A Hybrid Learnable Fusion of ConvNeXt and Swin Transformer for Optimized Image Classification
Abstract
1. Introduction
2. Related Works
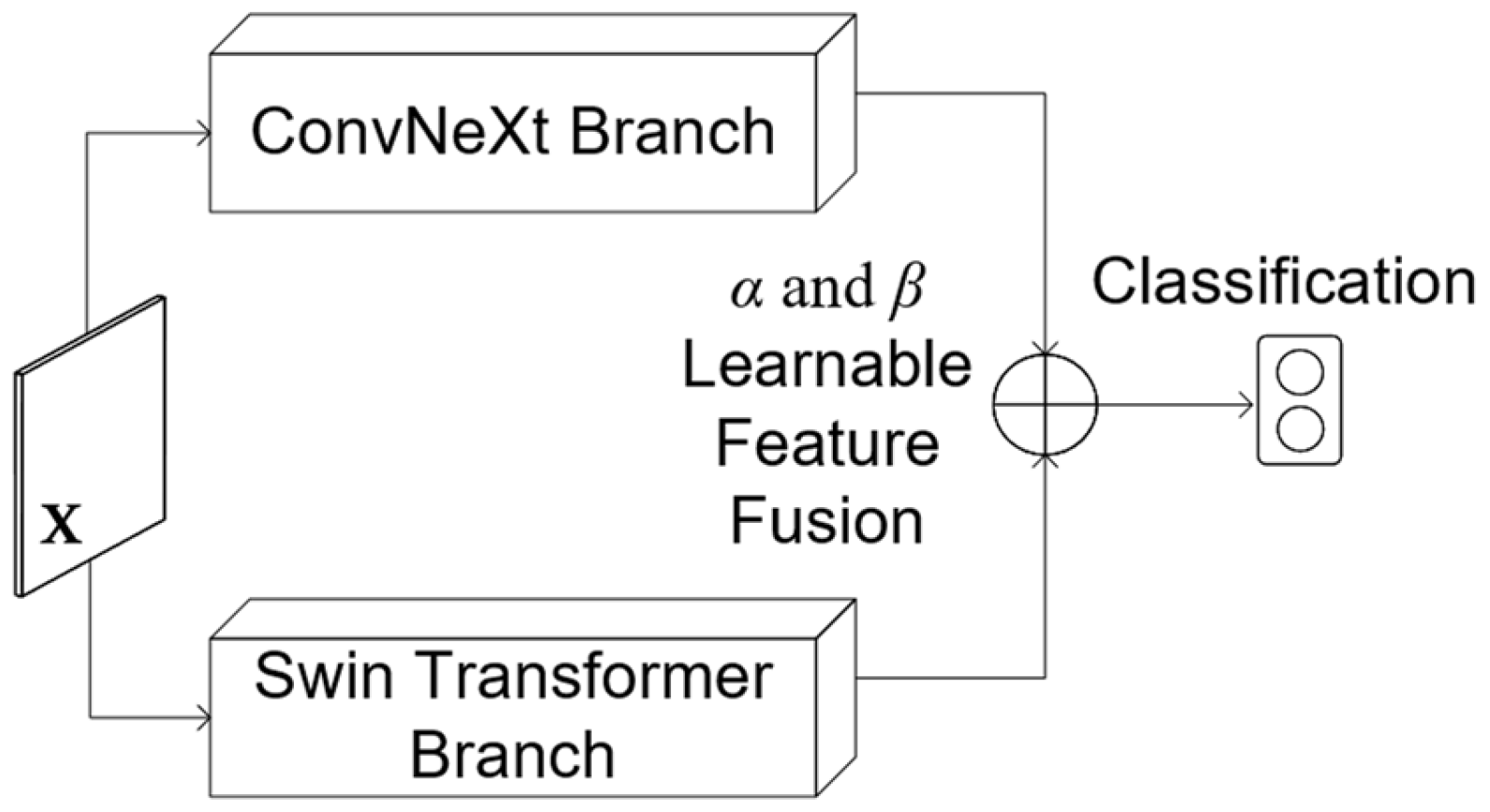
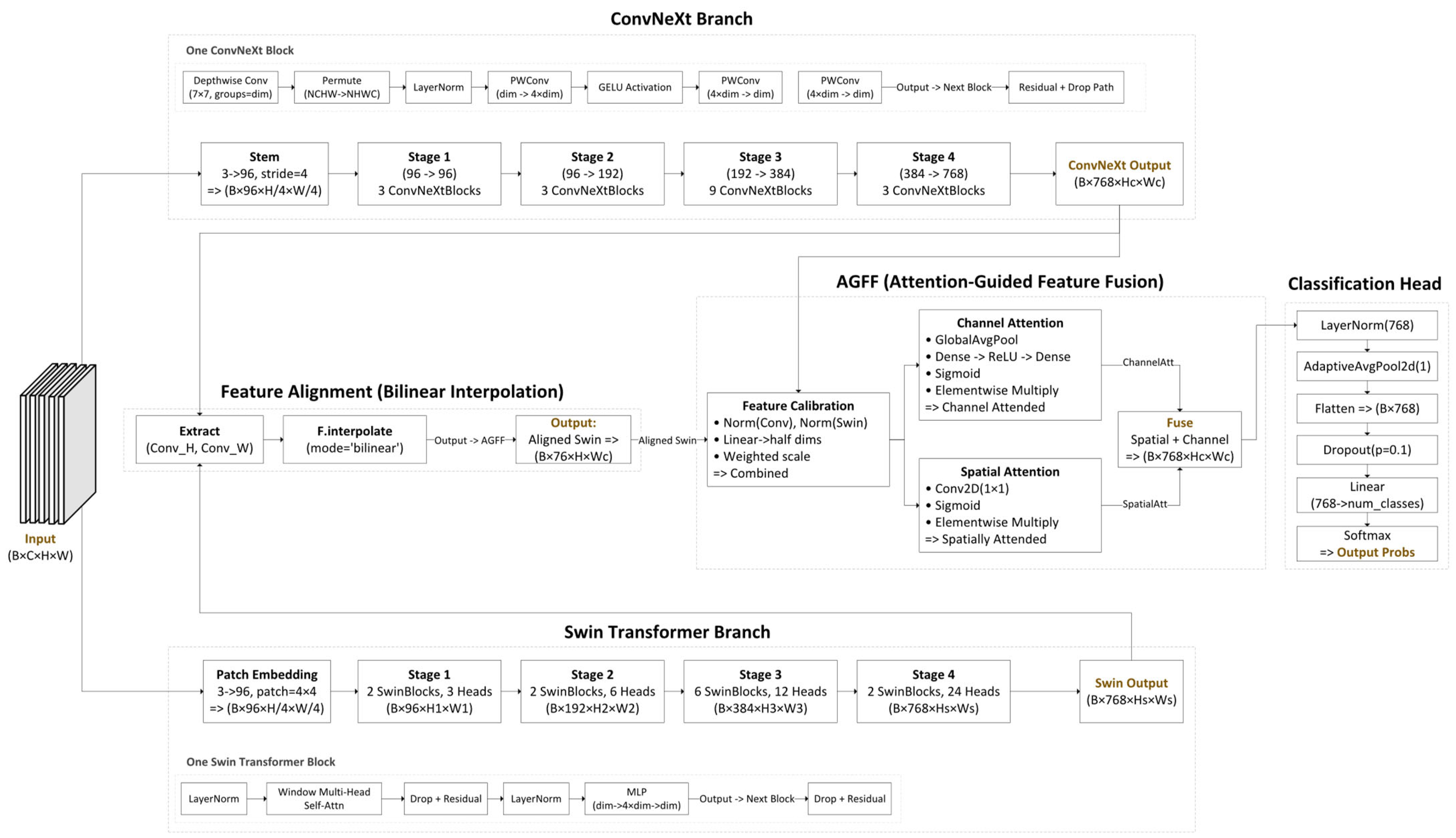
3. The Proposed Architecture
Delving Deeper into the Fusion
4. Experiments
4.1. Comparison Models
4.2. Datasets
4.2.1. Chest X-Ray (Pneumonia) Dataset
4.2.2. Maternal–Fetal US Dataset
4.2.3. Breast Ultrasound Images Dataset
4.2.4. Brain Tumor MRI Dataset
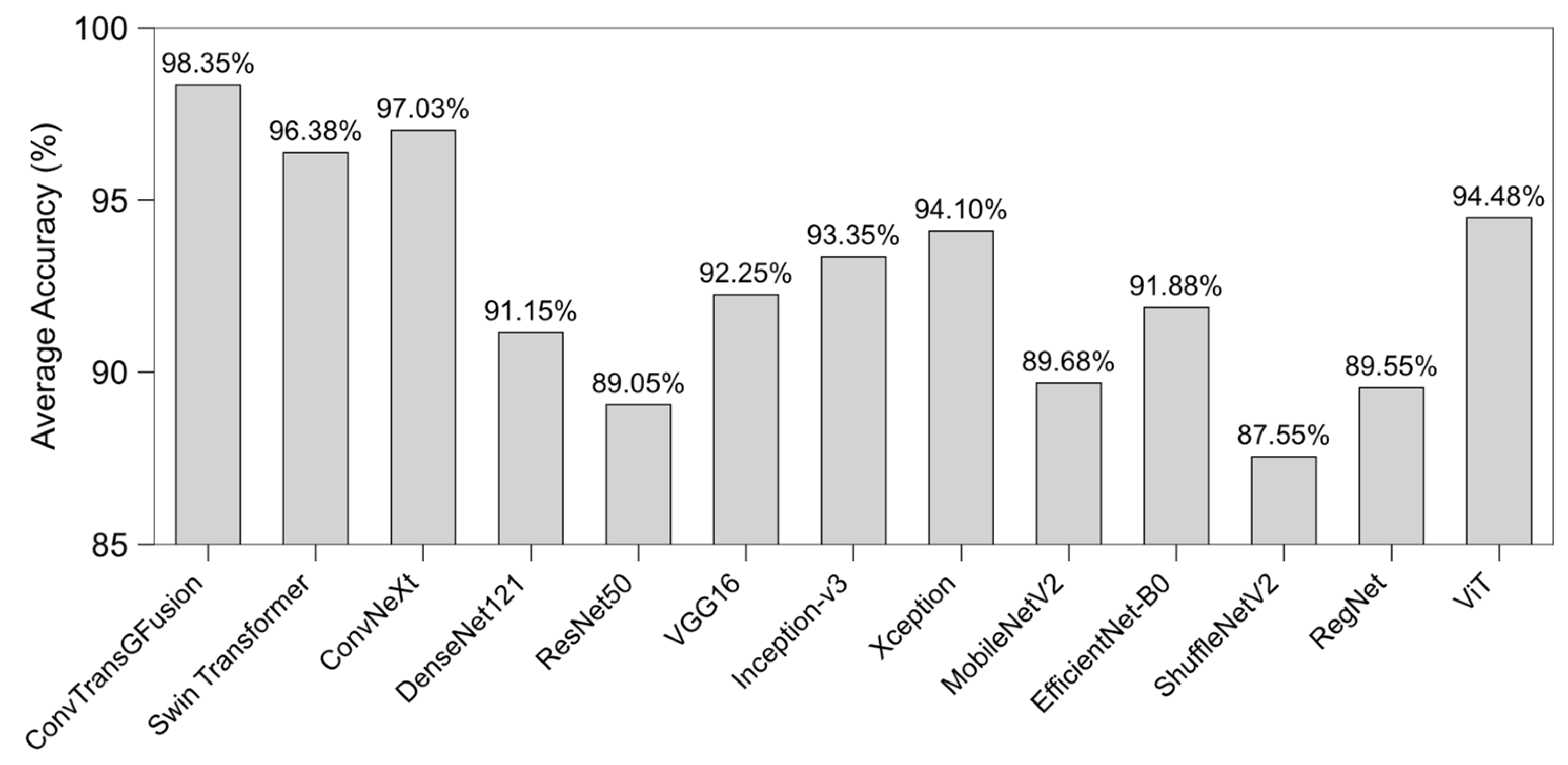
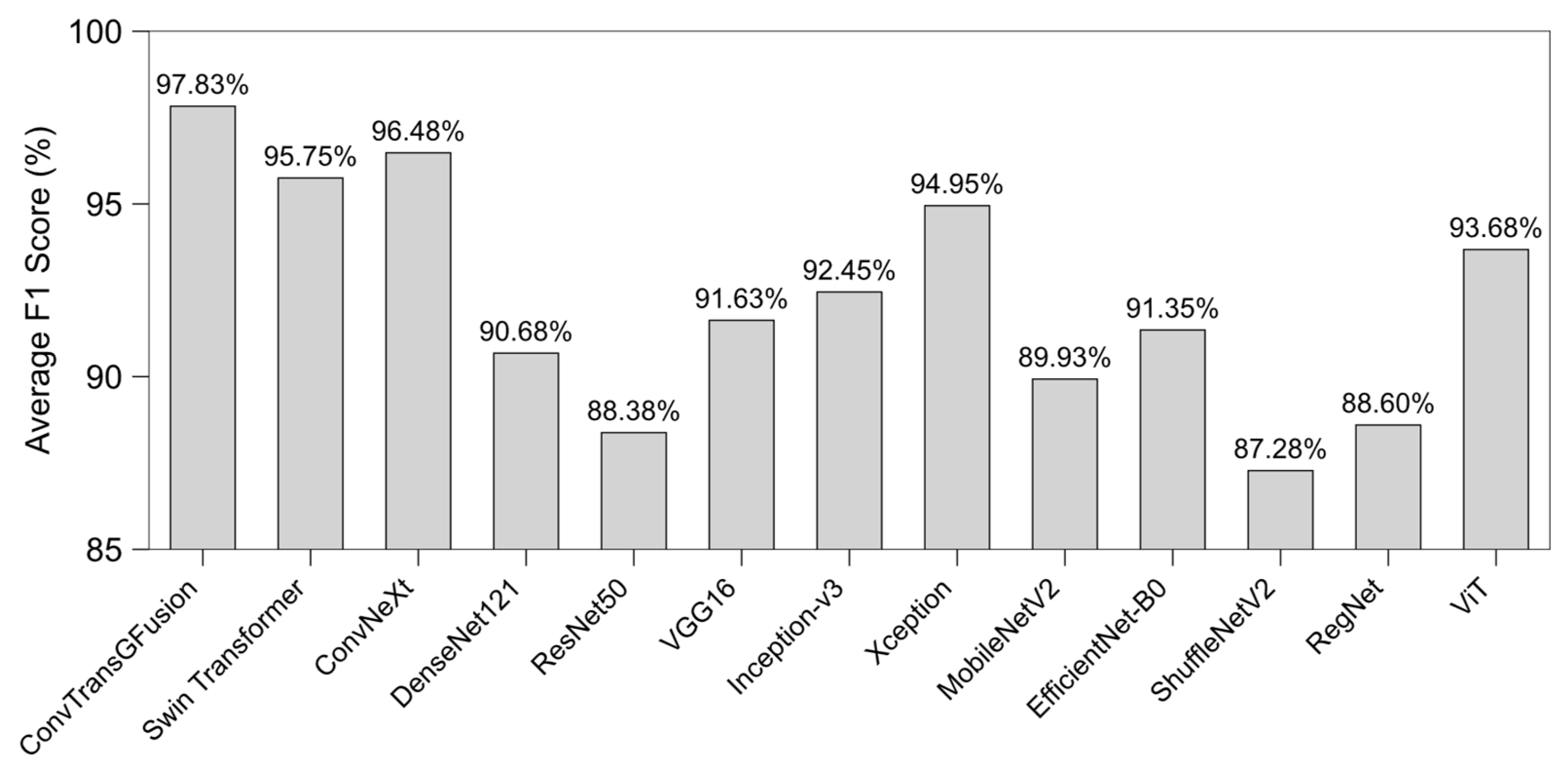
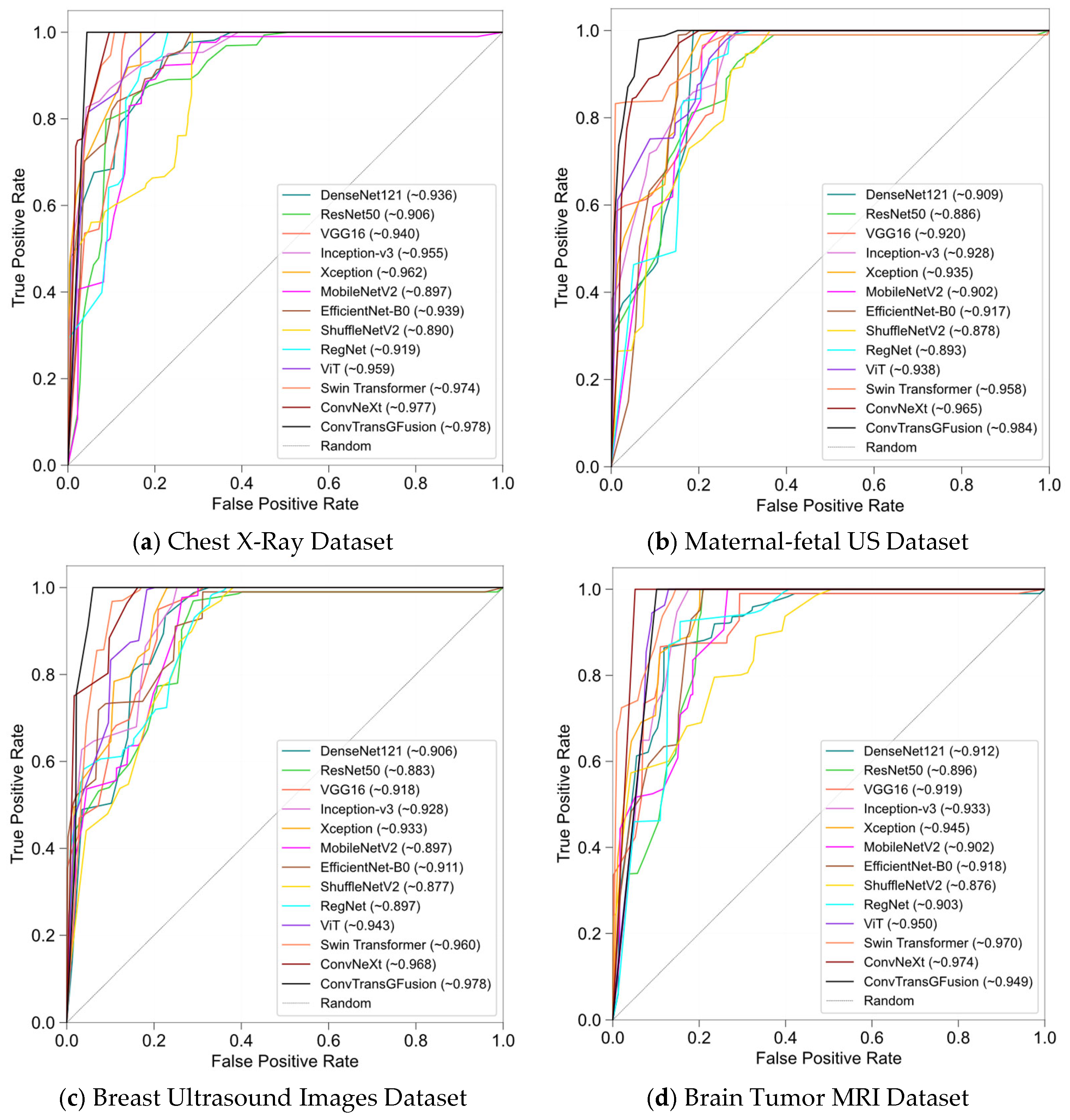
4.3. Results
4.3.1. Ablation Studies
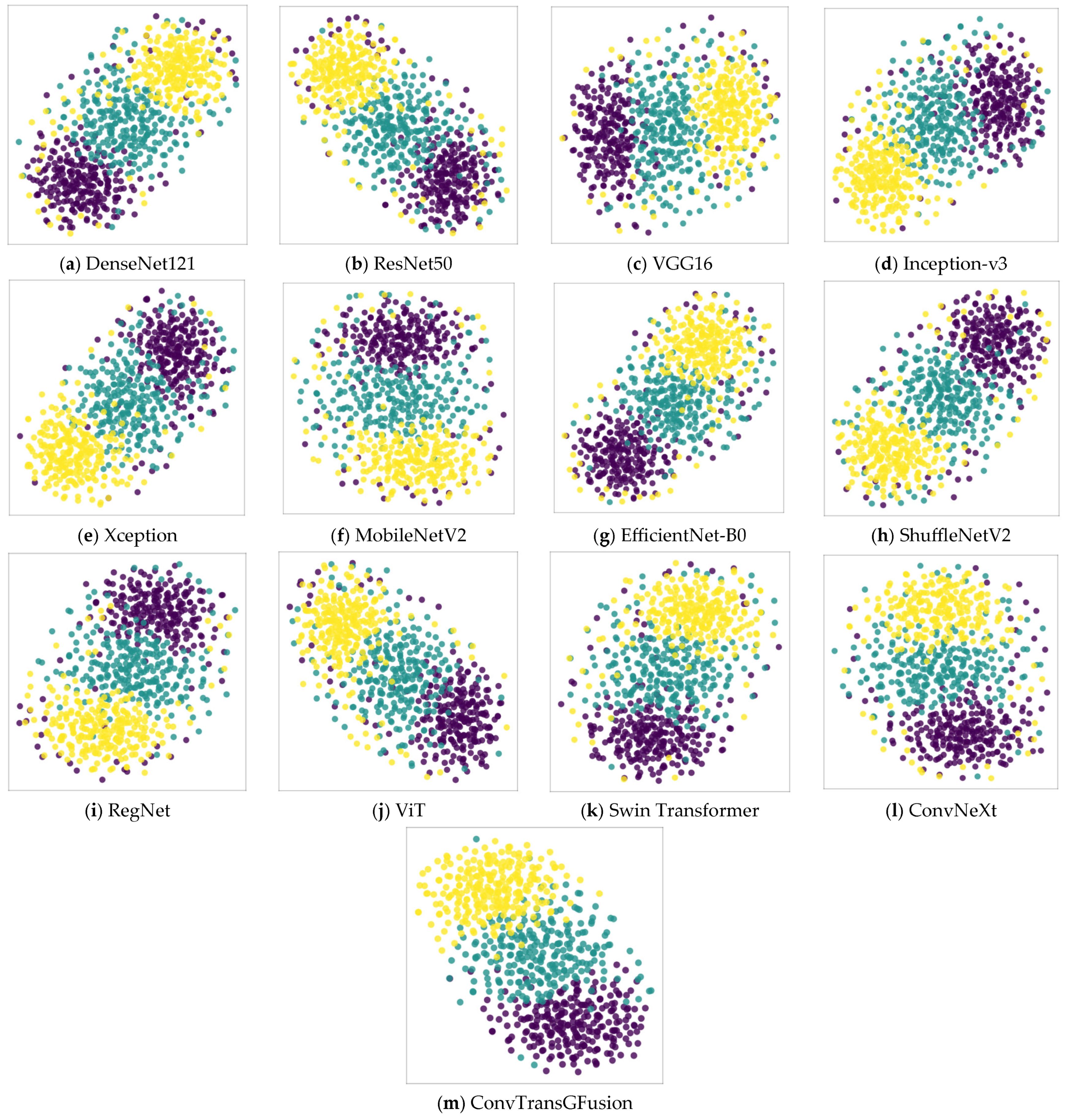
4.3.2. Statistical Validation and Interpretability
4.3.3. Calibration Performance
4.3.4. Robustness and Generalizability
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Model Configurations and Implementation
| Hyperparameter | Default Value | Dataset Variation | Tuning Method | Comments |
|---|---|---|---|---|
| Swin Window Size | 7 | - | Grid Search (5, 7, 9) | A window of size 7 balances local and global capture for typical 224 × 224 inputs. Larger window sizes may give slightly better global coverage but increase memory and training cost. |
| Attention Heads | 8 (initial stage) | Kept at 8 | Empirical + Memory Constraints | More attention heads enable richer feature representation. For higher-res datasets, additional heads capture complex global patterns but require more GPU memory. |
| ConvNeXt Kernel Size | 7 | None (uniform across all datasets) | Based on ConvNeXt Defaults | A large kernel (7) integrates more contextual information per layer, complementing the window-based self-attention in Swin. This synergy helps detect subtle lesions against broader anatomical context. |
| Layer Scale Init | 1e−6 | None | Set by the Literature | A small initialization value (1 × 10−6) stabilizes training, particularly when combining ConvNeXt blocks with transformer layers via fusion. |
| Feature Dimension | 768 | None | Architecture Design | Standard dimension for advanced backbones. Provides a sufficiently expressive feature space for dual-branch fusion without exploding parameter count. |
| Dropout Rate | 0.1 | Maternal–fetal US: 0.2 (if overfitting observed) | Monitored Validation Loss | Reduces overfitting risk, especially on smaller patches or highly repetitive data (ultrasound). For big-data scenarios, dropout can remain lower. |
| Patch Size | 224 × 224 | Maternal–fetal US: 256 × 256 | - | - |
| Brain MRI: 224 × 224 (kept default) | Preliminary Trials + GPU Limits | Larger patch sizes preserve critical details in high-res images. Maternal–fetal US benefits from slightly bigger patches for capturing fetal regions; 224 × 224 is standard to balance detail vs. speed. | - | - |
| Batch Normalization | Momentum = 0.9 | None | Adopted from Standard CNN Practices | Stabilizes updates for deeper networks. A high momentum (0.9) effectively smooths parameter updates in large-batch or small-batch scenarios. |
| Implementation Detail | Setting/Choice | Notes |
|---|---|---|
| Framework and language versions | PyTorch 2.7.0 + Python 3.13.0 | All experiments executed with this exact software stack (CUDA 12.8 backend). |
| Hardware | NVIDIA RTX 3090 (24 GB VRAM) + Intel Xeon Silver 4214 CPU | Single-GPU training; inference latency in Appendix A.2. |
| Train/Val/Test split | 70/10/20 (%), stratified by class | Same protocol for every dataset to ensure comparability. |
| Intensity normalization | Min-max [0, 1] for X-ray and maternal–fetal US datasets; z-score per volume for MRI | Per-image or per-volume scaling to remove scanner-specific intensity bias. |
| Resolution harmonization | Resize to 224 × 224 (MRI, X-ray); 256 × 256 (maternal–fetal US) | Bilinear interpolation; preserves aspect ratio via center crop/pad when necessary. |
| Slice re-orientation (MRI only) | Converted to RAS axial orientation | Ensures anatomical consistency before batching slices. |
| Augmentation pipeline | Random horizontal/vertical flip (p = 0.5); rotation ± 10°; brightness/contrast jitter ± 5%; Gaussian noise σ = 0.01–0.03 (robustness runs) | Implemented with torchvision.transforms. |
| Optimizer | Adam | Learning rate 1 × 10−4; chosen for fast convergence and lower memory overhead than SGD. |
| Learning Rate Decay | Step decay every 10 epochs | Reduces LR by factor of 0.1 to help stabilize training in later epochs. |
| Batch Size | Explored 8, 16, 32, and 64. | Adjusted based on GPU memory constraints; larger for smaller, lower-res images. |
| Total Epochs | 50 | Empirically sufficient for plateau in training and validation metrics. |
| Early Stopping | Patience = 5, Monitor: Val Loss | Stops training if validation loss does not improve for 5 consecutive epochs. |
Appendix A.2. Computational Analysis
| Model | Params (M) | FLOPs (G) | RTX 3090 Latency (ms/img) |
|---|---|---|---|
| ConvTransGFusion | 45.2 | 9.3 | 2.4 ± 0.1 |
| Swin Transformer-Tiny | 28.3 | 4.5 | 1.6 ± 0.1 |
| ConvNeXt-Tiny | 28.6 | 4.6 | 1.7 ± 0.1 |
| DenseNet121 | 25.6 | 4.1 | 1.5 ± 0.1 |
| ResNet50 | 8.0 | 4.0 | 1.4 ± 0.1 |
| VGG16 | 138.4 | 15.5 | 2.1 ± 0.1 |
| Inception-v3 | 23.9 | 11.5 | 1.8 ± 0.1 |
| Xception | 22.9 | 8.4 | 1.9 ± 0.1 |
| MobileNetV2 | 3.4 | 0.3 | 1.2 ± 0.1 |
| EfficientNet-B0 | 5.3 | 0.4 | 1.3 ± 0.1 |
| ShuffleNetV2 | 3.5 | 0.15 | 1.1 ± 0.1 |
| RegNet | 21.9 | 7.2 | 1.7 ± 0.1 |
| ViT-B/16 | 86.4 | 17.6 | 4.9 ± 0.2 |
References
- Huang, X.; Deng, Z.; Li, D.; Yuan, X.; Fu, Y. MISSFormer: An Effective Transformer for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 42, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 11976–11986. [Google Scholar]
- Ma, S.; Chen, C.; Zhang, L.; Yang, X.; Zhang, J.; Zhao, X. AMTrack:Transformer tracking via action information and mix-frequency features. Expert Syst. Appl. 2025, 261, 125451. [Google Scholar] [CrossRef]
- Dosovitskiy, A. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Sa, J.; Ryu, J.; Kim, H. ECTFormer: An efficient Conv-Transformer model design for image recognition. Pattern Recognit. 2025, 159, 111092. [Google Scholar] [CrossRef]
- Ji, X.; Chen, S.; Hao, L.-Y.; Zhou, J.; Chen, L. FBDPN: CNN-Transformer hybrid feature boosting and differential pyramid network for underwater object detection. Expert Syst. Appl. 2024, 256, 124978. [Google Scholar] [CrossRef]
- Manzari, O.N.; Ahmadabadi, H.; Kashiani, H.; Shokouhi, S.B.; Ayatollahi, A. MedViT: A robust vision transformer for generalized medical image classification. Comput. Biol. Med. 2023, 157, 106791. [Google Scholar] [CrossRef]
- Mehta, S.; Rastegari, M. Mobilevit: Light-weight, general-purpose, and mobile-friendly vision transformer. arXiv 2021, arXiv:2110.02178. [Google Scholar]
- D’Ascoli, S.; Touvron, H.; Leavitt, M.L.; Morcos, A.S.; Biroli, G.; Sagun, L. ConViT: Improving Vision Transformers with Soft Convolutional Inductive Biases. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Marina, M., Tong, Z., Eds.; PMLR: Birmingham, UK, 2021; pp. 2286–2296. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 10012–10022. [Google Scholar]
- Chu, X.; Tian, Z.; Wang, Y.; Zhang, B.; Ren, H.; Wei, X.; Xia, H.; Shen, C. Twins: Revisiting the design of spatial attention in vision transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 9355–9366. [Google Scholar]
- Gulsoy, T.; Baykal Kablan, E. FocalNeXt: A ConvNeXt augmented FocalNet architecture for lung cancer classification from CT-scan images. Expert Syst. Appl. 2025, 261, 125553. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Chen, J. Hybrid CNN-Transformer model for medical image segmentation with pyramid convolution and multi-layer perceptron. Biomed. Signal Process. Control 2023, 86, 105331. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, H.; Qin, W. SP-Det: Anchor-based lane detection network with structural prior perception. Pattern Recognit. Lett. 2025, 188, 60–66. [Google Scholar] [CrossRef]
- Nguyen, K.-D.; Zhou, Y.-H.; Nguyen, Q.-V.; Sun, M.-T.; Sakai, K.; Ku, W.-S. SILP: Enhancing skin lesion classification with spatial interaction and local perception. Expert Syst. Appl. 2024, 258, 125094. [Google Scholar] [CrossRef]
- Guo, B.; Qiao, Z.; Zhang, N.; Wang, Y.; Wu, F.; Peng, Q. Attention-based ConvNeXt with a parallel multiscale dilated convolution residual module for fault diagnosis of rotating machinery. Expert Syst. Appl. 2024, 249, 123764. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Q.; Yang, Z.; Li, Y.; Lee, C.H.; Shen, L. Recent progress in transformer-based medical image analysis. Comput. Biol. Med. 2023, 164, 107268. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; et al. A Survey on Vision Transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 87–110. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; PMLR: Birmingham, UK, 2021; pp. 10347–10357. [Google Scholar]
- Nie, X.; Jin, H.; Yan, Y.; Chen, X.; Zhu, Z.; Qi, D. ScopeViT: Scale-Aware Vision Transformer. Pattern Recognit. 2024, 153, 110470. [Google Scholar] [CrossRef]
- Diko, A.; Avola, D.; Cascio, M.; Cinque, L. ReViT: Enhancing vision transformers feature diversity with attention residual connections. Pattern Recognit. 2024, 156, 110853. [Google Scholar] [CrossRef]
- Tan, L.; Wu, H.; Zhu, J.; Liang, Y.; Xia, J. Clinical-inspired skin lesions recognition based on deep hair removal with multi-level feature fusion. Pattern Recognit. 2025, 161, 111325. [Google Scholar] [CrossRef]
- Wen, L.; Ye, Y.; Zuo, L. GAF-Net: A new automated segmentation method based on multiscale feature fusion and feedback module. Pattern Recognit. Lett. 2025, 187, 86–92. [Google Scholar] [CrossRef]
- Fu, J.; Ouyang, A.; Yang, J.; Yang, D.; Ge, G.; Jin, H.; He, B. SMDFnet: Saliency multiscale dense fusion network for MRI and CT image fusion. Comput. Biol. Med. 2025, 185, 109577. [Google Scholar] [CrossRef]
- Zhang, H.; Lian, J.; Yi, Z.; Wu, R.; Lu, X.; Ma, P.; Ma, Y. HAU-Net: Hybrid CNN-transformer for breast ultrasound image segmentation. Biomed. Signal Process. Control 2024, 87, 105427. [Google Scholar] [CrossRef]
- Tuncer, I.; Dogan, S.; Tuncer, T. MobileDenseNeXt: Investigations on biomedical image classification. Expert Syst. Appl. 2024, 255, 124685. [Google Scholar] [CrossRef]
- Raghaw, C.S.; Sharma, A.; Bansal, S.; Rehman, M.Z.U.; Kumar, N. CoTCoNet: An optimized coupled transformer-convolutional network with an adaptive graph reconstruction for leukemia detection. Comput. Biol. Med. 2024, 179, 108821. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yi, X.; Zhang, D.; Zhang, L.; Tian, Y.; Zhou, Z. ConvMedSegNet: A multi-receptive field depthwise convolutional neural network for medical image segmentation. Comput. Biol. Med. 2024, 176, 108559. [Google Scholar] [CrossRef]
- Maqsood, S.; Damaševičius, R.; Shahid, S.; Forkert, N.D. MOX-NET: Multi-stage deep hybrid feature fusion and selection framework for monkeypox classification. Expert Syst. Appl. 2024, 255, 124584. [Google Scholar] [CrossRef]
- Liu, Z.; Shen, L. CECT: Controllable ensemble CNN and transformer for COVID-19 image classification. Comput. Biol. Med. 2024, 173, 108388. [Google Scholar] [CrossRef]
- Liu, H.; Zhuang, Y.; Song, E.; Liao, Y.; Ye, G.; Yang, F.; Xu, X.; Xiao, X.; Hung, C.-C. A 3D boundary-guided hybrid network with convolutions and Transformers for lung tumor segmentation in CT images. Comput. Biol. Med. 2024, 180, 109009. [Google Scholar] [CrossRef]
- Li, J.; Feng, M.; Xia, C. DBCvT: Double Branch Convolutional Transformer for Medical Image Classification. Pattern Recognit. Lett. 2024, 186, 250–257. [Google Scholar] [CrossRef]
- Lau, K.W.; Po, L.-M.; Rehman, Y.A.U. Large Separable Kernel Attention: Rethinking the Large Kernel Attention design in CNN. Expert Syst. Appl. 2024, 236, 121352. [Google Scholar] [CrossRef]
- Khatri, U.; Kwon, G.-R. Diagnosis of Alzheimer’s disease via optimized lightweight convolution-attention and structural MRI. Comput. Biol. Med. 2024, 171, 108116. [Google Scholar] [CrossRef]
- Huo, X.; Tian, S.; Yang, Y.; Yu, L.; Zhang, W.; Li, A. SPA: Self-Peripheral-Attention for central–peripheral interactions in endoscopic image classification and segmentation. Expert Syst. Appl. 2024, 245, 123053. [Google Scholar] [CrossRef]
- Hu, Z.; Mei, W.; Chen, H.; Hou, W. Multi-scale feature fusion and class weight loss for skin lesion classification. Comput. Biol. Med. 2024, 176, 108594. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, Y.; Geng, X.; Tang, H.; Bhatti, U.A. PE-Transformer: Path enhanced transformer for improving underwater object detection. Expert Syst. Appl. 2024, 246, 123253. [Google Scholar] [CrossRef]
- Alharthi, A.G.; Alzahrani, S.M. Do it the transformer way: A comprehensive review of brain and vision transformers for autism spectrum disorder diagnosis and classification. Comput. Biol. Med. 2023, 167, 107667. [Google Scholar] [CrossRef] [PubMed]
- Qezelbash-Chamak, J.; Badamchizadeh, S.; Eshghi, K.; Asadi, Y. A survey of machine learning in kidney disease diagnosis. Mach. Learn. Appl. 2022, 10, 100418. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 4510–4520. [Google Scholar]
- Howard, A.G. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Barata, C.; Celebi, M.E.; Marques, J.S. Explainable skin lesion diagnosis using taxonomies. Pattern Recognit. 2021, 110, 107413. [Google Scholar] [CrossRef]
- Zeng, W.; Huang, J.; Wen, S.; Fu, Z. A masked-face detection algorithm based on M-EIOU loss and improved ConvNeXt. Expert Syst. Appl. 2023, 225, 120037. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]
- Chen, C.-F.R.; Fan, Q.; Panda, R. Crossvit: Cross-attention multi-scale vision transformer for image classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 357–366. [Google Scholar]
- Khan, M.A.; Rubab, S.; Kashif, A.; Sharif, M.I.; Muhammad, N.; Shah, J.H.; Zhang, Y.-D.; Satapathy, S.C. Lungs cancer classification from CT images: An integrated design of contrast based classical features fusion and selection. Pattern Recognit. Lett. 2020, 129, 77–85. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.E9. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Bonet-Carne, E.; Eixarch, E.; Crispi, F.; Gratacós, E. Evaluation of deep convolutional neural networks for automatic classification of common maternal fetal ultrasound planes. Sci. Rep. 2020, 10, 10200. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef] [PubMed]

| Model | Specialization |
|---|---|
| ConvTransGFusion | Proposed hybrid model that leverages ConvNeXt (for local convolutional features) and Swin Transformer (for global self-attention) via an AGFF mechanism. Designed to handle a wide range of image resolutions and mid-scale ultrasound/MRI data. |
| Swin Transformer | A hierarchical transformer utilizing window-based self-attention. Especially efficient for high-resolution chest X-rays and flexible enough for moderate-size maternal–fetal ultrasound or MRI slices. |
| ConvNeXt | A modernized CNN that integrates design elements from vision transformers (e.g., large kernel sizes, streamlined block structures). Shows competitive performance on medical imaging tasks like chest X-ray and breast ultrasound, offering a purely convolutional counterpart to transformers. |
| DenseNet121 | Uses dense connectivity, granting each layer direct access to gradients from every preceding layer. Highly effective in chest X-ray imaging, where feature reuse can aid convergence and performance. |
| ResNet50 | Introduces residual skip connections, enabling deeper CNNs without vanishing gradients. A widely recognized baseline for various medical tasks, including chest X-ray, brain MRI, and ultrasound-based classification, due to robust performance and ease of implementation. |
| VGG16 | A classical CNN with uniform 3 × 3 convolutions and deeper layers, historically essential in computer vision. Remains a go-to baseline for straightforward comparisons in chest X-ray (e.g., pneumonia) and mid-sized data (e.g., maternal–fetal ultrasound). Although parameter-heavy, it offers an older architecture benchmark. |
| Inception-v3 | Employs inception modules for multi-scale feature extraction. Known for handling large images. The parallel convolutional branches can also aid in capturing subtle structural variations in ultrasound (maternal–fetal, breast) or MRI data. |
| Xception | Leverages depthwise separable convolutions to decouple spatial and cross-channel operations, reducing parameters and improving efficiency. Widely used for brain tumor MRI data, where capturing morphological variations with fewer parameters is beneficial. |
| MobileNetV2 | A lightweight CNN with inverted residuals and depthwise separable convolutions. Performs well on low-resource scenarios, including maternal–fetal ultrasound clinics or portable chest X-ray systems. Useful to compare accuracy–efficiency trade-offs across tasks such as brain tumor MRI or breast ultrasound on limited hardware. |
| EfficientNet-B0 | Applies compound scaling (depth, width, resolution) for an optimal accuracy–efficiency balance. Frequently employed in medical imaging (e.g., chest X-ray, breast ultrasound) where varying input sizes are common. Its scaling strategy suits multi-resolution data while maintaining strong performance. |
| ShuffleNetV2 | Focuses on extreme efficiency using channel shuffle and group convolutions. A popular option for real-time or resource-constrained medical imaging tasks, such as remote maternal–fetal ultrasound on limited hardware, due to low-latency inference. |
| RegNet | Derived from design space exploration, providing a family of models covering a broad spectrum of complexities. Exhibits stable performance across different image scales—moderate ultrasound and other large medical images—and helps assess parameter efficiency systematically. |
| Vision Transformer (ViT) | A pure transformer that partitions images into fixed-size patches before applying multi-head self-attention. Scales effectively across imaging resolutions, whether brain tumor MRI or large chest X-rays. Useful for testing if fully transformer-based strategies can outdo or match hybrid/CNN solutions in biomedical contexts. |
| Model | Accuracy | Precision | Recall | Specificity | F1 Score | AUC |
|---|---|---|---|---|---|---|
| ConvTransGFusion | 98.9% | 98.5% | 98.2% | 98.7% | 98.3% | 99.0% |
| Swin Transformer | 96.6% | 96.0% | 95.7% | 96.7% | 95.8% | 97.4% |
| ConvNeXt | 97.1% | 96.4% | 96.2% | 97.0% | 96.3% | 97.8% |
| DenseNet121 | 91.8% | 91.2% | 90.4% | 92.3% | 90.8% | 93.6% |
| ResNet50 | 89.7% | 88.9% | 90.1% | 89.5% | 89.5% | 90.7% |
| VGG16 | 93.2% | 92.7% | 92.0% | 93.6% | 92.3% | 94.1% |
| Inception-v3 | 94.3% | 93.2% | 93.6% | 94.1% | 93.4% | 95.5% |
| Xception | 95.1% | 93.7% | 98.3% | 94.9% | 96.0% | 96.2% |
| MobileNetV2 | 88.6% | 87.9% | 88.5% | 88.8% | 88.2% | 89.8% |
| EfficientNet-B0 | 92.7% | 92.2% | 91.1% | 93.3% | 91.6% | 94.0% |
| ShuffleNetV2 | 87.5% | 88.3% | 86.8% | 87.7% | 87.5% | 88.9% |
| RegNet | 90.3% | 89.5% | 89.2% | 90.7% | 89.3% | 91.8% |
| ViT | 95.0% | 94.2% | 94.4% | 95.2% | 94.3% | 96.0% |
| Model | Accuracy | Precision | Recall | Specificity | F1 Score | AUC |
|---|---|---|---|---|---|---|
| ConvTransGFusion | 98.4% | 97.8% | 97.0% | 98.8% | 97.4% | 98.9% |
| Swin Transformer | 95.9% | 95.1% | 95.3% | 96.2% | 95.2% | 96.5% |
| ConvNeXt | 96.5% | 96.2% | 95.5% | 96.8% | 95.8% | 97.1% |
| DenseNet121 | 90.9% | 90.4% | 90.2% | 91.3% | 90.3% | 92.0% |
| ResNet50 | 88.6% | 88.2% | 87.9% | 88.9% | 88.1% | 89.7% |
| VGG16 | 92.1% | 91.3% | 90.8% | 92.6% | 91.0% | 93.2% |
| Inception-v3 | 92.9% | 92.1% | 91.5% | 93.3% | 91.8% | 94.3% |
| Xception | 93.4% | 92.0% | 94.1% | 92.8% | 93.0% | 94.7% |
| MobileNetV2 | 90.2% | 89.7% | 90.3% | 90.0% | 90.0% | 91.2% |
| EfficientNet-B0 | 91.7% | 91.0% | 90.5% | 92.1% | 90.7% | 93.0% |
| ShuffleNetV2 | 87.9% | 88.5% | 87.8% | 88.1% | 88.2% | 89.3% |
| RegNet | 89.3% | 88.8% | 88.5% | 89.6% | 88.6% | 90.2% |
| ViT | 93.7% | 93.2% | 92.4% | 94.2% | 92.8% | 95.1% |
| Model | Accuracy | Precision | Recall | Specificity | F1 Score | AUC |
|---|---|---|---|---|---|---|
| ConvTransGFusion | 97.9% | 97.1% | 97.4% | 98.1% | 97.2% | 98.6% |
| Swin Transformer | 96.0% | 95.5% | 95.8% | 96.2% | 95.6% | 97.3% |
| ConvNeXt | 96.9% | 96.3% | 96.5% | 97.0% | 96.4% | 97.5% |
| DenseNet121 | 90.6% | 90.2% | 89.8% | 90.9% | 90.0% | 92.1% |
| ResNet50 | 88.3% | 87.6% | 88.1% | 88.5% | 87.9% | 89.9% |
| VGG16 | 91.7% | 91.1% | 90.5% | 92.2% | 90.8% | 93.0% |
| Inception-v3 | 92.8% | 92.2% | 91.4% | 93.4% | 91.8% | 94.1% |
| Xception | 93.3% | 92.4% | 94.0% | 93.1% | 93.2% | 94.9% |
| MobileNetV2 | 89.8% | 88.8% | 89.7% | 90.1% | 89.2% | 90.5% |
| EfficientNet-B0 | 91.2% | 90.7% | 90.3% | 91.6% | 90.5% | 92.8% |
| ShuffleNetV2 | 87.7% | 87.0% | 87.5% | 87.9% | 87.2% | 88.8% |
| RegNet | 89.4% | 88.9% | 88.6% | 89.8% | 88.7% | 90.3% |
| ViT | 94.2% | 93.6% | 93.7% | 94.5% | 93.6% | 95.2% |
| Model | Accuracy | Precision | Recall | Specificity | F1 Score | AUC |
|---|---|---|---|---|---|---|
| ConvTransGFusion | 98.2% | 97.9% | 97.4% | 98.6% | 97.7% | 98.8% |
| Swin Transformer | 97.0% | 96.4% | 96.1% | 97.2% | 96.2% | 97.8% |
| ConvNeXt | 97.6% | 97.0% | 96.7% | 97.4% | 96.8% | 98.2% |
| DenseNet121 | 91.3% | 90.9% | 90.0% | 91.9% | 90.4% | 92.4% |
| ResNet50 | 89.6% | 88.8% | 89.1% | 90.1% | 88.9% | 90.2% |
| VGG16 | 92.0% | 91.4% | 90.9% | 92.6% | 91.1% | 93.3% |
| Inception-v3 | 93.4% | 92.3% | 92.8% | 93.8% | 92.5% | 94.6% |
| Xception | 94.6% | 93.7% | 95.2% | 94.3% | 94.4% | 95.5% |
| MobileNetV2 | 90.1% | 89.3% | 89.8% | 90.4% | 89.6% | 91.1% |
| EfficientNet-B0 | 91.9% | 91.5% | 90.7% | 92.5% | 91.1% | 93.0% |
| ShuffleNetV2 | 87.1% | 86.9% | 87.0% | 87.3% | 86.9% | 88.7% |
| RegNet | 89.2% | 88.7% | 88.1% | 89.5% | 88.4% | 90.0% |
| ViT | 95.0% | 94.4% | 94.2% | 95.3% | 94.3% | 95.9% |
| Model Variant | Channel Attention | Spatial Attention | Learnable Scalars (α,β) | Accuracy | F1 Score |
|---|---|---|---|---|---|
| Baseline (No AGFF) | ✘ | ✘ | ✘ | 88.71% | 90.03% |
| +Channel Attention Only | ✓ | ✘ | ✘ | 90.23% | 91.31% |
| +Spatial Attention Only | ✘ | ✓ | ✘ | 91.40% | 92.12% |
| +Channel and Spatial | ✓ | ✓ | ✘ | 92.39% | 93.10% |
| +Channel and Spatial +Fixed Scalars | ✓ | ✓ | Fixed | 94.18% | 94.05% |
| Full Model (Channel, Spatial, Learnable) | ✓ | ✓ | ✓ | 98.43% | 97.75% |
| Model | Mean Accuracy ± SD | p-Value (vs. ResNet50) | 95% CI of Accuracy | Attention Map/Explanation |
|---|---|---|---|---|
| ResNet50 (Baseline) | 89.32% ± 1.1% | N/A | [88.36%, 90.28%] | N/A (Baseline model) |
| DenseNet121 | 91.27% ± 1.0% | 0.024 | [90.39%, 92.15%] | No specific attention mechanism; highlights edges but misses context. |
| VGG16 | 92.42% ± 1.2% | 0.015 | [91.37%, 93.47%] | Standard Grad-CAM usage; focuses on high-contrast regions. |
| Inception-v3 | 93.40% ± 1.0% | 0.007 | [92.52%, 94.28%] | Multi-scale filters localize certain lesions but can blur boundaries. |
| Xception | 94.10% ± 1.1% | 0.003 | [93.14%, 95.06%] | Depthwise separable convolutions yield moderate interpretability maps. |
| MobileNetV2 | 89.88% ± 1.3% | 0.032 | [88.74%, 91.02%] | Lightweight model; attention maps are coarser, focusing on large areas. |
| EfficientNet-B0 | 91.98% ± 0.9% | 0.010 | [91.19%, 92.77%] | Balanced multi-scale approach with moderate attention detail. |
| ShuffleNetV2 | 87.55% ± 1.4% | 0.084 | [86.32%, 88.78%] | Extremely efficient but less precise focus; attention visuals less clear. |
| RegNet | 89.62% ± 1.1% | 0.020 | [88.66%, 90.58%] | Shows consistent edges in attention but lacks deeper context. |
| ViT | 94.63% ± 0.9% | 0.001 | [93.84%, 95.42%] | Pure attention approach; heatmaps highlight wide image regions. |
| Swin Transformer | 96.52% ± 0.8% | 0.001 | [95.82%, 97.22%] | Window-based attention yields strong context maps, especially for large images. |
| ConvNeXt | 97.20% ± 0.8% | <0.001 | [96.50%, 97.90%] | Convolutional design with large kernels, attention-like receptive fields. |
| ConvTransGFusion | 98.43% ± 0.7% | <0.001 | [97.82%, 99.04%] | Dual-attention fusion maps emphasize both global layout and local lesions. |
| Model | Chest X-Ray | Ultrasound | ||
|---|---|---|---|---|
| Brier Score | ECE (%) | Brier Score | ECE (%) | |
| ConvTransGFusion | 0.048 | 2.1 | 0.045 | 2.0 |
| Swin Transformer | 0.067 | 3.0 | 0.061 | 2.7 |
| ConvNeXt | 0.059 | 2.8 | 0.054 | 2.5 |
| DenseNet121 | 0.103 | 5.3 | 0.108 | 5.6 |
| ResNet50 | 0.101 | 5.4 | 0.102 | 5.3 |
| VGG16 | 0.115 | 6.4 | 0.112 | 6.1 |
| Inception-v3 | 0.087 | 4.3 | 0.082 | 4.0 |
| Xception | 0.075 | 3.4 | 0.071 | 3.2 |
| MobileNetV2 | 0.120 | 6.2 | 0.116 | 6.1 |
| EfficientNet-B0 | 0.094 | 4.7 | 0.089 | 4.4 |
| ShuffleNetV2 | 0.124 | 6.8 | 0.121 | 6.5 |
| RegNet | 0.109 | 5.7 | 0.106 | 5.5 |
| ViT | 0.073 | 3.3 | 0.069 | 3.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qezelbash-Chamak, J.; Hicklin, K. A Hybrid Learnable Fusion of ConvNeXt and Swin Transformer for Optimized Image Classification. IoT 2025, 6, 30. https://doi.org/10.3390/iot6020030
Qezelbash-Chamak J, Hicklin K. A Hybrid Learnable Fusion of ConvNeXt and Swin Transformer for Optimized Image Classification. IoT. 2025; 6(2):30. https://doi.org/10.3390/iot6020030
Chicago/Turabian StyleQezelbash-Chamak, Jaber, and Karen Hicklin. 2025. "A Hybrid Learnable Fusion of ConvNeXt and Swin Transformer for Optimized Image Classification" IoT 6, no. 2: 30. https://doi.org/10.3390/iot6020030
APA StyleQezelbash-Chamak, J., & Hicklin, K. (2025). A Hybrid Learnable Fusion of ConvNeXt and Swin Transformer for Optimized Image Classification. IoT, 6(2), 30. https://doi.org/10.3390/iot6020030






