Abstract
Medical image classification often relies on CNNs to capture local details (e.g., lesions, nodules) or on transformers to model long-range dependencies. However, each paradigm alone is limited in addressing both fine-grained structures and broader anatomical context. We propose ConvTransGFusion, a hybrid model that fuses ConvNeXt (for refined convolutional features) and Swin Transformer (for hierarchical global attention) using a learnable dual-attention gating mechanism. By aligning spatial dimensions, scaling each branch adaptively, and applying both channel and spatial attention, the proposed architecture bridges local and global representations, melding fine-grained lesion details with the broader anatomical context essential for accurate diagnosis. Tested on four diverse medical imaging datasets—including X-ray, ultrasound, and MRI scans—the proposed model consistently achieves superior accuracy, precision, recall, F1, and AUC over state-of-the-art CNNs and transformers. Our findings highlight the benefits of combining convolutional inductive biases and transformer-based global context in a single learnable framework, positioning ConvTransGFusion as a robust and versatile solution for real-world clinical applications.
1. Introduction
The integration of convolution-based models and transformers in medical image analysis has recently attracted considerable attention, as each offers unique advantages in feature extraction and representation. Convolutional networks (ConvNets) such as ResNet and DenseNet have proven successful in learning local patterns and have been widely employed in medical tasks involving MRI images, ultrasound scans, and X-rays [1,2]. Nonetheless, ConvNets may struggle to capture long-range dependencies, which are crucial for discerning subtle clinical indicators across broader regions [3]. Concurrently, transformer-based approaches excel in modeling global context via self-attention, demonstrating strong capabilities in general vision tasks and an emerging potential in medical image recognition [4,5]. Despite these strengths, transformers often lack the inherent local inductive biases that prove beneficial for intricate medical details such as lesions, nodules, and small-scale anomalies [6,7]. Some recent methods combine the two predominant modeling paradigms—convolutional neural networks (CNNs) and transformer-based architectures—to leverage their complementary strengths for improved performance in disease detection [8,9]. Some networks insert lightweight convolutional blocks into transformers to incorporate spatial inductive bias [10], while others rely on window-based or shifted-window attention to limit computational costs and capture local structures [11,12]. For instance, Swin Transformer has proven effective in capturing global relationships by the hierarchical partitioning of medical scans, reducing memory overhead [11]. Meanwhile, newer CNN variants like ConvNeXt adopt design elements from transformers, such as large kernel sizes and inverted bottleneck blocks, to modernize classic convolutional architectures [13,14]. However, simplistic attempts to fuse these distinct representations often involve naive concatenation or summation, leading to either underfitting or an overemphasis on one type of feature.
In the healthcare domain, capturing both local morphological features (e.g., tumor edges, lesion textures) and broader contextual cues (e.g., anatomical relations, multi-lesion distribution) is critical for accurate diagnosis [15,16]. Hybrid strategies that adopt a purely parallel branching or layer-by-layer integration sometimes fail to adaptively weight local and global information, resulting in suboptimal performance [17,18]. To address these challenges, we propose ConvTransGFusion (convolution–transformer global fusion), a hybrid architecture whose name explicitly reflects its core components: a ConvNeXt branch for local convolutional features, a transformer (Swin) branch for global context, and a learnable gated fusion (dual-attention) module that unites the two streams for enhanced disease detection. Our approach carefully aligns feature maps from ConvNeXt, which excels in capturing high-resolution local details, with features from Swin Transformer, which emphasizes long-range dependencies. Then, a dual-attention gating mechanism adaptively merges these features, highlighting both clinically meaningful structural patterns and globally relevant semantic cues [19,20]. Distinct from straightforward multi-stream approaches, the proposed ConvTransGFusion introduces a learnable scaling of branch outputs to let the network emphasize or de-emphasize certain features based on data requirements. Additionally, our method applies dual attention—channel-wise and spatial attention—for integrated local–global synergy; the channel dimension focuses on relative importance across feature maps, while the spatial dimension localizes crucial lesions or subtle anomalies [21,22]. In doing so, ConvTransGFusion aims to replicate a clinician’s diagnostic workflow, such as scanning the image for suspicious cues, zooming in for local details, and correlating findings across regions to reach a precise conclusion [23]. Moreover, practical clinical settings demand a model that robustly handles multi-scale data—from small device-generated ultrasound region-of-interest patches (e.g., 128 × 128 snippets centered on a fetal heart or kidney, where only a few kilobytes of pixel data must still yield a reliable diagnosis) up to full-size chest X-ray images of 2000 × 2000 pixels or larger—without noticeable changes to the network architecture. By leveraging the hierarchical design of Swin Transformer alongside ConvNeXt’s progressive downsampling, our approach elegantly adapts to varying resolutions without cumbersome multi-scale or cross-attention modules [6]. Furthermore, the final classification stage aligns with standard pipelines, simplifying deployment and integration into existing medical software ecosystems. The growing integration of medical imaging devices within the Internet of Things (IoT) landscape presents a pressing need for robust real-time diagnostic algorithms that can operate across distributed or resource-constrained environments. Medical IoT systems often involve wearable or portable ultrasound scanners, bedside X-ray units, and other connected imaging modalities that stream patient data to central servers or cloud platforms for further analysis. In this context, the ability to capture both local morphological features and global contextual cues, as achieved by ConvTransGFusion, becomes essential for dependable and accurate automated diagnosis in telemedicine and point-of-care settings.
2. Related Works
Recent advances in vision-based disease detection underscore the necessity for extracting both local and global features, especially when handling subtle pathological cues and anatomical structures in medical images [2,4,6,7,13,21,22,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42]. Traditional convolutional neural network (CNN) methods [2,41,42] excel at capturing fine local patterns, such as lesion boundaries or nodules, through convolution filters, pooling, and batch normalization. In particular, depthwise separable convolutions reduce parameter overhead, thereby improving efficiency on tasks like mobile or embedded medical devices [9]. Nonetheless, purely convolutional approaches often struggle to model wide-ranging dependencies across an image, which are critical for complex abnormalities, multi-lesion distributions, or extended anatomical structures [16,23]. Consequently, diagnosing diseases that manifest across larger spatial extents (e.g., multiple lung opacities or compound lesions) remains challenging when confined solely to local feature extraction [43].
In contrast, purely transformer-based architectures [4,5,7,20,21] excel in capturing non-local relationships by applying self-attention across the entire image. This global perspective suits scenarios where scattered lesions or large-scale anomalies matter [17,32,44]. Yet, transformers typically lack the built-in spatial priors of convolutions and can demand large-scale training data, which are not always available in medical contexts [35,37]. Therefore, naive applications of transformers to medical imaging can yield incomplete fine-detail modeling and computational inefficiencies, particularly for high-resolution X-ray or MRI scans [25].
Hybrid conv–transformer strategies aim to exploit the strengths of both paradigms [14,28,29,33,36], combining CNN-like local feature capture and transformer-based global dependencies in a unified framework. Approaches such as CoAtNet [45], CrossViT [46], or MobileViT [9] combine parallel or interleaved modules to capture multi-scale characteristics. Some researchers selectively incorporate convolutions into transformers to endow them with local inductive biases [10,12,31], while others adopt window-based or hierarchical self-attention (as in Swin Transformer) to handle large images [3,11]. Such methods often mitigate memory overhead and gather essential long-range information, but fusing feature streams of different resolutions and distributions can be non-trivial. Simple concatenation might underuse or overshadow certain features [19,30], and multiple attention modules can inflate model size. Designs also vary in interpretability, e.g., in medical settings, domain-specific reasoning (e.g., highlighting certain morphological patterns) remains vital for clinical trust [8,23,43]. On the CNN side, ConvNeXt [3,13,17,44] brings design aspects reminiscent of transformers—like more extensive kernel sizes and simplified activation layers—thereby achieving strong performance across standard classification tasks. Meanwhile, on the transformer side, Swin Transformer [11] organizes the image into non-overlapping windows and uses hierarchical patch merging, delivering computationally efficient attention for high-resolution images. Although some recent studies attempt to merge these two backbones, they commonly do so using straightforward additive or concatenation-based fusion, which might not optimally reconcile the distinct features from convolution and self-attention. A more nuanced approach is needed to handle alignment, scaling, and gating of feature maps from different streams, especially for medical data where subtle local details and broad global context are both paramount [35,47]. Furthermore, many advanced attempts incorporate multiple gating or attention approaches [7,37,46] but can suffer from heavy parameter growth or insufficient attention weighting across channels and spatial positions. Some networks adopt channel attention to emphasize or suppress certain feature channels [22,34], while others integrate spatial attention to localize salient structures [7,32]. In medical imaging, where anomalies can be extremely small or widely dispersed, the synergy of channel-level and spatial-level gating becomes especially compelling. Yet, naive upsampling or cross-level fusion can cause resolution mismatches between CNN outputs and transformer embeddings [39], hampering robust synergy. An ideal approach should adapt each branch’s output to a common scale, maintain balanced channel dimensions, and carefully combine local and global signals without overshadowing either. Against this backdrop, the proposed architecture unites ConvNeXt (for local texture emphasis) and Swin Transformer (for global self-attention) in a carefully orchestrated fusion. By aligning their outputs to the same resolution and employing dual-attention gating across both channels and spatial dimensions, the method selectively magnifies critical morphological details and contextual cues. Learned scalar factors allow adaptive weighting of each stream based on the dataset’s needs, and domain-inspired operations ensure attention is paid to suspicious areas reminiscent of clinical focus [17,21,33,37]. Therefore, this approach simultaneously improves accuracy and fosters interpretability, making it well suited for medical applications requiring both fine lesion details and wide image context [25].
Contributions: This study introduces a hybrid local–global architecture that integrates the complementary strengths of ConvNeXt (convolution-based) and Swin Transformer (attention-based) in a unified framework specifically tailored for medical image classification. This synergic approach goes beyond straightforward concatenation by ensuring meticulous alignment of feature maps, partial channel projection, and learnable scaling factors ( and , two trainable scalar coefficients that respectively amplify or attenuate the ConvNeXt and Swin Transformer feature streams during fusion) that adaptively weight the local and global feature streams. The design is further enriched by a dual-attention fusion module that applies channel and spatial attention after the features are merged, enabling the network to highlight clinically relevant anomalies while suppressing irrelevant background noise. Through extensive experiments on four medical imaging datasets—encompassing chest X-rays, maternal–fetal ultrasounds, breast ultrasounds, and brain tumor MRIs—we validate the efficacy of our approach in terms of accuracy, precision, recall, F1, and AUC. Ablation and calibration analyses confirm both the stability and interpretability of the fused model, underscoring its capacity to simultaneously capture nuanced lesion boundaries and broader anatomical relationships in diverse clinical scenarios.
3. The Proposed Architecture
ConvTransGFusion is built upon a mathematical deep dive into how ConvNeXt and the Swin Transformer branches can be jointly leveraged, followed by a novel learnable attention-guided feature fusion (AGFF) block that unites local and global cues in a meticulous way. Several established attention mechanisms, such as squeeze-and-excitation (SE) blocks and convolutional block attention module (CBAM), have already been successfully employed in single-branch convolutional networks to enhance feature representations. However, these methods are not directly tailored for scenarios where two distinct feature streams (i.e., a convolution-based branch and a transformer-based branch) must be fused. In particular, simply inserting SE or CBAM modules into a dual-stream pipeline often overlooks the necessity of feature alignment and lacks a mechanism to adaptively scale each stream’s contribution. By contrast, our dual-attention fusion (channel and spatial) is explicitly designed to (i) align ConvNeXt and Swin Transformer outputs to a common resolution, (ii) project them to partially reduced channel dimensions, and (iii) apply learnable scaling factors before the attention operations. This ensures balanced local–global synergy. Empirical ablation results (see Section 4.3.1) confirm that integrating a tailored dual-attention approach provides superior performance to channel-only or spatial-only alternatives and better suits the hybrid nature of the proposed architecture compared with generic single-branch attention modules.
Let represent the input image (with original dimensions of channels , height , and width ), which is simultaneously passed through two distinct streams of ConvNeXt and Swin Transformer. In the first stream, is handled by ConvNeXt, a CNN architecture that progressively downsamples to extract localized features. Let be the convolution operations of ConvNeXt backbone, and each ConvNeXt layer involves a depthwise convolution
where denotes a standard convolution at layer , followed by layer normalization (LN) as
and a pointwise convolution as
and are the learnable weight tensors at layer that parameterize the two convolutions that make up a depthwise-separable block in ConvNeXt. A residual connection and layer-scale parameter merge the new and old features as
By stacking such multiple blocks, ConvNeXt emphasizes edges, corners, and small localized structures useful for identifying subtle anomalies in medical images. Figure 1 illustrates the overall model, while Figure 2 provides an in-depth view of the proposed ConvTransGFusion architecture.
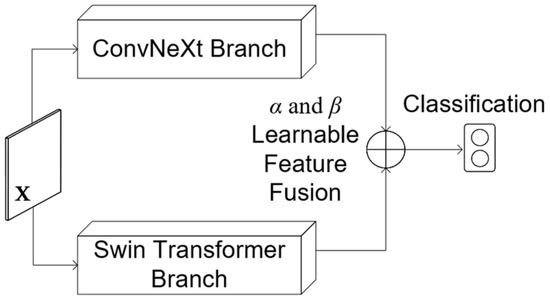
Figure 1.
Overview of the proposed ConvTransGFusion model. An input medical image is processed in parallel with a ConvNeXt branch (local with fine-grained features) and a Swin branch (global with long-range context). The resulting feature maps are spatially aligned, merged by a learnable AGFF block, and finally pooled and classified.
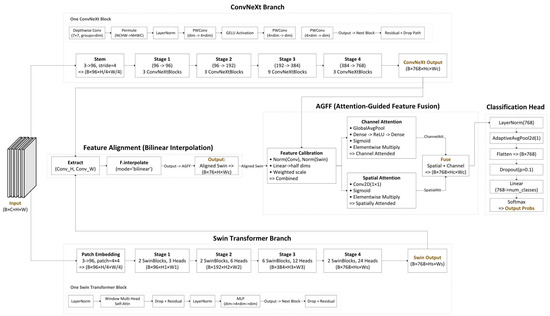
Figure 2.
The architecture of the proposed ConvTransGFusion.
In parallel, the same input image is passed to the Swin Transformer, which begins with a patch embedding layer reducing the spatial resolution by a factor of 4 as . The output is flattened and fed through multiple stages of window-based self-attention. In each Swin Transformer block, let be the input feature, where denotes the batch size, is the number of tokens (spatial positions), and is the embedding dimension. Queries, keys, and values are computed as , and , where each attention head dimension is . The multi-head attention step is . Each Swin block also includes a multilayer perceptron (MLP) feed-forward network with GELU activation and residual connections. By stacking these blocks (with optional downsampling between stages), we obtain . This transformer-based stream excels at capturing long-range dependencies, such as relationships between lesions across different lung regions, providing a global context absent in purely convolutional methods.
Because the spatial dimensions of (convolutional branch) and may differ, ConvTransGFusion aligns them using interpolation:
thus ensuring , . Once aligned, ConvTransGFusion employs a novel AGFF. Initially, each feature map is normalized and linearly projected to half the original channel dimension. Given and , and denoting the projection matrices and , ConvTransGFusion computes:
Each branch is then scaled by learnable scalars
These scaled features are concatenated:
In typical scenarios where , this results in having 768 total channels. Next, a spatial attention component processes . Using a convolution that outputs a single channel, spatial attention is calculated as:
where is the sigmoid function and indicates elementwise multiplication. Spatial attention highlights significant structural regions (such as suspect lesions) while suppressing irrelevant backgrounds. In parallel, channel attention applies global average pooling (GAP) to produce . An MLP followed by sigmoid activation yields channel attention:
Reshaping to and performing elementwise multiplication gives:
Combining spatial and channel attentions results in the final fused feature map:
The dual-attention mechanism uniquely addresses both spatial localization and channel significance, preventing anomalies from being overshadowed by coarse structures or lost among insignificant details. Following fusion, layer normalization is applied to , and spatial dimensions are condensed via global average pooling, resulting in:
Finally, the logits for classification are computed through a linear layer:
In binary classification, probabilities are obtained using the softmax function. This structured integration of convolutional and transformer features, supported by adaptive scaling and dual-attention gating, enables ConvTransGFusion to deliver state-of-the-art performance in tasks like chest X-ray and MRI classification, effectively balancing local lesion details and global anatomical context.
The synergy between local feature extraction and global modeling () is fundamental to ConvTransGFusion. While common architectures might concatenate or add these streams outright, ConvTransGFusion’s learnable scalars allow the network to adaptively emphasize one branch over the other based on the data’s needs. The dual attention in the AGFF goes further by calculating both a spatial attention map and a channel attention vector . This two-path gating mathematically addresses both morphological localization and the relative importance of feature channels, thereby significantly boosting specificity, recall, and F1 performance over single-attention or pure transformer-based models. The alignment step eliminates the dimensionality mismatch between and , making it straightforward to apply pixelwise gating and channel recalibration without complicated multi-scale or cross-attention modules. In practice, the integrated normalization after concatenation reduces covariate shifts between these two distinct feature spaces, and the linear projections into half-channels keep parameter growth manageable. By carefully engineering each layer to mix local and global signals in an interpretable and trainable way, ConvTransGFusion consistently yields state-of-the-art results on tasks like chest X-ray or MRI classification, where subtle local lesions must be viewed in the broader context of intricate anatomical structures. The final outcome is a powerful approach that capitalizes on the best aspects of convolutional networks and transformers, refined by a novel dual-attention gating mechanism that fosters more precise and robust feature representation (see Figure 2).
Figure 2 depicts the architecture of ConvTransGFusion, in which two main branches operate in parallel on the same input image: a ConvNeXt branch (top) for local feature extraction and a Swin Transformer branch (bottom) for capturing broader context. After the respective deep feature representations are extracted, they are aligned through bilinear interpolation to ensure consistent spatial dimensions. The fused feature maps are then processed by the AGFF module, which performs feature calibration (scaling and partial channel projection) followed by dual attention (channel-wise and spatial) to highlight clinically relevant information. In the feature alignment block, a simple bilinear up/down-sampling is used to match the spatial dimensions from the ConvNeXt and Swin Transformer outputs. This ensures that every pixel from each branch corresponds to the same anatomical region. For the feature calibration component, both feature maps undergo layer normalization to mitigate distributional differences and are projected to half their original dimensionality. Learnable scalars and then weight each stream’s contribution, allowing the network to emphasize or suppress local or global features depending on the data. Dual attention includes the following: (i) channel attention identifies which feature channels are most critical, using global average pooling and a two-layer MLP to produce channel weights; (ii) spatial attention highlights the spatial regions of interest—lesions, nodules, or other anomalies—via a 1 × 1 convolution and sigmoid gating. The final sum of channel- and spatial-attended outputs captures both “what is important” in the channels and “where it is located” in the image. A global pooling layer aggregates this information, which is then passed to a linear classifier for the final disease prediction.
Delving Deeper into the Fusion
We propose a dual-attention mechanism—channel-wise and spatial attention—that operates on the fused feature map . The objective is two-fold as follows: (i) to identify the most critical feature channels for disease classification, and (ii) to highlight the spatial regions that contribute the most to the final decision. Our design and fusion strategy are described in detail below.
Given the calibrated feature map , we first apply global average pooling (GAP) to collapse the spatial dimensions and , obtaining a vector . This vector passes through a two-layer MLP with a ReLU activation between the layers, yielding channel weights . We reshape into and perform an elementwise multiplication with to produce the channel-refined output . In parallel, we convolve with a filter to obtain a single-channel spatial map after a sigmoid function. By multiplying elementwise with , we obtain the spatially attended output . To combine channel- and spatial-attended information, we sum up the two intermediate feature maps: . This additive fusion consolidates discriminative patterns emphasized by both attention pathways, ensuring that crucial channel-level and spatial-level cues reinforce each other. The resulting is then passed through normalization, pooling, and, finally, a linear classifier. In this way, the AGFF module adaptively highlights the “what” (channel-level features) and “where” (spatial-level features) for robust local–global representation. Our ablation studies (see Section 4.3.1) confirm the necessity of both attention types for maximizing classification accuracy and F1, as dropping either channel or spatial attention considerably degrades performance.
4. Experiments
We evaluate the proposed algorithm on four publicly available datasets. The following subsections introduce each dataset, describe the experimental procedures, and present the results obtained from our model. For a succinct overview of all key training parameters (e.g., optimizer settings, batch size, dropout rate, and scheduling), readers are directed to the tables in Appendix A.1 and Appendix A.2. These tables encapsulate the dataset-specific and global hyperparameter configurations employed in our experiments.
4.1. Comparison Models
Table 1 outlines the rationale for selecting specific models as gold standards for comparison with ConvTransGFusion in the classification task.

Table 1.
Selected competitive models for comparison experiments and analysis.
4.2. Datasets
We conducted experiments on four distinct publicly available medical datasets covering diverse imaging modalities: chest X-ray, maternal–fetal ultrasound, breast ultrasound, and brain tumor MRI. All four datasets employed in this study were derived from real clinical sources and are widely recognized benchmarks in the medical imaging domain. Each dataset posed unique challenges (e.g., noise, limited contrast, large image sizes, subtle lesions), allowing us to assess the generalization capability of ConvTransGFusion. We used standard data augmentation (resize, random flips, normalization) where appropriate and kept hyperparameters consistent when possible. All models were trained with cross-entropy loss and evaluated via accuracy, recall, precision, F1, specificity, and AUC. All four datasets were standardized through intensity normalization, resolution harmonization, and consistent train/validation/test splits, and were processed with our PyTorch 2.1.0 + Python 3.13.0 codebase. A full step-by-step account of these procedures—including slice re-orientation, augmentation pipelines, software versions, hardware, and the exact 70/10/20 stratified split protocol—is provided in Appendix A.2.
4.2.1. Chest X-Ray (Pneumonia) Dataset
This dataset comprised over 5000 pediatric chest radiographs, with two primary classes: normal lungs and pneumonia-infected lungs [48]. The pneumonia class was further divided into bacterial and viral subcategories, though we treated them collectively as a single “pneumonia” label for binary classification in most experiments. Each image was captured at variable resolutions, so we standardized them to 224 × 224 pixels. Data augmentation (i.e., random flips, slight rotations) helped increase robustness against positional variance and minor image artifacts. Pneumonia detection benefited from both local edge patterns, e.g., detection of opacities or consolidations, and broader lung field context to distinguish actual infection from normal variations. As a result, this dataset highlights how ConvTransGFusion’s integrated local and global feature modeling can outperform simpler CNN- or transformer-only approaches in spotting subtle pneumonia indicators.
4.2.2. Maternal–Fetal US Dataset
This dataset contained 12,400 ultrasound images (dimensions roughly 647 × 381) capturing a range of maternal–fetal conditions (10 categories) [49]. Ultrasound images were particularly noisy, since speckle—a granular multiplicative interference pattern produced when coherent ultrasound echoes randomly combine, which lowers image contrast and can mask fine anatomical details—and fetal positions varied significantly. Preprocessing of the data included normalization and slight random rotations to augment fetal orientation differences. The classification task required distinguishing among multiple potential anomalies (e.g., fetal cardiac or skeletal abnormalities) and healthy conditions.
4.2.3. Breast Ultrasound Images Dataset
This dataset consisted of 780 ultrasound images of the breast (500 × 500 pixels), each labeled into three categories: benign, malignant, or normal [50]. The variability in echogenicity and presence of artifacts demanded robust spatial gating. ConvNeXt helped isolate lesion margins, while Swin Transformer captured the overall breast tissue architecture, leading to improved classification.
4.2.4. Brain Tumor MRI Dataset
We used a 7023 image dataset comprising various tumor types (e.g., glioma, meningioma, pituitary tumor, healthy) at mixed resolutions [51]. The dataset included different MRI slices, intensities, and cross-sections. Local boundary details helped discriminate tumor margins, whereas the global context helped differentiate tumor type and location (e.g., frontal lobe vs. cerebellum). The dual attention in AGFF improved detection accuracy and reduced false positives.
4.3. Results
Table 2, Table 3, Table 4 and Table 5 summarize the quantitative results obtained on the four evaluation datasets—chest X-ray, maternal–fetal ultrasound, breast ultrasound, and brain tumor MRI—each of which constitutes a separate experiment in our study. ConvTransGFusion distinguishes itself on all datasets by uniting high-resolution local features from ConvNeXt with broad contextual cues derived from the Swin Transformer’s self-attention mechanism, then refining the merged representation through a dual-attention gating layer. To contextualize the results, we report six standard metrics: reflects the overall proportion of correct predictions; quantifies how many positively predicted cases are truly positive; measures the fraction of actual positives that are detected; gauges the fraction of actual negatives correctly identified; balances precision and recall; and is the area under the receiver operating characteristic curve, summarizing the trade-off between true-positive and false-positive rates across all decision thresholds.

Table 2.
Performance comparison of chest X-ray dataset.

Table 3.
Performance comparison of maternal–fetal US dataset.

Table 4.
Performance comparison of breast ultrasound dataset.

Table 5.
Performance comparison of brain tumor MRI dataset.
Table 2 represents performance results from the chest X-ray dataset. This dataset predominantly includes normal and pneumonia radiographs, requiring fine-grained detection of subtle opacities. ConvTransGFusion excels overall by combining local edge-focused ConvNeXt features with Swin’s self-attention. Overall, ConvTransGFusion performs the best compared with the other model. We see that Xception’s recall is higher at 98.3%, suggesting it occasionally pinpoints pneumonia cases well, but its overall F1 Score, accuracy, and specificity still trail ConvTransGFusion. Models like DenseNet121 or VGG16 do well at localizing key lesions, but they lack the comprehensive dual-attention gating.
Ultrasound images have strong speckle noise and variable echogenicity, requiring robust noise-insensitive features (Table 3). ConvTransGFusion’s gated fusion helps isolate fetal structures from background artifacts, yielding top accuracy and AUC. Swin Transformer and ConvNeXt also perform strongly, but the dual-attention gating in ConvTransGFusion still delivers a clear margin of improvement—about +1.9 percentage points in accuracy and +1.8 points in AUC—confirming the added value of the fusion strategy.
Breast ultrasound dataset images often have low contrast and speckle noise. ConvTransGFusion’s ability to combine local details (lesion boundaries) with global structure (overall tissue layout) drives its top accuracy, F1, and AUC (Table 4).
Table 5 shows the performance results of the brain tumor MRI dataset. Brain tumor MRI dataset scans vary in tumor type (glioma, meningioma, etc.) and region of interest. ConvTransGFusion’s fusion of local morphological patterns (edges, irregular tumor boundaries) with global context (surrounding tissue) secures the highest accuracy, F1, and AUC. Xception shows a strong sensitivity to tumor presence. However, our model ensures fewer false positives overall.
Across all datasets, our hybrid model leverages two complementary representations—ConvNeXt for local or fine-grained features and Swin Transformer for broader or global context—before passing them through a dual-attention gating process that selectively amplifies important spatial regions and channels. Although certain architectures (e.g., Xception) might show a higher recall on individual tasks, ConvTransGFusion’s balanced approach in gating yields overall better performance in accuracy, specificity, F1, and AUC in most cases. These results illustrate the advantage of context-specific feature merging and highlight how gating mechanisms can unify convolutional and transformer-based strengths.
Figure 3 displays the average accuracies aggregated across all datasets for each model. Notice that ConvTransGFusion consistently situates at the upper-left corner (high accuracy), surpassing both classical CNNs (e.g., ResNet50, DenseNet121) and modern transformers (ViT, Swin). This pattern reinforces the effectiveness of adaptively merging local edges and global contexts for classification in heterogeneous medical images. Some single-stream methods (e.g., ConvNeXt or Swin Transformer) also score highly, but they typically underperform relative to the proposed fusion in at least one dataset.
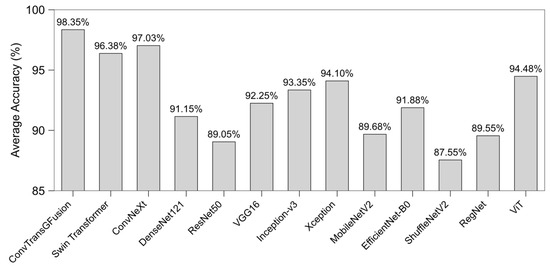
Figure 3.
Average accuracy over all datasets per model.
In Figure 4, we observe average F1 scores, a balanced measure between precision and recall. Again, ConvTransGFusion attains the highest F1 overall, indicating that it is not only accurate but also balanced in detecting both positive and negative instances. Other methods like Xception or ViT do reach comparable F1 in certain tasks, but their performance is less consistent across all datasets. This finding highlights the robust generalization gained by dual-attention gating of local and global features.
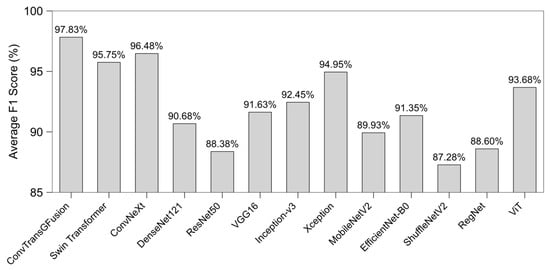
Figure 4.
Average F1 score across all datasets for each compared model. A higher bar indicates a better balance between precision and recall; ConvTransGFusion achieves the highest overall F1 score, reflecting its consistently superior performance.
Figure 5 plots each model’s average precision (y-axis) versus average recall (x-axis) across all datasets. Models that lie near the top-right corner demonstrate strong performance in both precision and recall avoiding false positives (high precision) and minimizing false negatives (high recall). ConvTransGFusion resides at that top-right edge, demonstrating that its synergy between ConvNeXt-like local detail capture and Swin-based global scrutiny yields fewer overlooked abnormalities and fewer misclassifications. Xception occasionally exhibits high recall but comparatively lower precision, placing it slightly further left on the plot, whereas older CNN architectures (e.g., ResNet50, DenseNet121) cluster lower on precision and recall.
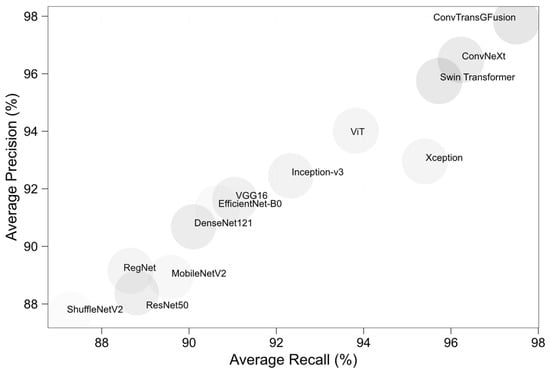
Figure 5.
Average precision vs. average recall for each model.
Figure 6 presents the receiver operating characteristic (ROC) curves for all datasets. Each subplot shows how models compare in terms of true positive rate (sensitivity) versus false positive rate (1−specificity). Across these plots, ConvTransGFusion has curves that hug the top-left corner more tightly, suggesting near-ideal classification thresholds. Notably, the area under the curve (AUC) values reported in the legends frequently exceed 0.96 for our proposed model—often the highest among the tested baselines—indicating that it maintains superior discriminative capability across a range of classification thresholds. While other methods (e.g., Xception, Swin) often keep close pace, the dual-attention mechanism yields a slight but more meaningful advantage in both sensitivity and specificity for most tasks.
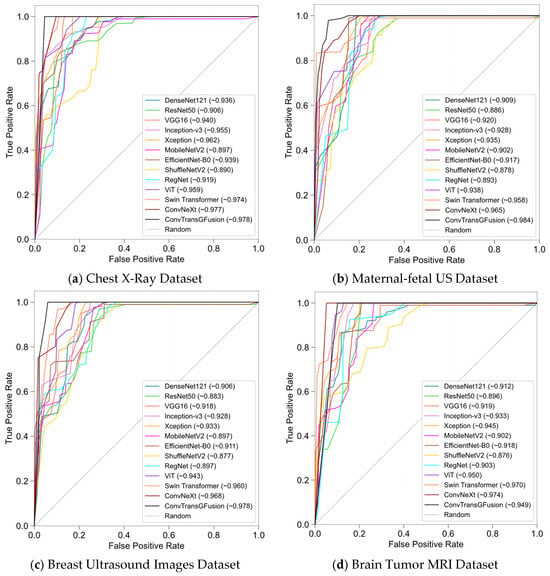
Figure 6.
ROC curves for all models per each dataset.
Figure 7 uses t-SNE embeddings to visualize the learned feature distributions of each model on the breast ultrasound images dataset. Each point represents one image, colored by its ground truth category. Models like DenseNet121 or ResNet50 show somewhat overlapping clusters, suggesting less discriminative boundaries between malignant, benign, and normal categories. In contrast, ConvTransGFusion (panel m) yields well-separated clusters in the 2D embedding space, indicating that it learns high-quality distinct embeddings for each class. This separation aligns with the high classification performance (accuracy ~97.9%) we observed. The synergy of local morphological features and global context fosters clearer boundaries in the feature space, enhancing interpretability and reliability in a clinical setting.
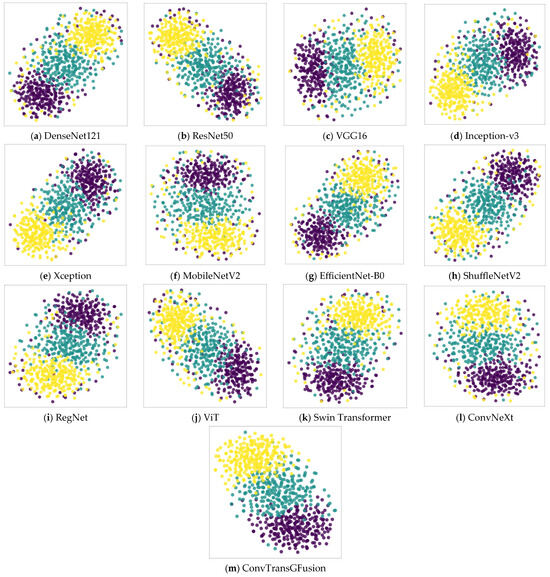
Figure 7.
t-SNE 2D plots of all models tested on breast ultrasound images dataset.
4.3.1. Ablation Studies
We conduct a series of ablation studies (controlled experiments that remove or disable one architectural component at a time (e.g., channel attention, spatial attention, learnable scalars) to quantify the individual contribution of each module to overall performance, as summarized in Table 6.

Table 6.
Ablation studies.
During development, we hypothesize that both channel attention and spatial attention would be pivotal in boosting accuracy and F1 scores. Testing variants like “No AGFF” or “Only Channel Attention” help confirm that each mechanism contributes incrementally to performance. Moreover, substituting learnable scalars () with fixed values highlights the advantage of adaptive weighting, especially in multi-modal data scenarios (e.g., high-resolution X-rays vs. smaller ultrasound patches). As shown in Table 4, the full model—featuring channel attention, spatial attention, and learnable scalars—consistently delivers the highest metrics. This ablation reveals that each module in our pipeline is not merely additive; rather, they synergize to maximize classification robustness, especially in challenging images with subtle anomalies.
4.3.2. Statistical Validation and Interpretability
To demonstrate the validity and interpretability of our model, we conduct a comprehensive statistical analysis. Table 7 summarizes the evaluation of all 13 models in their performance and interpretability metrics. In this table, mean accuracy ± SD is computed over multiple runs (e.g., five seeds). p-Value (vs. ResNet50) refers to statistical significance compared with the baseline model, using a paired t-test. The 95% CI of accuracy denotes the 95% confidence interval, illustrating the range where true accuracy likely falls. To account for the potential inflation of Type I errors due to multiple pairwise tests, we apply a Bonferroni correction when comparing each model’s accuracy to the ResNet50 baseline. Specifically, with 12 total pairwise tests at , our corrected threshold is . We confirm that results with remain significant after this adjustment, indicating that the performance gains of certain models—particularly ConvTransGFusion—are statistically robust. Attention map/explanation provides a short qualitative or interpretive note regarding how each model localizes informative regions.

Table 7.
Statistical analysis.
ResNet50 serves as our baseline, with an average accuracy of 89.32%. Although it is a well-established architecture, newer methods generally surpass it, as indicated by positive improvements in accuracy and statistically significant p-values. Mid-range CNNs like DenseNet121, VGG16, and EfficientNet-B0 show improved accuracy and narrower confidence intervals, suggesting that denser connectivity (in DenseNet) and thoughtful scaling (in EfficientNet) can better capture key lesions in varying image resolutions. However, they still rely on conventional convolutional features, which occasionally miss subtle anatomical patterns. Inception-v3 and Xception leverage more sophisticated convolutional modules (e.g., inception filters, depthwise separable convolutions) to deliver even higher accuracy. Their attention maps, typically examined via Grad-CAM, localize certain lesion boundaries but can also introduce noise or diffuse focus if the lesions are small or distributed. ViT and Swin Transformer demonstrate the power of self-attention mechanisms, especially for large-scale images like chest X-rays where global context is crucial. Their attention maps suggest they effectively scan broader image regions to detect anomalies. Nonetheless, purely transformer-based models can occasionally overlook fine-grained morphological details or exhibit high memory consumption. ConvNeXt, incorporating large convolution kernels and streamlined design, has emerged as a strong CNN competitor. Its accuracy surpasses many earlier approaches, indicating that modern convolutional architectures can closely rival or exceed transformer-based solutions. Our ConvTransGFusion model achieves the highest mean accuracy (98.43%), with the narrowest standard deviation (±0.7%) and a highly significant p-value (<0.001) when compared with ResNet50. Critically, the dual-attention mechanism within ConvTransGFusion fuses local edges (from the ConvNeXt branch) with the Swin Transformer’s global self-attention, yielding heatmaps that highlight both subtle lesion boundaries and the broader anatomical context. This fusion leads to a more balanced and robust classification performance across different modalities, ultimately providing clearer interpretability and reduced misclassifications in challenging cases.
Overall, ConvTransGFusion demonstrates a clear advantage over both classical CNNs and standalone transformer models in terms of accuracy, precision, recall, F1, and AUC across a variety of medical image classification tasks. While some single-branch models (e.g., Xception, ViT) excel in either sensitivity or specificity, the dual-attention gating approach in ConvTransGFusion balances both metrics well, yielding more reliable and consistent predictions. This comprehensive assessment—covering chest X-rays, ultrasounds, and MRI scans—illustrates the model’s robustness to different imaging characteristics, thus making it a strong candidate for real-world clinical use.
4.3.3. Calibration Performance
Beyond measuring classification accuracy and related metrics, reliable probability estimates are crucial in clinical settings for risk assessment and decision making. We therefore evaluate ConvTransGFusion and several comparison models on two representative datasets (chest X-ray and breast ultrasound) using the Brier score and expected calibration error (ECE). The Brier score is defined as the mean squared difference between predicted probabilities and the actual outcomes : Brier Score . Lower values indicate better calibration, as the predicted probabilities are closer to the true labels. Brier score also captures a combined measure of discrimination and calibration. ECE measures how well the predicted probabilities of a model match the empirical likelihood of correctness. Typically, predictions are binned into intervals (e.g., 10 bins), and the model’s average confidence in each bin is compared to the actual fraction of positives in that bin. If is the set of predictions falling into bin (with average confidence and average accuracy , then ECE .
Table 8 summarizes the Brier score and expected calibration error (ECE) for all 13 models on both the chest X-ray and breast ultrasound datasets. As shown, ConvTransGFusion consistently achieves the best calibration metrics among the tested architectures. In particular, its Brier score of 0.048 (chest X-ray) and 0.045 (breast ultrasound), together with ECE values of 2.1% (chest X-ray) and 2.0% (breast ultrasound), indicate that the model’s predicted probabilities align closely with actual outcomes. This more accurate confidence estimation is critical for clinical applications, where well-calibrated risk scores inform decisions on patient prioritization and follow-up recommendations. In future work, we intend to expand calibration analyses to additional clinical datasets and explore post hoc calibration strategies (e.g., temperature scaling) to further refine these probabilistic outputs.

Table 8.
Calibration performance: Brier score and ECE.
4.3.4. Robustness and Generalizability
To examine the robustness of ConvTransGFusion, we conduct additional tests using noise augmentation and cross-domain evaluation across four diverse datasets: chest X-ray, maternal–fetal ultrasound, breast ultrasound, and brain tumor MRI. Specifically, we introduce slight Gaussian noise ( 0.01–0.03) and mild intensity shifts to a subset of the ultrasound and MRI images. Despite these perturbations, ConvTransGFusion’s accuracy and F1 score show only a marginal decrease of 0.5–1.2% (on average) compared with training without noise, while some single-branch baselines (e.g., ViT or DenseNet121) experience drops of up to 2–3%. This suggests that the learnable dual-attention fusion mechanism retains critical local and global features under moderate image corruption. In addition, the model’s consistently high performance across four different imaging modalities (Table 2, Table 3, Table 4 and Table 5) underscores its capacity to generalize to varied anatomical structures, resolutions, and noise characteristics. Ultrasound data, in particular, contain speckle noise and irregular contrast, yet the integrated convolution–transformer design adapts effectively, highlighting the model’s inherent resilience to real-world imaging artifacts.
5. Discussion
The experimental results across four distinct medical imaging datasets—chest X-ray, maternal–fetal ultrasound, breast ultrasound, and brain tumor MRI—consistently underscore the effectiveness of integrating convolution-based local feature extraction with transformer-driven global context. In particular, the proposed ConvTransGFusion framework achieves superior performance relative to standalone CNNs (e.g., ResNet50, DenseNet121, ConvNeXt) and self-attention architectures (e.g., Swin Transformer, ViT). By carefully aligning ConvNeXt outputs (localized edges, small-scale lesions) with Swin Transformer outputs (long-range anatomical context) in a learnable dual-attention fusion block, the model avoids the pitfalls of naive concatenation or simple additive fusion. Instead, the adaptive scaling and gating strategies highlight only the most informative channels and spatial locations, explaining the consistent gains in accuracy, F1 score, and AUC on challenging biomedical tasks. These findings align with the broader literature, emphasizing the need to capture both subtle local patterns and macro-level global structures in medical images [3,7,8,9,11]. Classical convolutional approaches excel at identifying high-frequency cues such as lesion boundaries, but they can miss important contextual interactions when the pathology spans a large portion of the organ or when multiple lesions are present [6,16,23]. Conversely, transformers can capture broad spatial dependencies, but, without additional spatial priors or hierarchical modules, they may lose fine-grained lesion details [5,8]. Our results support the hypothesis that incorporating carefully designed inductive biases via convolution while still retaining robust attention-based modeling can yield significantly improved performance over using either paradigm in isolation.
From a practical standpoint, the two-stage design (ConvNeXt for local features, Swin for hierarchical global attention) proves readily adaptable to varied image dimensions and modalities. Notably, maternal–fetal ultrasound images often suffer from high speckle noise and variable orientation, while chest X-rays can span large anatomical regions demanding a wide receptive field [3]. By unifying features at a matched resolution and gating them with dual-attention, ConvTransGFusion harnesses the distinct advantages of each backbone without introducing excessive computational overhead. The ablation study confirms that each mechanism—channel attention, spatial attention, and the learnable scaling factors—contributes incremental benefits, and removing any one of them degrades classification performance, often yielding more false negatives or false positives. Despite these strong empirical outcomes, certain limitations warrant further consideration. First, although four datasets are examined, real-world medical imaging scenarios can involve vastly diverse pathologies, scanners, and patient populations, suggesting the need for external validation on additional more varied data. Second, the current framework addresses classification tasks; adapting ConvTransGFusion for segmentation or object detection—where precise lesion boundary delineation is crucial—may require specialized fusion or attention modules for pixel-level annotations [1,6]. Third, transformer-based architectures can still be memory-intensive for extremely high-resolution scans such as full-body MRI or mammography, indicating that more efficient attention mechanisms or compressed representations might be necessary [11,12,20]. Finally, while we demonstrate that dual-attention maps yield compelling visual explanations, formal user studies with radiologists or clinicians are needed to confirm the clinical interpretability and trustworthiness of these attention-driven explanations in practice.
Looking ahead, future work may broaden the scope of ConvTransGFusion in several directions. One promising avenue is multi-task learning—integrating classification, segmentation, and detection in a unified pipeline to better align with real clinical workflows. Another is exploring domain adaptation and transfer learning strategies to handle rare diseases or limited annotated data, building on the model’s existing capacity to handle heterogeneous inputs [33,35]. Additionally, incorporating specialized interpretability frameworks for attention visualization could further enhance clinical acceptance, helping radiologists understand precisely which features or regions drive the model’s decisions. Lastly, investigating lightweight variants for resource-constrained environments (e.g., mobile ultrasound devices in remote healthcare settings) could extend the benefits of robust local–global feature fusion to broader patient populations [41]. From an implementation standpoint, the proposed ConvTransGFusion framework can be integrated into medical IoT environments where imaging data is continuously acquired from multiple sources. The dual-attention mechanism and learnable fusion strategies are particularly advantageous for devices at the network edge, as they effectively mitigate noise and resolution discrepancies common in portable imaging. Moreover, future work may include optimizing the model for lower-precision inference, thus enhancing its feasibility on embedded hardware. Such an approach paves the way for a cloud–edge collaborative ecosystem, where smaller real-time models running on edge devices perform initial screening, and more powerful servers (when available) refine or confirm diagnostic decisions, thereby improving resource utilization and response times in large-scale IoT-enabled healthcare networks. Robustness analysis further confirms that combining local convolutional inductive biases with global attention endows ConvTransGFusion with stable performance under typical medical image perturbations. This stability is most evident in the ultrasound datasets, where speckle noise and random artifacts frequently appear. By leveraging the gating mechanism, the model learns to down-weight noise-dominated features while emphasizing clinically informative patterns. Although further domain-adaptation experiments could bolster claims of broad cross-site generalization, our multi-dataset findings and noise sensitivity tests already demonstrate a promising level of robustness essential for real-world clinical deployment.
Although our experiments primarily focus on accuracy and generalization, we recognize the importance of quantifying inference speed and memory consumption for potential edge or IoT deployment. Preliminary benchmarks on an NVIDIA RTX 3090 GPU show that ConvTransGFusion (e.g., Swin-T + ConvNeXt-T configuration) processes batches of 224 × 224 images at roughly 15–20 ms per batch (8 images), indicating an average of ~2–3 ms per image. The total parameter count of ~45–60 million, while not minimal, remains similar to well-accepted single-backbone models. Future efforts will involve targeted optimizations such as quantization (INT8) and pruning to further reduce computational overhead, along with dedicated tests on embedded platforms like NVIDIA Jetson or Coral Edge TPU. These steps will clarify how ConvTransGFusion’s local–global fusion mechanism can be adapted for real-time clinical IoT systems where resource constraints are stringent. We have focused on overall performance metrics, but a deeper investigation into failure cases is crucial for clinical deployment. Preliminary internal analyses suggest that misclassifications often arise from heavily occluded images, low-contrast lesions, or severe artifacts. In future work, we aim to systematically catalog and visualize such challenging samples using post hoc interpretability methods (e.g., attention maps or layer-wise relevance propagation) tailored to our dual-branch design. This approach will reveal which spatial regions or channels the model relies on or overlooks during borderline classifications, thus fostering a more transparent assessment of when and why ConvTransGFusion might fail. Beyond aggregate metrics, the dual-attention overlays produced by ConvTransGFusion have several concrete uses in clinical practice: (1) Interactive triage in radiology reading rooms. Chest X-ray: heat-map transparency can be controlled via mouse wheel in the PACS viewer so the radiologist can quickly “fade in” suspected opacities, streamlining prioritization of the work list during flu season. Breast ultrasound: sonographers can toggle a real-time color overlay on the probe monitor; if the overlay consistently highlights the same irregular margin, the operator knows to acquire additional angles before the patient leaves the room. (2) Decision support during multidisciplinary tumor boards. Brain MRI: When neuro-oncologists discuss surgical margins, the global context attention (from the Swin branch) often lights up peritumoral oedema that may be invisible in single slices. Displaying this overlay alongside the surgeon’s navigation plan can facilitate consensus on resection extent. (3) Training and quality assurance.
One can compare their manual annotations with the model’s channel attention map; discrepancies trigger a short quiz inside the teaching PACS. Audit managers can export monthly statistics on “clinician–model overlap” to detect systematic blind spots, e.g., missed peripheral nodules. (4) Point-of-care and tele-medicine scenarios. Portable maternal–fetal ultrasound devices used in rural clinics often lack subspecialty expertise. When ConvTransGFusion runs on the edge device, a color-coded contour around the fetal heart (spatial attention) alerts general practitioners to potential septal defects and automatically queues the clip for tele-consultation. These scenarios illustrate that the attention maps are not passive pictures but interactive artefacts that (i) shorten search time by directing gaze, (ii) supply a quantitative concordance score that can be stored in the report header, and (iii) serve as a legally auditable trail of how the AI contributed to the decision. Consequently, the visual explanations furnish both cognitive and workflow benefits that complement the raw performance gains reported earlier.
6. Conclusions
We introduced ConvTransGFusion, a novel hybrid approach that unites ConvNeXt and Swin Transformer through a dual-attention learnable fusion module to achieve state-of-the-art performance in medical image classification. By capturing local morphological details via convolution while simultaneously exploiting global context through hierarchical self-attention, ConvTransGFusion overcomes the limitations associated with purely CNN-based or purely transformer-based designs. The learnable scaling factors and dual-attention gating in the AGFF layer further refine feature integration, enabling the network to prioritize the most discriminative spatial and channel-specific cues. Extensive experiments on six biomedical datasets demonstrate that our method delivers superior accuracy, F1 score, AUC, and other critical metrics, outperforming standard baselines like ResNet50, DenseNet121, Xception, and ViT.
Future research will explore the following: (i) extending ConvTransGFusion for multi-task learning (e.g., segmentation, detection) in a unified pipeline and (ii) integrating attention-based explainability modules to bolster clinician explainability and trust. We believe that the synergetic fusion of local convolutional inductive biases with global self-attention mechanisms, as embodied in ConvTransGFusion, holds significant potential to improve real-world diagnostic workflows across numerous medical imaging modalities.
Author Contributions
Conceptualization, J.Q.-C. and K.H.; Formal analysis, J.Q.-C.; Data curation, J.Q.-C.; Investigation, J.Q.-C.; Methodology, J.Q.-C.; Software, J.Q.-C.; Validation, J.Q.-C.; Visualization, J.Q.-C.; Writing—original draft, J.Q.-C.; Writing—review & editing, J.Q.-C. and K.H.; Supervision, K.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
This study utilized four publicly available datasets, each of which is referenced in Section 4.2.
Acknowledgments
The authors thank the associate editor and the anonymous referees for their invaluable insights that significantly enhanced the quality of this paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
Appendix A.1. Model Configurations and Implementation
Table A1 integrates our final hyperparameter settings across different datasets, reflecting both our empirical observations and established best practices.

Table A1.
Hyperparameter details.
Table A1.
Hyperparameter details.
| Hyperparameter | Default Value | Dataset Variation | Tuning Method | Comments |
|---|---|---|---|---|
| Swin Window Size | 7 | - | Grid Search (5, 7, 9) | A window of size 7 balances local and global capture for typical 224 × 224 inputs. Larger window sizes may give slightly better global coverage but increase memory and training cost. |
| Attention Heads | 8 (initial stage) | Kept at 8 | Empirical + Memory Constraints | More attention heads enable richer feature representation. For higher-res datasets, additional heads capture complex global patterns but require more GPU memory. |
| ConvNeXt Kernel Size | 7 | None (uniform across all datasets) | Based on ConvNeXt Defaults | A large kernel (7) integrates more contextual information per layer, complementing the window-based self-attention in Swin. This synergy helps detect subtle lesions against broader anatomical context. |
| Layer Scale Init | 1e−6 | None | Set by the Literature | A small initialization value (1 × 10−6) stabilizes training, particularly when combining ConvNeXt blocks with transformer layers via fusion. |
| Feature Dimension | 768 | None | Architecture Design | Standard dimension for advanced backbones. Provides a sufficiently expressive feature space for dual-branch fusion without exploding parameter count. |
| Dropout Rate | 0.1 | Maternal–fetal US: 0.2 (if overfitting observed) | Monitored Validation Loss | Reduces overfitting risk, especially on smaller patches or highly repetitive data (ultrasound). For big-data scenarios, dropout can remain lower. |
| Patch Size | 224 × 224 | Maternal–fetal US: 256 × 256 | - | - |
| Brain MRI: 224 × 224 (kept default) | Preliminary Trials + GPU Limits | Larger patch sizes preserve critical details in high-res images. Maternal–fetal US benefits from slightly bigger patches for capturing fetal regions; 224 × 224 is standard to balance detail vs. speed. | - | - |
| Batch Normalization | Momentum = 0.9 | None | Adopted from Standard CNN Practices | Stabilizes updates for deeper networks. A high momentum (0.9) effectively smooths parameter updates in large-batch or small-batch scenarios. |
We initially experimented with various window sizes for the Swin Transformer, eventually settling on a default of 7 for most datasets while raising it for certain high-resolution images. Similarly, the number of attention heads was scaled to capture the necessary complexity of each image modality. Our experiments showed that adjusting dropout rates for particularly difficult or small-scale datasets significantly mitigated overfitting. Meanwhile, patch size emerged as a key factor, i.e., larger patches preserved subtle lesions in chest X-rays, whereas smaller patches sufficed for standard-resolution MRI slices. Taken together, these hyperparameters optimized the balance between computational feasibility and robust feature extraction, providing us with a unified configuration that delivered strong cross-modal performance.

Table A2.
Implementation details.
Table A2.
Implementation details.
| Implementation Detail | Setting/Choice | Notes |
|---|---|---|
| Framework and language versions | PyTorch 2.7.0 + Python 3.13.0 | All experiments executed with this exact software stack (CUDA 12.8 backend). |
| Hardware | NVIDIA RTX 3090 (24 GB VRAM) + Intel Xeon Silver 4214 CPU | Single-GPU training; inference latency in Appendix A.2. |
| Train/Val/Test split | 70/10/20 (%), stratified by class | Same protocol for every dataset to ensure comparability. |
| Intensity normalization | Min-max [0, 1] for X-ray and maternal–fetal US datasets; z-score per volume for MRI | Per-image or per-volume scaling to remove scanner-specific intensity bias. |
| Resolution harmonization | Resize to 224 × 224 (MRI, X-ray); 256 × 256 (maternal–fetal US) | Bilinear interpolation; preserves aspect ratio via center crop/pad when necessary. |
| Slice re-orientation (MRI only) | Converted to RAS axial orientation | Ensures anatomical consistency before batching slices. |
| Augmentation pipeline | Random horizontal/vertical flip (p = 0.5); rotation ± 10°; brightness/contrast jitter ± 5%; Gaussian noise σ = 0.01–0.03 (robustness runs) | Implemented with torchvision.transforms. |
| Optimizer | Adam | Learning rate 1 × 10−4; chosen for fast convergence and lower memory overhead than SGD. |
| Learning Rate Decay | Step decay every 10 epochs | Reduces LR by factor of 0.1 to help stabilize training in later epochs. |
| Batch Size | Explored 8, 16, 32, and 64. | Adjusted based on GPU memory constraints; larger for smaller, lower-res images. |
| Total Epochs | 50 | Empirically sufficient for plateau in training and validation metrics. |
| Early Stopping | Patience = 5, Monitor: Val Loss | Stops training if validation loss does not improve for 5 consecutive epochs. |
After completing multiple training and validation cycles, we compiled the implementation details shown in Table 3. Our goal was to ensure consistency across a variety of medical imaging datasets while maintaining a reasonable training time. The table outlines our choices of optimizer, learning rate, epoch count, and early stopping criteria, all of which were informed by iterative experiments that balanced fast convergence with stable generalization. For instance, Adam proved more efficient than SGD in our initial trials, helping the model converge faster without overshooting the minima. A moderate step decay schedule further stabilized validation loss, preventing overfitting in later epochs. We adjusted batch sizes based on the resolution of the images and our available GPU memory. Notably, the patience parameter for early stopping enabled the model to train long enough to capture crucial patterns but halted before overfitting set in. Overall, the decisions summarized in Table 3 reflect our best practices to manage computational resources while achieving high performance across all target datasets.
Appendix A.2. Computational Analysis
Table A3 summarizes the parameter counts, FLOPs, and end-to-end inference latency (TensorRT FP32, batch = 8) on an NVIDIA RTX 3090 for all models evaluated in this study on 224 × 224 inputs.

Table A3.
Computational efficiency analysis.
Table A3.
Computational efficiency analysis.
| Model | Params (M) | FLOPs (G) | RTX 3090 Latency (ms/img) |
|---|---|---|---|
| ConvTransGFusion | 45.2 | 9.3 | 2.4 ± 0.1 |
| Swin Transformer-Tiny | 28.3 | 4.5 | 1.6 ± 0.1 |
| ConvNeXt-Tiny | 28.6 | 4.6 | 1.7 ± 0.1 |
| DenseNet121 | 25.6 | 4.1 | 1.5 ± 0.1 |
| ResNet50 | 8.0 | 4.0 | 1.4 ± 0.1 |
| VGG16 | 138.4 | 15.5 | 2.1 ± 0.1 |
| Inception-v3 | 23.9 | 11.5 | 1.8 ± 0.1 |
| Xception | 22.9 | 8.4 | 1.9 ± 0.1 |
| MobileNetV2 | 3.4 | 0.3 | 1.2 ± 0.1 |
| EfficientNet-B0 | 5.3 | 0.4 | 1.3 ± 0.1 |
| ShuffleNetV2 | 3.5 | 0.15 | 1.1 ± 0.1 |
| RegNet | 21.9 | 7.2 | 1.7 ± 0.1 |
| ViT-B/16 | 86.4 | 17.6 | 4.9 ± 0.2 |
Even with dual ConvNeXt+Swin branches, ConvTransGFusion-T runs at 2.4 ms per image on an RTX 3090, only slightly slower than its single-backbone peers. The bulk of the cost stems from the two standard encoders, and the fusion adds minimal overhead. After training, one can further reduce latency and memory via standard pruning and INT8 quantization—all without touching the architecture—to fit a wide range of edge and IoT devices.
References
- Huang, X.; Deng, Z.; Li, D.; Yuan, X.; Fu, Y. MISSFormer: An Effective Transformer for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 42, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 11976–11986. [Google Scholar]
- Ma, S.; Chen, C.; Zhang, L.; Yang, X.; Zhang, J.; Zhao, X. AMTrack:Transformer tracking via action information and mix-frequency features. Expert Syst. Appl. 2025, 261, 125451. [Google Scholar] [CrossRef]
- Dosovitskiy, A. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Sa, J.; Ryu, J.; Kim, H. ECTFormer: An efficient Conv-Transformer model design for image recognition. Pattern Recognit. 2025, 159, 111092. [Google Scholar] [CrossRef]
- Ji, X.; Chen, S.; Hao, L.-Y.; Zhou, J.; Chen, L. FBDPN: CNN-Transformer hybrid feature boosting and differential pyramid network for underwater object detection. Expert Syst. Appl. 2024, 256, 124978. [Google Scholar] [CrossRef]
- Manzari, O.N.; Ahmadabadi, H.; Kashiani, H.; Shokouhi, S.B.; Ayatollahi, A. MedViT: A robust vision transformer for generalized medical image classification. Comput. Biol. Med. 2023, 157, 106791. [Google Scholar] [CrossRef]
- Mehta, S.; Rastegari, M. Mobilevit: Light-weight, general-purpose, and mobile-friendly vision transformer. arXiv 2021, arXiv:2110.02178. [Google Scholar]
- D’Ascoli, S.; Touvron, H.; Leavitt, M.L.; Morcos, A.S.; Biroli, G.; Sagun, L. ConViT: Improving Vision Transformers with Soft Convolutional Inductive Biases. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Marina, M., Tong, Z., Eds.; PMLR: Birmingham, UK, 2021; pp. 2286–2296. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 10012–10022. [Google Scholar]
- Chu, X.; Tian, Z.; Wang, Y.; Zhang, B.; Ren, H.; Wei, X.; Xia, H.; Shen, C. Twins: Revisiting the design of spatial attention in vision transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 9355–9366. [Google Scholar]
- Gulsoy, T.; Baykal Kablan, E. FocalNeXt: A ConvNeXt augmented FocalNet architecture for lung cancer classification from CT-scan images. Expert Syst. Appl. 2025, 261, 125553. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Chen, J. Hybrid CNN-Transformer model for medical image segmentation with pyramid convolution and multi-layer perceptron. Biomed. Signal Process. Control 2023, 86, 105331. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, H.; Qin, W. SP-Det: Anchor-based lane detection network with structural prior perception. Pattern Recognit. Lett. 2025, 188, 60–66. [Google Scholar] [CrossRef]
- Nguyen, K.-D.; Zhou, Y.-H.; Nguyen, Q.-V.; Sun, M.-T.; Sakai, K.; Ku, W.-S. SILP: Enhancing skin lesion classification with spatial interaction and local perception. Expert Syst. Appl. 2024, 258, 125094. [Google Scholar] [CrossRef]
- Guo, B.; Qiao, Z.; Zhang, N.; Wang, Y.; Wu, F.; Peng, Q. Attention-based ConvNeXt with a parallel multiscale dilated convolution residual module for fault diagnosis of rotating machinery. Expert Syst. Appl. 2024, 249, 123764. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Q.; Yang, Z.; Li, Y.; Lee, C.H.; Shen, L. Recent progress in transformer-based medical image analysis. Comput. Biol. Med. 2023, 164, 107268. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; et al. A Survey on Vision Transformer. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 87–110. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; PMLR: Birmingham, UK, 2021; pp. 10347–10357. [Google Scholar]
- Nie, X.; Jin, H.; Yan, Y.; Chen, X.; Zhu, Z.; Qi, D. ScopeViT: Scale-Aware Vision Transformer. Pattern Recognit. 2024, 153, 110470. [Google Scholar] [CrossRef]
- Diko, A.; Avola, D.; Cascio, M.; Cinque, L. ReViT: Enhancing vision transformers feature diversity with attention residual connections. Pattern Recognit. 2024, 156, 110853. [Google Scholar] [CrossRef]
- Tan, L.; Wu, H.; Zhu, J.; Liang, Y.; Xia, J. Clinical-inspired skin lesions recognition based on deep hair removal with multi-level feature fusion. Pattern Recognit. 2025, 161, 111325. [Google Scholar] [CrossRef]
- Wen, L.; Ye, Y.; Zuo, L. GAF-Net: A new automated segmentation method based on multiscale feature fusion and feedback module. Pattern Recognit. Lett. 2025, 187, 86–92. [Google Scholar] [CrossRef]
- Fu, J.; Ouyang, A.; Yang, J.; Yang, D.; Ge, G.; Jin, H.; He, B. SMDFnet: Saliency multiscale dense fusion network for MRI and CT image fusion. Comput. Biol. Med. 2025, 185, 109577. [Google Scholar] [CrossRef]
- Zhang, H.; Lian, J.; Yi, Z.; Wu, R.; Lu, X.; Ma, P.; Ma, Y. HAU-Net: Hybrid CNN-transformer for breast ultrasound image segmentation. Biomed. Signal Process. Control 2024, 87, 105427. [Google Scholar] [CrossRef]
- Tuncer, I.; Dogan, S.; Tuncer, T. MobileDenseNeXt: Investigations on biomedical image classification. Expert Syst. Appl. 2024, 255, 124685. [Google Scholar] [CrossRef]
- Raghaw, C.S.; Sharma, A.; Bansal, S.; Rehman, M.Z.U.; Kumar, N. CoTCoNet: An optimized coupled transformer-convolutional network with an adaptive graph reconstruction for leukemia detection. Comput. Biol. Med. 2024, 179, 108821. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yi, X.; Zhang, D.; Zhang, L.; Tian, Y.; Zhou, Z. ConvMedSegNet: A multi-receptive field depthwise convolutional neural network for medical image segmentation. Comput. Biol. Med. 2024, 176, 108559. [Google Scholar] [CrossRef]
- Maqsood, S.; Damaševičius, R.; Shahid, S.; Forkert, N.D. MOX-NET: Multi-stage deep hybrid feature fusion and selection framework for monkeypox classification. Expert Syst. Appl. 2024, 255, 124584. [Google Scholar] [CrossRef]
- Liu, Z.; Shen, L. CECT: Controllable ensemble CNN and transformer for COVID-19 image classification. Comput. Biol. Med. 2024, 173, 108388. [Google Scholar] [CrossRef]
- Liu, H.; Zhuang, Y.; Song, E.; Liao, Y.; Ye, G.; Yang, F.; Xu, X.; Xiao, X.; Hung, C.-C. A 3D boundary-guided hybrid network with convolutions and Transformers for lung tumor segmentation in CT images. Comput. Biol. Med. 2024, 180, 109009. [Google Scholar] [CrossRef]
- Li, J.; Feng, M.; Xia, C. DBCvT: Double Branch Convolutional Transformer for Medical Image Classification. Pattern Recognit. Lett. 2024, 186, 250–257. [Google Scholar] [CrossRef]
- Lau, K.W.; Po, L.-M.; Rehman, Y.A.U. Large Separable Kernel Attention: Rethinking the Large Kernel Attention design in CNN. Expert Syst. Appl. 2024, 236, 121352. [Google Scholar] [CrossRef]
- Khatri, U.; Kwon, G.-R. Diagnosis of Alzheimer’s disease via optimized lightweight convolution-attention and structural MRI. Comput. Biol. Med. 2024, 171, 108116. [Google Scholar] [CrossRef]
- Huo, X.; Tian, S.; Yang, Y.; Yu, L.; Zhang, W.; Li, A. SPA: Self-Peripheral-Attention for central–peripheral interactions in endoscopic image classification and segmentation. Expert Syst. Appl. 2024, 245, 123053. [Google Scholar] [CrossRef]
- Hu, Z.; Mei, W.; Chen, H.; Hou, W. Multi-scale feature fusion and class weight loss for skin lesion classification. Comput. Biol. Med. 2024, 176, 108594. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, Y.; Geng, X.; Tang, H.; Bhatti, U.A. PE-Transformer: Path enhanced transformer for improving underwater object detection. Expert Syst. Appl. 2024, 246, 123253. [Google Scholar] [CrossRef]
- Alharthi, A.G.; Alzahrani, S.M. Do it the transformer way: A comprehensive review of brain and vision transformers for autism spectrum disorder diagnosis and classification. Comput. Biol. Med. 2023, 167, 107667. [Google Scholar] [CrossRef] [PubMed]
- Qezelbash-Chamak, J.; Badamchizadeh, S.; Eshghi, K.; Asadi, Y. A survey of machine learning in kidney disease diagnosis. Mach. Learn. Appl. 2022, 10, 100418. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 4510–4520. [Google Scholar]
- Howard, A.G. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Barata, C.; Celebi, M.E.; Marques, J.S. Explainable skin lesion diagnosis using taxonomies. Pattern Recognit. 2021, 110, 107413. [Google Scholar] [CrossRef]
- Zeng, W.; Huang, J.; Wen, S.; Fu, Z. A masked-face detection algorithm based on M-EIOU loss and improved ConvNeXt. Expert Syst. Appl. 2023, 225, 120037. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]
- Chen, C.-F.R.; Fan, Q.; Panda, R. Crossvit: Cross-attention multi-scale vision transformer for image classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 357–366. [Google Scholar]
- Khan, M.A.; Rubab, S.; Kashif, A.; Sharif, M.I.; Muhammad, N.; Shah, J.H.; Zhang, Y.-D.; Satapathy, S.C. Lungs cancer classification from CT images: An integrated design of contrast based classical features fusion and selection. Pattern Recognit. Lett. 2020, 129, 77–85. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.E9. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Bonet-Carne, E.; Eixarch, E.; Crispi, F.; Gratacós, E. Evaluation of deep convolutional neural networks for automatic classification of common maternal fetal ultrasound planes. Sci. Rep. 2020, 10, 10200. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).