oxygens at C-9 position are speculated to form hydrogen bonds to His323 (H5) and Ser289 (H3) or His449 (H10/11) and Tyr473 (H12) whose residues form the AF-2 pocket of PPARγ [8,19]. (b) Speculated binding of eupatilin with GW9662 to the Ω-pocket by the docking simulation analysis (Figure 5). The result of docking simulation (Figure 5) is schematically described. Hydrogen bonds between the oxygen at C-5, C-6 and C-7 and the amine on the backbone of Glu343 (around b3), the hydrogen on the side chain of Arg288 (H3) and carbonyl on the backbone of Leu340 (b3), respectively, are described with dotted lines.
4. Discussion
Artemisia campestris is known for its medicinal and culinary properties. A PPARγ ligand, capillartemisin B, identified in the extract of Artemisia campestris in this study (Figure 1a) may partially support the anti-diabetic activity of this plant [3], since PPARγ is involved in the regulation of glucose and lipid metabolism that is related to diabetes [4]. Capillartemisin B stimulated lipid accumulation in differentiated 3T3-L1 cells similar to a PPARγ full agonist, troglitazone, a kind of thiazolidine derivative (TZD) (Figure 2), but eupatilin and santaflavone, that were also identified in the extract of Artemisia campestris as PPARγ ligands with or without GW9662, did not stimulate lipid accumulation (Figure 4e–h), although these compounds stimulated PPARγ-dependent reporter expression (Figure 3c). As described below, the structure of the complex between PPARγ LBD and each ligand determines the biological function of each PPARγ ligand.
PPARγ ligands, TZDs, such as pioglitazone and rosiglitazone, have been utilized as clinical medicine for diabetes, but adverse effects, such as weight gain and heart attack risk, have been noted. Safer PPARγ agonists for the treatment of diabetes are needed. The release of corepressors from PPARγ or recruitment of coactivators to a ligand–PPARγ complex plays an important role in the transactivation of target genes of PPARγ. For this recruitment of coactivators, the structure of the PPARγ–ligand complex determines what coactivators bind to the complex. Waku et al. demonstrated that PPARγ possesses a Y-shaped ligand-binding domain (LBD) which is divided into two sub-pockets by Cys285 and Met364 [8]. 5-methoxy-indole acetate (MIA), a metabolite of serotonin (5-HT), binds to the sub-pocket (AF-2 pocket) by directly interacting with Tyr473 of the PPARγ LBD, while a fatty acid metabolite, 15-oxo-eicosatetraenoic acid (15-oxoETE), that works as a PPARγ ligand, binds to the distinct sub-pocket (Ω-pocket) [8].
In this study, capillartemisin B, identified in the extract of Artemisia campestris as a PPARγ ligand, stimulated lipid accumulation in 3T3-L1 cells similar to the PPARγ full agonist troglitazone (Figure 1 and Figure 2). PPARγ has a Y-shaped ligand-binding domain that consists of the AF-2 sub-pocket and the Ω-sub-pocket. From the result of the docking simulation between PPARγ LBD and artepillin C, whose structure is identical to capillartemisin B except for the absence of a hydroxy group at 5′-C position substituted by a hydrogen according to Ikeda et al. [19], two oxygens of capillartemisin B at C-9 position are speculated to form hydrogen bonds with Ser289 on helix H3, His323 on helix H5, His449 on helix H10/11 and Tyr473 on helix H12 in the AF-2 pocket, similar to artepillin C (Figure 6a). Structural change around helix H12 between transcriptionally repressive and active conformations of PPARγ LBD by the binding of its ligand is known to be critical for the release of a corepressor and recruitment of a coactivator that contributes to the regulation of PPARγ target gene expression [20]. Capillartemisin B, that activated PPARγ-dependent reporter expression (Figure 1b), stimulated lipid accumulation of the differentiated 3T3-L1 adipocytes similar to a PPARγ full agonist, troglitazone (Figure 2).
A PPARγ antagonist, GW9662 or T0070907, covalently binds to Cys285, preventing an agonist of TZDs, such as rosiglitazone or pioglitazone, from entering the AF-2 site, while there was an open space among helix H3, the Ω-loop and the β-sheet (Ω-pocket) in the GW9662-bound PPARγ- LBD (Figure 6b), also termed as an alternative binding site [21]. In the previous study, we described that ethyl-p-methoxycinnamate or its hydrolyzed product p-methoxycinnamic acid exhibited a cooperative effect with GW9662 on PPARγ transactivation [12]. Luteolin, a flavonoid that has hydroxy groups at C-6, C-3′ and C-4 positions instead of a methoxy group compared to eupatilin, also cooperatively activated PPARγ-dependent reporter activity [12]. By using X-ray crystal structural analysis, Puhl et al. reported that luteolin occupied the Ω-pocket and formed two hydrogen bonds with Lys265 and His266 both located at the Ω-loop that links helix H2 and H3 and hydrophobic interactions with Phe264, Gly284, Phe287, Cys285, Arg288, Leu330, Ser342, Ile341, Met364 and Val339 with PPARγ LBD [18]. Several residues of the above were suggested to interact with luteolin and eupatilin in GW9662-bound PPARγ LBD in our previous study [12] and by the present results of the docking simulation of luteolin and eupatilin to GW9662-bound PPARγ LBD. Luteolin induced the expression of the Glut4 gene but did not induce that of the aP2 and adiponectin genes among the PPARγ target genes, suggesting that a subtle difference of conformation of PPARγ LBD caused by each ligand determines the recruitment of coactivators that regulate target gene expression. Eupatilin was reported to inhibit adipogenesis in 3T3-L1 cells through the suppression of PPARγ [22] and this study showed that eupatilin with or without GW9662 did not stimulate lipid accumulation (Figure 4e–h) but that eupatilin cooperatively activated PPARγ-dependent reporter expression (Figure 3c), suggesting that a combination of eupatilin and GW9662 or a single molecule with a hybrid structure of eupatilin and GW9662 may be useful as a strong PPARγ antagonist in lipid accumulation. The other studies suggested that eupatilin is an agonist of PPARα that inhibits the development of oxazolone-induced atopic dermatitis [23] and TNF α-induced metalloproteinase signaling [24]. Santaflavone is a kind of hydroxylated polymethoxyflavone (HPMF) that exists in the Citrus genus and mixtures of PMFs showed chemopreventive activity for colonic tumorigenesis [25] and suppressed the accumulation of lipid droplets in adipocytes [26].
In this study, we described two types of PPARγ ligands that bind to the different sub-pockets of PPARγ LBD from the extracts of Artemisia campestris, one is capillartemisin B that is supposed to bind to the AF-2 pocket of PPARγ LBD (Figure 6a) and the other one includes eupatilin and santaflavone and was predicted to bind to the Ω-pocket (Figure 6b). Capillartemisin B stimulated lipid accumulation while eupatilin and santaflavone with or without GW9662 did not, suggesting that eupatilin or santaflavone activated PPARγ as a partial agonist in a helix H12-independent manner [27] or as antagonists. Since we found several other compounds whose precise structures were not completely identified, but activated PPARγ reporter in the extract of Artemisia campestris, there are many compounds that can be used alone or be used for further chemical decoration to afford more potent compounds through some drug design approaches, including fragment-based drug discovery [28].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/chemistry3020045/s1. Figure S1: Purification scheme and the luciferase reporter system.
Author Contributions
Conceptualization, M.N. (Masaya Nagao) and Y.M.; methodology, M.N. (Masaya Nagao) and Y.M.; software, M.N. (Masaya Nagao) and Y.M.; validation, T.H., M.O. and M.N. (Masaya Nagao); formal analysis, T.H., M.O. and M.N. (Masaya Nagao); investigation, T.H., M.O., Y.M., K.N. and K.I.; resources, K.K. and M.N. (Mohamed Neffati); data curation, M.N. (Masaya Nagao); writing—original draft preparation, M.N. (Masaya Nagao) and Y.M.; supervision, M.N. (Masaya Nagao); project administration, H.I.; funding acquisition, M.N. (Masaya Nagao). All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA)’s Science and Technology Research Partnership for Sustainable Development (SATREPS) Projects: “Valorization of Bio-resources in Semi-Arid and Arid Land for Regional Development” and “Valorization of Bioresources based on Scientific Evidence in Semi and Arid Land for Creation of New Industry” (JPMJSA1506).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
The data presented in this study are available in this article and the Supplementary Materials.
Acknowledgments
pUAS-tk-luc was a gift from Peter J. Ratcliffe, Oxford University, UK, and pact-βgal was a gift from Kazumitsu Ueda, Kyoto University, Japan.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
The docking data are available from the authors.
References
- Dib, I.; El Alaoui-Faris, F.E. Artemisia campestris L.: Review on taxonomical aspects, cytogeography, biological activities and bioactive compounds. Biomed. Pharm. 2019, 109, 1884–1906. [Google Scholar] [CrossRef]
- Megdiche-Ksouri, W.; Trabelsi, N.; Mkadmini, K.; Bourgou, S.; Noumi, A.; Snoussi, M.; Barbria, R.; Tebourbi, O.; Ksouri, R. Artemisia campestris phenolic compounds have antioxidant and antimicrobial activity. Ind. Crop. Prod. 2015, 63, 104–113. [Google Scholar] [CrossRef]
- Sefi, M.; Fetoui, H.; Soudani, N.; Chtourou, Y.; Makni, M.; Zeghal, N. Artemisia campestris leaf extract alleviates early diabetic nephropathy in rats by inhibiting protein oxidation and nitric oxide end products. Pathol. Res. Pr. 2012, 208, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARγamma. Annu Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.V.; Brüne, B.; von Knethen, A. The nuclear hormone receptor PPARγ as a therapeutic target in major diseases. Sci. World J. 2010, 10, 2181–2197. [Google Scholar] [CrossRef] [PubMed]
- Rubenstrunk, A.; Hanf, R.; Hum, D.W.; Fruchart, J.C.; Staels, B. Safety issues and prospects for future generations of PPAR modulators. Biochim. Biophys Acta 2007, 1771, 1065–1081. [Google Scholar] [CrossRef]
- Higgins, L.S.; Depaoli, A.M. Selective peroxisome proliferator-activated receptor gamma (PPARγamma) modulation as a strategy for safer therapeutic PPARγamma activation. Am. J. Clin. Nutr. 2010, 91, 267s–272s. [Google Scholar] [CrossRef] [PubMed]
- Waku, T.; Shiraki, T.; Oyama, T.; Maebara, K.; Nakamori, R.; Morikawa, K. The nuclear receptor PPARγ individually responds to serotonin- and fatty acid-metabolites. Embo. J. 2010, 29, 3395–3407. [Google Scholar] [CrossRef]
- Nolte, R.T.; Wisely, G.B.; Westin, S.; Cobb, J.E.; Lambert, M.H.; Kurokawa, R.; Rosenfeld, M.G.; Willson, T.M.; Glass, C.K.; Milburn, M.V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 1998, 395, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.; Schwabe, J.W. Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 2004, 29, 317–324. [Google Scholar] [CrossRef]
- Leesnitzer, L.M.; Parks, D.J.; Bledsoe, R.K.; Cobb, J.E.; Collins, J.L.; Consler, T.G.; Davis, R.G.; Hull-Ryde, E.A.; Lenhard, J.M.; Patel, L.; et al. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry 2002, 41, 6640–6650. [Google Scholar] [CrossRef]
- Ohtera, A.; Miyamae, Y.; Yoshida, K.; Maejima, K.; Akita, T.; Kakizuka, A.; Irie, K.; Masuda, S.; Kambe, T.; Nagao, M. Identification of a New Type of Covalent PPARγ Agonist using a Ligand-Linking Strategy. Acs Chem. Biol. 2015, 10, 2794–2804. [Google Scholar] [CrossRef]
- Ohtera, A.; Miyamae, Y.; Nakai, N.; Kawachi, A.; Kawada, K.; Han, J.; Isoda, H.; Neffati, M.; Akita, T.; Maejima, K.; et al. Identification of 6-octadecynoic acid from a methanol extract of Marrubium vulgare L. as a peroxisome proliferator-activated receptor γ agonist. Biochem. Biophys Res. Commun. 2013, 440, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, H.; Ogawa, W.; Matsumoto, M.; Kuroda, S.; Takata, M.; Sugimoto, T.; Spiegelman, B.M.; Kasuga, M. Posttranscriptional control of adipocyte differentiation through activation of phosphoinositide 3-kinase. J. Biol. Chem. 1998, 273, 28945–28952. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, I.; Fukuda, Y.; Yoshihara, M.; Yamahara, J.; Yoshikawa, M. Capillartemisin A and B, two new choleretic principles from Artemisiae Capillaris Herba. Chem Pharm Bull. 1983, 31, 352–355. [Google Scholar] [CrossRef]
- Akkal, S.; Benayache, F.; Bentamene, A.; Medjroubi, K.; Seguin, E.; Tillequin, F. Flavonoid Aglycones from Centaurea napifolia. Chem. Nat. Compd. 2003, 39, 219–220. [Google Scholar] [CrossRef]
- Deng, Y.-r.; Song, A.; Wang, H. Chemical Components of Seriphidium santolium Poljak. J. Chin. Chem. Soc. 2004, 51, 629–636. [Google Scholar] [CrossRef]
- Puhl, A.C.; Bernardes, A.; Silveira, R.L.; Yuan, J.; Campos, J.L.; Saidemberg, D.M.; Palma, M.S.; Cvoro, A.; Ayers, S.D.; Webb, P.; et al. Mode of peroxisome proliferator-activated receptor γ activation by luteolin. Mol. Pharm. 2012, 81, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, R.; Yanagisawa, M.; Takahashi, N.; Kawada, T.; Kumazawa, S.; Yamaotsu, N.; Nakagome, I.; Hirono, S.; Tsuda, T. Brazilian propolis-derived components inhibit TNF-α-mediated downregulation of adiponectin expression via different mechanisms in 3T3-L1 adipocytes. Biochim. Biophys. Acta 2011, 1810, 695–703. [Google Scholar] [CrossRef]
- Shang, J.; Mosure, S.A.; Zheng, J.; Brust, R.; Bass, J.; Nichols, A.; Solt, L.A.; Griffin, P.R.; Kojetin, D.J. A molecular switch regulating transcriptional repression and activation of PPARγ. Nat. Commun. 2020, 11, 956. [Google Scholar] [CrossRef]
- Hughes, T.S.; Giri, P.K.; de Vera, I.M.; Marciano, D.P.; Kuruvilla, D.S.; Shin, Y.; Blayo, A.L.; Kamenecka, T.M.; Burris, T.P.; Griffin, P.R.; et al. An alternate binding site for PPARγ ligands. Nat. Commun. 2014, 5, 3571. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Lee, S.G.; Min, K.; Kwon, T.K.; Kim, H.J.; Nam, J.O. Eupatilin inhibits adipogenesis through suppression of PPARγ activity in 3T3-L1 cells. Biomed. Pharm. 2018, 103, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kim, J.C.; Park, N.J.; Bong, S.K.; Lee, S.; Jegal, H.; Jin, L.T.; Kim, S.M.; Kim, Y.K.; Kim, S.N. Eupatilin, an activator of PPARα, inhibits the development of oxazolone-induced atopic dermatitis symptoms in Balb/c mice. Biochem. Biophy.s Res. Commun 2018, 496, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kim, J.C.; Choi, Y.; Lee, S.; Kang, K.S.; Kim, Y.K.; Kim, S.N. Eupatilin with PPARα agonistic effects inhibits TNFα-induced MMP signaling in HaCaT cells. Biochem. Biophys. Res. Commun. 2017, 493, 220–226. [Google Scholar] [CrossRef]
- Lai, C.S.; Tsai, M.L.; Cheng, A.C.; Li, S.; Lo, C.Y.; Wang, Y.; Xiao, H.; Ho, C.T.; Wang, Y.J.; Pan, M.H. Chemoprevention of colonic tumorigenesis by dietary hydroxylated polymethoxyflavones in azoxymethane-treated mice. Mol. Nutr. Food Res. 2011, 55, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.S.; Ho, M.H.; Tsai, M.L.; Li, S.; Badmaev, V.; Ho, C.T.; Pan, M.H. Suppression of adipogenesis and obesity in high-fat induced mouse model by hydroxylated polymethoxyflavones. J. Agric. Food Chem. 2013, 61, 10320–10328. [Google Scholar] [CrossRef]
- Bruning, J.B.; Chalmers, M.J.; Prasad, S.; Busby, S.A.; Kamenecka, T.M.; He, Y.; Nettles, K.W.; Griffin, P.R. Partial agonists activate PPARγamma using a helix 12 independent mechanism. Structure 2007, 15, 1258–1271. [Google Scholar] [CrossRef]
- Scott, D.E.; Coyne, A.G.; Hudson, S.A.; Abell, C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry 2012, 51, 4990–5003. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).