Abstract
Fusarium ear rot (FER) is a common disease in maize caused by the pathogen Fusarium verticillioides. Because of the quantitative nature of the disease, scoring disease severity is difficult and nuanced, relying on various ways to quantify the damage caused by the pathogen. Towards the goal of designing a system with greater objectivity, reproducibility, and accuracy than subjective scores or estimations of the infected area, a system of semi-automated image acquisition and subsequent image analysis was designed. The tool created for image acquisition, “The Ear Unwrapper”, successfully obtained images of the full exterior of maize ears. A set of images produced from The Ear Unwrapper was then used as an example of how machine learning could be used to estimate disease severity from unannotated images. A high correlation (0.74) was found between the methods estimating the area of disease, but low correlations (0.47 and 0.28) were found between the number of infected kernels and the area of disease, indicating how different methods can result in contrasting severity scores. This study provides an example of how a simplified image acquisition tool can be built and incorporated into a machine learning pipeline to measure phenotypes of interest. We also present how the use of machine learning in image analysis can be adapted from open-source software to estimate complex phenotypes such as Fusarium ear rot.
1. Introduction
Imaging of plant material for the purpose of phenotypic analysis continues to gain popularity in fields such as plant breeding, plant pathology, and plant systematics [1,2,3,4]. On a larger scale, drone, airplane, and satellite imagery are used in large-scale agriculture for monitoring a myriad of agronomic traits [5,6]. Imaging technology and the subsequent analysis allow for a reduction in labor and time-related costs while maintaining, if not improving, accuracy. The use of machine learning (ML) pipelines can decrease bias from human subjectivity, improve the efficiency of analysis pipelines, and increase the accuracy of predictions [7,8]. However, obtaining and preprocessing enough high-quality training data often creates a barrier to entry that is too high for labs or individuals to develop a robust, accurate, and efficient pipeline of image acquisition and analysis.
In plant pathology, accurate disease phenotyping results are used to guide day-to-day agricultural production (choice of plant variety, applications of pesticides, rogueing of plants), long-term breeding programs, and molecular research [9,10,11]. To acquire images for analysis, the type and scale of the image were based on the purpose of the study. Those attempting to identify regions in a field under stress and image acquisition from drones, satellites, or other aerial tools are the most common [12]. Conversely, studying individual plants requires close-up and high-resolution images that are either compiled by hand or by using another device close to the target. Most studies using ML in plant pathology attempt to accurately identify (or classify) a microorganism; therefore, images are often obtained with microscopes or hand-held tools [13,14]. Quantitative disease severity estimations that are required by plant breeding programs are not limited to local image acquisition but instead depend on the scale of the study. Regardless, quantification of biotic or abiotic stressors is generally achieved by breaking down an image or set of images into smaller component parts (such as pixels) and using classification methods on those smaller parts before they are quantified [15,16]. This is often calculated or presented as a ratio to provide a consistent scale that is associated with plant health.
Maize (Zea mays L.) is a cereal crop that is grown worldwide and is the primary calorie source for livestock. As it is an international staple crop, maize has been the subject of ML for image analysis in a variety of studies where image acquisition methods are standardized [17,18,19,20,21]. The disease addressed in this work is Fusarium ear rot (FER) of maize caused by the pathogen Fusarium verticillioides (Sacc.) Nirenberg (synonym F. moniliforme Sheldon, teleomorph Gibberella moniliformis Wineland), and the investigation focuses on the area of infected tissue on the ear of the maize plant. FER is an important disease internationally, as the mycotoxin produced by F. verticillioides, fumonisin, causes diseases in humans and livestock; therefore, breeding to reduce FER is desirable. Conventional phenotyping of FER severity has been highly subjective, relying on a researcher to quantify the severity by quickly inspecting each ear and deciding on a severity score. Breeding efforts to reduce FER, or any such disease, require accurate phenotyping to determine resistant and susceptible varieties in a population. Only minimal gains towards breeding FER resistant lines have been achieved, likely due to the difficulty in obtaining accurate phenotypic data. As mentioned, image analysis can improve objectivity and reliability of phenotyping generally, but it has not been performed in the FER pathosystem. FER severity cannot be estimated without removing the husk of the ear and viewing the whole ear to observe the damage caused by the pathogen [22,23,24]. Thus, traditional imaging of one side of a harvested ear is not sufficient to capture information from the entire ear. In addition, high-quality image acquisition of the ear is required to be able to distinguish between different types of damage or disease that may be present on the ear, whether for an ML pipeline or for traditional phenotyping.
The goal of the system presented here was to obtain an image of a full ear of maize in such a way that it could be used to quantify disease severity following inoculation. Our expectation would be that if we could “unwrap” the ear, such as one might remove the label from a can of food in order to convert a cylindrical object to a rectangular image, we would be able to accurately measure lesion size from all parts of the ear. As diseases (particularly FER) often occur in multiple places on a single ear or wraps around the ear, obtaining a full view of the ear is necessary for accurate phenotyping. The images produced by the machine presented here, named “The Ear Unwrapper”, were then analyzed with an ML pipeline estimating disease severity using probabilistic pixel classification as an example of how the images could be used in larger studies requiring high-throughput phenotyping. This paper does not attempt to show improved performance compared to other methods of severity estimation, as molecular genetics are required for validation. Though we present an example of how the images can be processed using an ML pipeline, there are additional uses for image-based-phenotyping, such as having a visual record for subsequent researchers to use. The Ear Unwrapper can be applied to many studies investigating maize ear phenotypes that require a 360° of the ear.
2. Materials and Methods
2.1. FER Disease Inoculation
A diversity panel of 66 sweetcorn and 2 field corn varieties of maize was grown in Citra, FL, from March–June of 2022. Thirty ears were harvested approximately 21 days after 50% silking was observed in each variety. The ears were brought to the inoculation location before the husk was wiped down with 70% ethanol. Circles were drawn on the husk in the center of each ear to indicate the inoculation location. An 18-gauge needle was then used to make a hole through the husk to the cob. A Prima-tech® vaccinator was then primed with a spore solution, and another 18-gauge needle was attached. Prior to inoculation, spore solutions were created by growing isolates of Fusarium verticillioides in potato dextrose agar plates, taking five 1 cm2 mycelial plugs from the plates, and placing them in ½ strength potato dextrose broth to shake for five days. After five days, spore solutions were filtered through sterile cheesecloth and concentrated or diluted to equal 1 × 105 conidia per ml. Once the spore solution was primed into the vaccinator, the needle was then placed into the existing hole in the ear, and 0.75 mL of the spore solution was injected. Once inoculated, the ears were placed in bags for humidity and isolation before being incubated at 27 °C for seven days. After seven days, the ears were removed from the bags, husked, shanks were removed, and the ears were individually placed on The Ear Unwrapper stepper motor. Often, maize ears would have damage from other sources, primarily on the top of the ear. In these cases, the affected part of the ear was removed before being mounted on The Ear Unwrapper to minimize the confounding factors that would impact subsequent image analysis.
2.2. Hardware
To “unwrap” each ear of corn, the approach was to rotate the ear around its axis (the cob) and, using a camera, take a continuous set of images to extract a single row of pixels from each to form one image. The motor used was a Nema 17 stepper motor that controls the position and rotation of the motor (and the object mounted on the motor) at a fraction of a degree. Here, each pulse of the stepper motor represents 0.45 degrees thus, 800 pulses are required to spin 360 degrees. The camera used was a See3CAM_24CUG camera (Figure 1B). It was chosen because it has UVC driver compatibility that facilitates integration into an operating system as a USB webcam and because it has an external shutter trigger. The external trigger allows the microcontroller to control the shutter for synchronization with the stepper motor. As the motor is sent a pulse to rotate 0.45 degrees, the camera is also sent a pulse to capture an image. By syncing the pulses of the camera and the motor, one can control the speed while maintaining the accuracy of the image that is being produced via stitching. The pulses that are sent to the motor and the camera come from an Arduino UNO (SparkFun RedBoard, Niwot, CO, USA) (Figure 1C). Although reducing the time it took to obtain a single image was important, obtaining a clear, well-lit, and focused image were all factors that reduced the frame rate. The USB bandwidth and stability of the corn ear when rotating also forced a reduction in the frame rate.
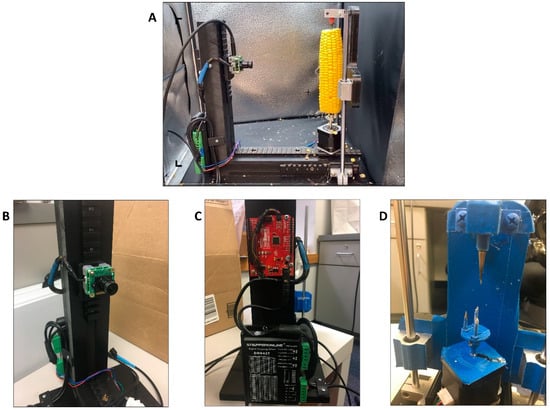
Figure 1.
Overview of The Ear Unwrapper. (A) Example of setup for imaging a maize ear on The Ear Unwrapper. (B) See3CAM_24CUG camera. (C) Arduino UNO (SparkFun RedBoard). (D) Modified Stepper motor, screw mounts, and aluminum conical spike.
The frame for the machine was built with nylon and high-density polyethylene (HDPE) to provide durability for mounting the other components and allowing room for the ear to rotate less than one foot from the camera lens. The camera and motor were mounted on Picatinny rails. The stepper motor mount that came with the device was removed and replaced with two spikes (sharpened screws) that screwed into the existing stepper motor mount holes. Two spikes were used to provide the leverage required to rotate the ear on the sliding rails. A single conical spike of aluminum was fixed directly above the center of the modified stepper motor. This conical spike provided additional stability to the maize ear when mounted on the modified stepper spikes and allowed the ear to rotate freely (Figure 1D).
To prepare the ears to produce an image, the ears were husked, the shanks were cut or snapped off, silks were removed, and the unpollinated tips of the ears were removed. The Ear Unwrapper was placed in a photobox (Fotodiox™) to obtain images with consistent lighting and background (Figure 1A). The ears were then mounted in The Ear Unwrapper by gently pressing the base of the ear onto the modified stepper motor spikes until it would stand without assistance and then sliding down the conical spike to the tip of the ear. The door to the photobox was closed, and a custom Python script was used to capture, process, and output images.
2.3. Calibration and Error Correction
The 24CUG camera uses a wide-angle lens with focal length 3.0 mm and aperture f/2.8, a benefit because the camera could be positioned closer to the maize ear while keeping the full ear in view. To correct for the “fisheye” effect, a correction algorithm was used to remove this distortion and extract the rectangular image. This was done in Python with the OpenCV library and calibrated by printing out a checkerboard and photographing it with the camera at a fixed distance from the camera to the motor. This also allowed for the determination of pixels/cm measurements. The de-warped image of the checkerboard was used to produce a correction matrix, which was then applied to every image captured by the camera. To more efficiently use the sensor of the 24CUG camera default 1280 × 720 resolution, we rotated the camera 90 degrees so that the 1280 resolution was vertical and could better accommodate the geometry of the corn ears. When an image was taken, a single vertical line of 1280 pixels was obtained before the motor turned 0.45 degrees and another image was taken, and the next vertical line was stitched to the previous line of pixels. This was repeated 800 times until the ear completed a 360-degree rotation. This value was based on an estimation that the average ear of corn is roughly 800 pixels or 2 inches in diameter when viewed at a specified distance and resolution from the camera lens. Although there are substantial morphological differences in the ears of corn that could cause the image to be stretched or compressed, particularly when using a diversity panel, this was addressed by capturing an additional four images, each 200 pulses (or 90 degrees) apart. These four additional images captured the full field of view rather than a line of pixels. Segmenting the ear from these four images allowed us to calculate an estimate of the ear diameter, and correct distortions in the ‘unwrapped’ image incurred when imaging ears that were significantly wider or narrower than the 2” ear diameter the image dimensions were calibrated for.
2.4. Image Processing
A custom Python application was built to interface with the UVC driver, control the camera shutter and the stepper motor (via the Arduino), and capture the images. This application also provided a command–line interface to allow user control for photo capture and file naming. The photos were captured, named, and saved according to the date, time, and ID number entered by the user as a unique identifier. Images were saved as PNG files and then filtered and sorted based on a set of criteria. First, images of ears with <50% pollination, significant damage that was not caused by F. verticillioides, or images that were blurred were removed as these factors influence the ability to measure disease severity. After removing and filtering the images, 59 images remained before 10 images were selected to train the model. When assigning images to the training folder, the images were chosen based on morphological and phenotypic diversity to provide a range of disease severity, ear size, and a diversity of lesion colors and shapes. Images used for training were also selected so that the visual differentiation between healthy and diseased tissues was clear. The testing set consisted of the remaining 49 images.
2.5. Pixel Classification
To build and train the classification model, we used the interactive machine learning software, “Ilastik” [25]. This software provides a pre-defined feature space across several workflows for biological image analysis, reducing both the amount of computing time and training data required to develop an accurate classifier. We used the ‘pixel classification’ workflow to semantically segment the images according to our three class labels: background, healthy tissue, and diseased tissue. The background consisted of all image areas that were not ear tissue, and all maize ear tissues without fungal hyphae were considered healthy. Any tissue that contained visually apparent disease symptoms was annotated as diseased tissue (Supplemental Figure S2). We used the default Random Forest classifier and feature selection settings of σ3 (Sigma of 1.60) for Color/Intensity, Edge, and Texture. The ten training images were imported to Ilastik, and each image was manually annotated with the class labels until the real-time classifier predictions became stable (i.e., when additional annotation caused either no visible change or minimal change in the model feedback). Once the training was complete, the remaining 49 testing/validation images were imported for batch processing. The model was used to generate a probability map for each image, i.e., an image with identical dimensions, where each pixel is colored by the classifier according to the class label that it most likely represents in the original image (Figure 2B).
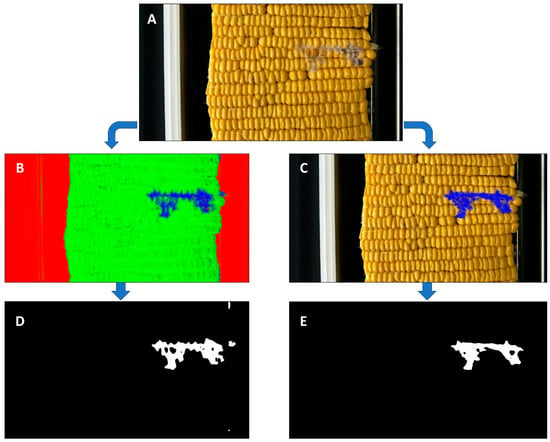
Figure 2.
Image analysis pipeline for a single ear of maize. (A) Raw image produced from The Ear Unwrapper. (B) Ilastik prediction output, where red is background, green is healthy tissue, and blue is diseased tissue. (C) Manual annotation in Microsoft Paint of disease region. (D) Binary mask showing disease pixels in white from Ilastik prediction output. (E) Binary mask showing disease pixels in white from Paint prediction output.
2.6. ImageJ and OpenCV Processing
Outputs from the testing dataset were processed in ImageJ (FIJI) and OpenCV using a custom Python script [26,27]. Probability files were converted from TIFF to PNG, and a median blur function (Ksize = 11) was applied to each image in ImageJ. In OpenCV, the RGB images were converted to an HSV (hue, saturation, and value) representation and thresholded to create a mask for pixels classified as likely to be diseased. This mask image was converted to binary (black and white), where the disease is represented as white pixels (Figure 2D,E). These white pixels were then counted using a Python script and written out to a CSV file with their corresponding unique ID numbers for reference to their manually recorded scores.
2.7. Scoring Methods and Correlations
The ears that were imaged were also scored in two different ways. The first was a user with experience with FER infections, counting the kernels that had visual symptoms consistent with FER. The second was by manually annotating each image in the program Microsoft Paint (on Windows 10) by coloring the disease pixels with the same blue color used in Ilastik, using the same methods to blur, threshold, and mask the image as in the Ilastik output images (Figure 2C,E). It should also be noted that the expert counting diseased kernels was the same person manually annotating the images on Paint to provide continuity. To correlate the infected kernel counting scores with the predicted pixel counts from Ilastik and the manually annotated pixel counts from Paint, the log scores of each were calculated and compared.
3. Results
3.1. Image Acquisition and Descriptive Statistics
Each maize ear, once mounted on the modified stepper motor, required 10 s for a full image to be taken. After size correction and de-warping, images were roughly 1280 × 450–600 depending on the diameter of the ear. Over 3000 ears were imaged using The Ear Unwrapper, which showed high reproducibility and no errors in image acquisition.
3.2. Correlation of Severity Scores
Using the index of each photo, the disease severity scores were adjusted and correlated in different ways. Log adjusted counts of white pixels (representing disease) from the manually annotated Paint output were compared to the log adjusted pixel counts from the Ilastik prediction software, generating an R2 = 0.7409 (Figure 3A). Next, log values of the pixel count for both Paint and Ilastik predictions were calculated and then correlated with the log adjusted infected kernel counts. The log values of the Paint counts compared with the log adjusted infected kernel counts generated an R2 = 0.4677 (Figure 3B), and the log values of the Ilastik scores compared with the log infected kernel counts generated an R2 = 0.2831 (Figure 3C). Additional statistics from the output of all scoring methods can be found in Table 1.
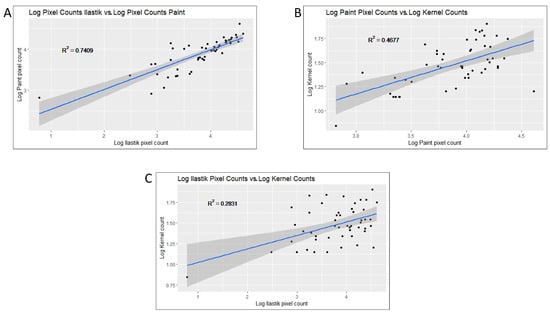
Figure 3.
Graphical display of correlations between the phenotyping counts. (A) Log adjusted pixel counts produced from Ilastik vs. log adjusted pixel counts produced from Paint: R2 = 0.74. (B) Log adjusted pixel counts produced from Paint vs. log adjusted kernel counts: R2 = 0.47. (C) Log adjusted pixel counts produced from Ilastik vs. log adjusted kernel counts: R2 = 0.28.

Table 1.
Statistics from the output of infected kernel counts, Ilastik predictions, and Paint annotations.
4. Discussion
The technology presented in this study demonstrates a method for quantifying disease symptoms by imaging maize ears via “unwrapping” the ear exterior to be viewed in a single rectangular image for different image analysis objectives. The Ear Unwrapper method of image stitching and de-warping of the different sized and shaped ears showed reliable, consistent, and accurate results in the downstream analysis. As phenotypic data acquisition is often the bottleneck that most breeding programs face, this study provides an example of how to remove this bottleneck by pairing commercially available hardware with machine learning. Here, we used images produced by The Ear Unwrapper in a machine learning (ML) pipeline to quantify the severity of damage caused by the Fusarium ear rot (FER) pathogen. The ML pipeline and results shown here are not meant as a validation of the phenotyping method, nor confirmation of the ML software “Ilastik” in FER phenotyping, but rather as an example application for images produced by The Ear Unwrapper. As FER is an important disease in maize, accurate disease severity estimates are necessary to improve disease resistance in maize breeding programs. The Ear Unwrapper platform could also potentially be used for other diseases that infect maize ears or for other investigations regarding the morphology and phenotypes of the maize ear.
Though the sample size of the images chosen in this study was small (testing set N = 49), correlations between the results of different phenotyping methods were consistently positively correlated. While correlations between the log adjusted infected kernel counts and the log adjusted pixel counts were low (R2 = 0.4677 and R2 = 0.2831), this assumes a linear relationship between scoring methods. It is important to note that within the FER pathosystem, several methods of measuring disease severity exist, though none are universally used and accepted. However, when comparing the two methods that used the same images generated by The Ear Unwrapper, the correlation was much higher (R2 = 0.7409), suggesting that the ML prediction pipeline is more accurate for estimating the disease area. By contrasting this high correlation with the lower correlation values between infected kernel counting and pixel counts, it could provide evidence that infected kernel counting does not correlate well with the lesion area. For example, the same size lesion on ears with different sized kernels would generate two different kernel counts but the same area of disease (or number of pixels). The most common method of FER severity phenotyping is visual estimation between 1–10 or 1–7 but does not use quantitative measures for these estimates. The correlations in the data table are provided to show how different phenotyping methods could produce variable results and subsequent selection changes in what varieties are used for breeding. No method is accepted as superior for use in phenotyping disease severity, so additional work to determine the genetic basis (heritability) of these different disease severity estimates should be performed.
Regardless, the high positive correlation between the images manually annotated in Paint and those generated from Ilastik demonstrate how high-quality image production is a valuable tool with or without building an ML pipeline, but that even with small data sets ML pipelines can measure complex traits of interest such as FER infection severity using probabilistic pixel classification. As image segmentation for quantitative analysis is usually performed by complex deep learning neural networks, this evaluation of a quantitative trait using a classification model provides an example of how more user-friendly software and other data-driven models can be used to obtain reasonable results [28,29,30]. There are several options that may be used to improve this method. Increasing the number of images used for training will provide better resolution around the quality of the results and possibly improve the calibration to provide more accurate predictions. Using more highly customized pixel classification methods or applying more computationally intensive neural network models could possibly provide better results; however, generating quantifiable data that can be objectively validated would give greater weight to images that increase correlations between predicted and actual scores.
While The Ear Unwrapper can only image a single ear at a time, the same framework and image acquisition techniques could be applied using multiple ears/motors with only a slightly higher cost. This technology could also be adapted for applications using other wavelengths outside the visual spectrum, as the pipeline is mostly hardware-agnostic, provided that the camera can accept external triggers for shutter control and supports UVC device standards. Other imaging devices may be used if driver-level interfacing or hardware modifications are provided. Similar technologies have been used for kernel analysis (counting and sorting), ear morphology (size, shape, and color), and molecular traits. The Ear Unwrapper, given the appropriate downstream image analysis tools, can be applied for these traits as well. While currently built to image maize ears, other cylindrical products that require the analysis of the full image are easily adapted using the same framework. In this study, only ears with high (>90%) seed set were included, but ears with low seed set and irregular morphologies were also imaged and produced high-quality results (Supplemental Figure S1).
5. Conclusions
In this paper, we describe how, by using off-the-shelf parts and open-source software, a semi-automated image acquisition machine was built, and the produced images were used in a pixel classification model to estimate disease severity from the maize disease Fusarium ear rot. We also calculated the correlation between the different methods of severity estimation to demonstrate how these methods could change the results and subsequent actions taken by disease resistance breeders.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriengineering5030077/s1, Supplemental Figure S1. Examples of morphologically diverse maize sample images produced by The Ear Unwrapper. Supplemental Figure S2. Example of annotation in Ilastik. Yellow lines are drawn on the background. Blue lines are drawn on healthy tissue, and red lines are drawn on diseased tissue. Supplemental Table S1. All raw data values were used to generate results.
Author Contributions
J.B. and O.H. initiated the project. O.H. acquired the data. O.H., C.B. and D.H. contributed to the design of The Ear Unwrapper. C.B. and D.H. built the hardware and accompanying software for The Ear Unwrapper. J.B. oversaw the project and secured all funding. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Florida and the United States Department of Agriculture–National Institute of Food and Agriculture Tactical Sciences for Agricultural Biosecurity Institute of Food and Agricultural Sciences Project 13117320.
Data Availability Statement
The raw dataset used to generate these results can be found in Supplemental Table S1. The codebase for The Ear Unwrapper can be found at (https://github.com/ohudson1/The-Ear-Unwrapper-.git (accessed on 24 May 2023)). All code used is open source and code bases for both Ilastik (https://github.com/ilastik (accessed on 1 January 2023)).
Acknowledgments
Marcio Resende and the Sweetcorn Lab for assistance with the field trials.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| FER | Fusarium ear rot |
| ML | Machine Learning |
| HDPE | high-density polyethylene |
References
- Das Choudhury, S.; Samal, A.; Awada, T. Leveraging Image Analysis for High-Throughput Plant Phenotyping. Front. Plant Sci. 2019, 10, 508. [Google Scholar] [CrossRef]
- Hartmann, A.; Czauderna, T.; Hoffmann, R.; Stein, N.; Schreiber, F. HTPheno: An Image Analysis Pipeline for High-Throughput Plant Phenotyping. BMC Bioinform. 2011, 12, 148. [Google Scholar] [CrossRef]
- Fida, F.; Talib, M.R.; Fida, F.; Iqbal, N.; Aqeel, M.; Iqbal, M.N.; Noman, A. Leaf Image Recognition Based Identification of Plants: Supportive Framework for Plant Systematics. PSM Biol. Res. 2018, 3, 125–131. [Google Scholar]
- Gobalakrishnan, N.; Pradeep, K.; Raman, C.; Ali, L.J.; Gopinath, M. A Systematic Review on Image Processing and Machine Learning Techniques for Detecting Plant Diseases. In Proceedings of the 2020 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 28–30 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 0465–0468. [Google Scholar]
- Zhang, C.; Marzougui, A.; Sankaran, S. High-Resolution Satellite Imagery Applications in Crop Phenotyping: An Overview. Comput. Electron. Agric. 2020, 175, 105584. [Google Scholar] [CrossRef]
- van der Merwe, D.; Burchfield, D.R.; Witt, T.D.; Price, K.P.; Sharda, A. Drones in Agriculture. Adv. Agron. 2020, 162, 1–30. [Google Scholar]
- Katarya, R.; Raturi, A.; Mehndiratta, A.; Thapper, A. Impact of Machine Learning Techniques in Precision Agriculture. In Proceedings of the 2020 3rd International Conference on Emerging Technologies in Computer Engineering: Machine Learning and Internet of Things (ICETCE), Jaipur, India, 7–8 February 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–6. [Google Scholar]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine Learning Applications for Precision Agriculture: A Comprehensive Review. IEEE Access 2020, 9, 4843–4873. [Google Scholar] [CrossRef]
- Mutka, A.M.; Bart, R.S. Image-Based Phenotyping of Plant Disease Symptoms. Front. Plant Sci. 2015, 5, 734. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, N.; Lee, S.; Mockler, T.C. High Throughput Phenotyping to Accelerate Crop Breeding and Monitoring of Diseases in the Field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors–Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Gago, J.; Douthe, C.; Coopman, R.E.; Gallego, P.P.; Ribas-Carbo, M.; Flexas, J.; Escalona, J.; Medrano, H. UAVs Challenge to Assess Water Stress for Sustainable Agriculture. Agric. Water Manag. 2015, 153, 9–19. [Google Scholar] [CrossRef]
- Cheshkova, A. A Review of Hyperspectral Image Analysis Techniques for Plant Disease Detection and Identif Ication. Vavilov J. Genet. Breed. 2022, 26, 202. [Google Scholar] [CrossRef] [PubMed]
- Perez-Sanz, F.; Navarro, P.J.; Egea-Cortines, M. Plant Phenomics: An Overview of Image Acquisition Technologies and Image Data Analysis Algorithms. GigaScience 2017, 6, gix092. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Oberti, R.; West, J.; Mouazen, A.M.; Bochtis, D. Detection of Biotic and Abiotic Stresses in Crops by Using Hierarchical Self Organizing Classifiers. Precis. Agric. 2017, 18, 383–393. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Wiesner-Hanks, T.; Stewart, E.L.; Kaczmar, N.; DeChant, C.; Wu, H.; Nelson, R.J.; Lipson, H.; Gore, M.A. Image Set for Deep Learning: Field Images of Maize Annotated with Disease Symptoms. BMC Res. Notes 2018, 11, 440. [Google Scholar] [CrossRef]
- Leena, N.; Saju, K. Classification of Macronutrient Deficiencies in Maize Plant Using Machine Learning. Int. J. Electr. Comput. Eng. (IJECE) 2018, 8, 4197–4203. [Google Scholar]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling Maize Above-Ground Biomass Based on Machine Learning Approaches Using UAV Remote-Sensing Data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef]
- Tu, K.; Wen, S.; Cheng, Y.; Xu, Y.; Pan, T.; Hou, H.; Gu, R.; Wang, J.; Wang, F.; Sun, Q. A Model for Genuineness Detection in Genetically and Phenotypically Similar Maize Variety Seeds Based on Hyperspectral Imaging and Machine Learning. Plant Methods 2022, 18, 81. [Google Scholar] [CrossRef]
- Panigrahi, K.P.; Das, H.; Sahoo, A.K.; Moharana, S.C. Maize Leaf Disease Detection and Classification Using Machine Learning Algorithms; Springer: Berlin/Heidelberg, Germany, 2020; pp. 659–669. [Google Scholar]
- Munkvold, G.P.; Hellmich, R.L.; Showers, W. Reduced Fusarium Ear Rot and Symptomless Infection in Kernels of Maize Genetically Engineered for European Corn Borer Resistance. Phytopathology 1997, 87, 1071–1077. [Google Scholar] [CrossRef]
- Munkvold, G.P. Epidemiology of Fusarium Diseases and Their Mycotoxins in Maize Ears. Eur. J. Plant Pathol. 2003, 109, 705–713. [Google Scholar] [CrossRef]
- Mesterházy, Á.; Lemmens, M.; Reid, L.M. Breeding for Resistance to Ear Rots Caused by Fusarium spp. in Maize—A Review. Plant Breed. 2012, 131, 1–19. [Google Scholar] [CrossRef]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Bradski, G. The OpenCV Library. Dr Dobb’s J. Softw. Tools 2000, 25, 120–123. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Midtvedt, B.; Helgadottir, S.; Argun, A.; Pineda, J.; Midtvedt, D.; Volpe, G. Quantitative Digital Microscopy with Deep Learning. Appl. Phys. Rev. 2021, 8, 011310. [Google Scholar] [CrossRef]
- Van Valen, D.A.; Kudo, T.; Lane, K.M.; Macklin, D.N.; Quach, N.T.; DeFelice, M.M.; Maayan, I.; Tanouchi, Y.; Ashley, E.A.; Covert, M.W. Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments. PLoS Comput. Biol. 2016, 12, e1005177. [Google Scholar] [CrossRef]
- Ebrahim, S.A.; Poshtan, J.; Jamali, S.M.; Ebrahim, N.A. Quantitative and Qualitative Analysis of Time-Series Classification Using Deep Learning. IEEE Access 2020, 8, 90202–90215. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).