Structure and Metabolic Activity of the Gut Microbiota in Diarrhea-Predominant Irritable Bowel Syndrome Combined with Functional Dyspepsia
Abstract
1. Introduction
2. Results
2.1. Gut Microbiota Data
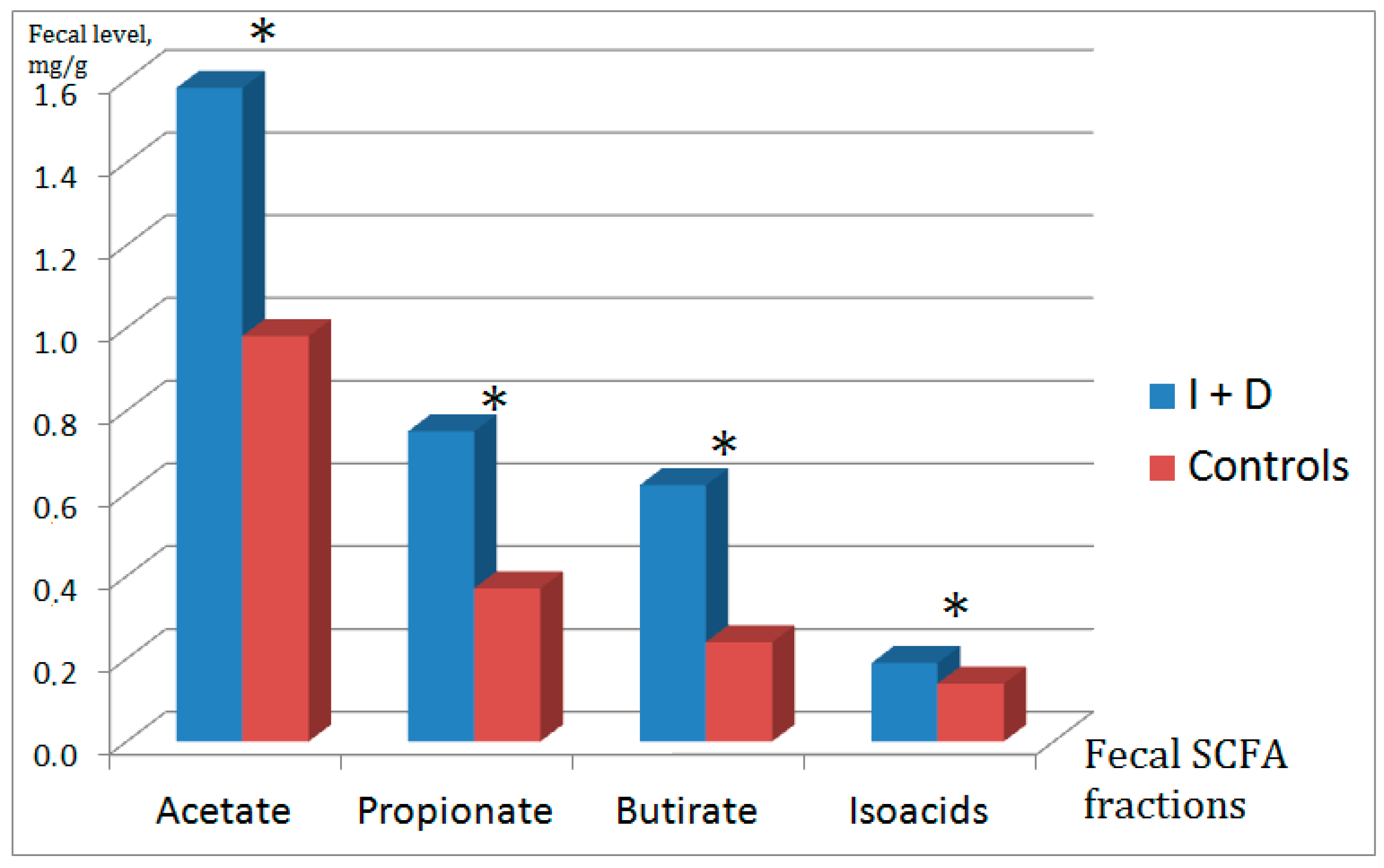
2.2. Fecal Short-Chain Fatty Acids
3. Materials and Methods
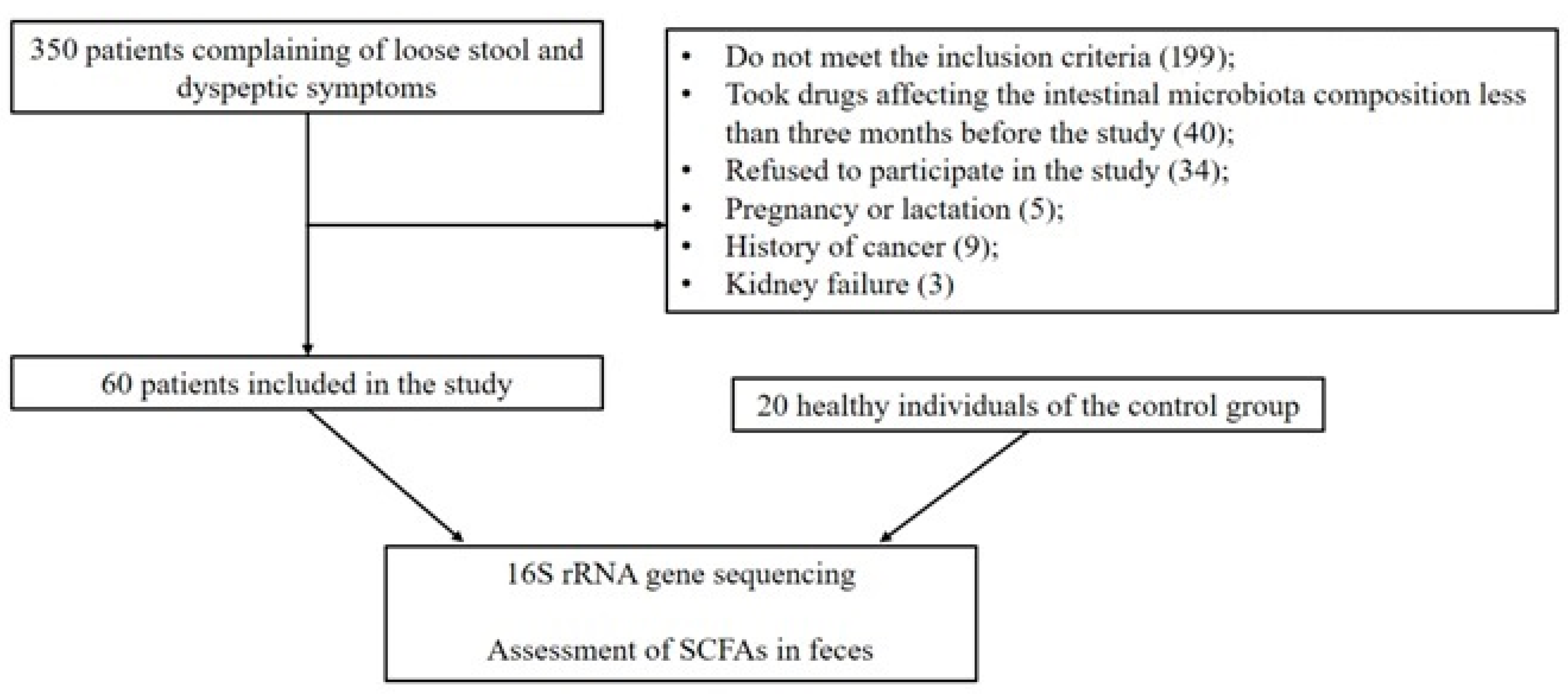
3.1. Patients
3.2. Severity of Symptoms Assessment
3.3. 16S rRNA Genes Library Preparartion and Sequencing
3.4. 16S rRNA Sequencing Data Analysis
3.5. Evaluation of the Content of Acetic, Propionic, Butyric, Acids and SCFA in Fecal Samples
3.6. Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lacy, B.E.; Mearin, F.; Chang, L.; Chey, W.D.; Lembo, A.J.; Simren, M.; Spiller, R. Bowel Disorders. Gastroenterology 2016, 150, 1393–1407. [Google Scholar] [CrossRef]
- Stanghellini, V.; Chan, F.K.; Hasler, W.L.; Malagelada, J.R.; Suzuki, H.; Tack, J.; Talley, N.J. Gastroduodenal Disorders. Gastroenterology 2016, 150, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Ivashkin, V.T.; Maev, I.V.; Shelygin, Y.A.; Baranskaya, E.K.; Belous, S.S.; Belousova, E.A.; Beniashvili, A.G.; Vasilyev, S.V.; Veselov, A.V.; Grigoryev, E.G.; et al. Diagnosis and Treatment of Irritable Bowel Syndrome: Clinical Recommendations of the Russian Gastroenterological Association and Association of Coloproctologists of Russia. Russ. J. Gastroenterol. Hepatol. Coloproctol 2021, 31, 74–95. [Google Scholar] [CrossRef]
- Jalanka-Tuovinen, J.; Salonen, A.; Nikkilä, J.; Immonen, O.; Kekkonen, R.; Lahti, L.; Palva, A.; de Vos, W.M. Intestinal Microbiota in Healthy Adults: Temporal Analysis Reveals Individual and Common Core and Relation to Intestinal Symptoms. PLoS ONE 2011, 6, e23035. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7618. [Google Scholar] [CrossRef] [PubMed]
- Ng, Q.X.; Soh, A.Y.S.; Loke, W.; Lim, D.Y.; Yeo, W.-S. The role of inflammation in irritable bowel syndrome (IBS). J. Inflamm. Res. 2018, 11, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Zolnikova, O.Y.; Potskhverashvili, N.D.; Kokina, N.I.; Trukhmanov, A.S.; Ivashkin, V.T. Intestinal Short-Chain Fatty Acids in Patients with Bronchial Asthma. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2019, 29, 53–59. [Google Scholar] [CrossRef]
- Ng, Q.X.; Yau, C.E.; Yaow, C.Y.L.; Chong, R.I.H.; Chong, N.Z.-Y.; Teoh, S.E.; Lim, Y.L.; Soh, A.Y.S.; Ng, W.K.; Thumboo, J. What Has Longitudinal ‘Omics’ Studies Taught Us about Irritable Bowel Syndrome? A Systematic Review. Metabolites 2023, 13, 484. [Google Scholar] [CrossRef]
- Sun, Q.; Jia, Q.; Song, L.; Duan, L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome. Medicine 2019, 98, e14513. [Google Scholar] [CrossRef] [PubMed]
- Farup, P.G.; Rudi, K.; Hestad, K. Faecal short-chain fatty acids - a diagnostic biomarker for irritable bowel syndrome? BMC Gastroenterol. 2016, 16, 51. [Google Scholar] [CrossRef] [PubMed]
- Ivashkin, V.; Sheptulin, A.; Lapina, T.; Trukhmanov, A.; Kartavenko, I.; Kiprianis, V.; Okhlobystina, O.; Mayev, I. Diagnosis and treatment of the functional dyspepsia: Clinical guidelines of the Russian Gastroenterological Association. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2017, 27, 50–61. [Google Scholar] [CrossRef]
- Kulich, K.R.; Madisch, A.; Pacini, F.; Piqué, J.M.; Regula, J.; Van Rensburg, C.J.; Újszászy, L.; Carlsson, J.; Halling, K.; Wiklund, I.K. Reliability and validity of the Gastrointestinal Symptom Rating Scale (GSRS) and Quality of Life in Reflux and Dyspepsia (QOLRAD) questionnaire in dyspepsia: A six-country study. Health Qual. Life Outcomes 2008, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Parikh, H.I.; Koparde, V.N.; Bradley, S.P.; Buck, G.A.; Sheth, N.U. MeFiT: Merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinform. 2016, 17, 491. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 4, D590–D596. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B.; Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-Like Differential Expression (ALDEx) Analysis for Mixed Population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef]
- Zhuang, X.; Xiong, L.; Li, L.; Li, M.; Chen, M. Alterations of gut microbiota in patients with irritable bowel syndrome: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2017, 32, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.N.; Wu, H.; Chen, Y.Z.; Chen, Y.J.; Shen, X.Z.; Liu, T.T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig. Liver Dis. 2017, 49, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Zhu, S.; Wang, B.; Duan, L. Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing. Clin. Transl. Gastroenterol. 2019, 10, e00012. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Tian, Z.; Li, L.; Zeng, Z.; Chen, M.; Xiong, L. Fecal Microbiota Alterations Associated With Diarrhea-Predominant Irritable Bowel Syndrome. Front. Microbiol. 2018, 9, 1600. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Liu, R.; Lee, A.; Long, Y.; Du, L.; Lai, S.; Chen, X.; Wang, L.; Si, J.; Owyang, C.; et al. Altered Intestinal Microbiota with Increased Abundance of Prevotella Is Associated with High Risk of Diarrhea-Predominant Irritable Bowel Syndrome. Gastroenterol. Res. Pr. 2018, 2018, 6961783. [Google Scholar] [CrossRef] [PubMed]
- Rangel, I.; Sundin, J.; Fuentes, S.; Repsilber, D.; de Vos, W.M.; Brummer, R.J. The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment. Pharmacol. Ther. 2015, 42, 1211–1221. [Google Scholar] [CrossRef]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 521–530. [Google Scholar] [CrossRef]
- Krogius-Kurikka, L.; Lyra, A.; Malinen, E.; Aarnikunnas, J.; Tuimala, J.; Paulin, L.; Mäkivuokko, H.; Kajander, K.; Palva, A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009, 9, 95. [Google Scholar] [CrossRef]
- Shukla, R.; Ghoshal, U.; Dhole, T.N.; Ghoshal, U.C. Fecal Microbiota in Patients with Irritable Bowel Syndrome Compared with Healthy Controls Using Real-Time Polymerase Chain Reaction: An Evidence of Dysbiosis. Dig. Dis. Sci. 2015, 60, 2953–2962. [Google Scholar] [CrossRef]
- Himmel, M.E.; Yao, Y.; Orban, P.C.; Steiner, T.S.; Levings, M.K. Regulatory T-cell therapy for inflammatory bowel disease: More questions than answers. Immunology 2012, 136, 115–122. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Ivashkin, V.; Poluektov, Y.; Kogan, E.; Shifrin, O.; Sheptulin, A.; Kovaleva, A.; Kurbatova, A.; Krasnov, G.; Poluektova, E. Disruption of the pro-inflammatory, anti-inflammatory cytokines and tight junction proteins expression, associated with changes of the composition of the gut microbiota in patients with irritable bowel syndrome. PLoS ONE 2021, 16, e0252930. [Google Scholar] [CrossRef] [PubMed]
- Mitselou, A.; Grammeniatis, V.; Varouktsi, A.; Papadatos, S.S.; Katsanos, K.; Galani, V. Proinflammatory cytokines in irritable bowel syndrome: A comparison with inflammatory bowel disease. Intest. Res. 2020, 18, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Ivashkin, V.T.; Zolnikova, O.Y. Irritable Bowel Syndrome in Terms of Changes in the Microbiota. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2019, 29, 68–76. [Google Scholar] [CrossRef]
- Ulyanin, A.; Poluektova, Y.; Pavlov, C.; Ivashkin, V. Potential of nutraceutical products for irritable bowel syndrome remission maintenance. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2018, 28, 101–108. [Google Scholar] [CrossRef]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Fredericks, E.; Theunissen, R.; Roux, S. Short chain fatty acids and monocarboxylate transporters in irritable bowel syndrome. Turk. J. Gastroenterol. 2020, 31, 840–847. [Google Scholar] [CrossRef]
- Gargari, G.; Taverniti, V.; Gardana, C.; Cremon, C.; Canducci, F.; Pagano, I.; Barbaro, M.R.; Bellacosa, L.; Castellazzi, A.M.; Valsecchi, C.; et al. Fecal Clostridiales distribution and short-chain fatty acids reflect bowel habits in irritable bowel syndrome. Environ. Microbiol. 2018, 20, 3201–3213. [Google Scholar] [CrossRef] [PubMed]
- Ringel-Kulka, T.; Choi, C.H.; Temas, D.; Kim, A.; Maier, D.M.; Scott, K.; A Galanko, J.; Ringel, Y. Altered Colonic Bacterial Fermentation as a Potential Pathophysiological Factor in Irritable Bowel Syndrome. Am. J. Gastroenterol. 2015, 110, 1339–1346. [Google Scholar] [CrossRef]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Kobayashi, M.; Mikami, D.; Kimura, H.; Kamiyama, K.; Morikawa, Y.; Yokoi, S.; Kasuno, K.; Takahashi, N.; Taniguchi, T.; Iwano, M. Short-chain fatty acids, GPR41 and GPR43 ligands, inhibit TNF-α-induced MCP-1 expression by modulating p38 and JNK signaling pathways in human renal cortical epithelial cells. Biochem. Biophys. Res. Commun. 2017, 486, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Park, B.-O.; Kang, J.S.; Paudel, S.; Park, S.G.; Park, B.C.; Han, S.-B.; Kwak, Y.-S.; Kim, J.-H.; Kim, S. Novel GPR43 Agonists Exert an Anti-Inflammatory Effect in a Colitis Model. Biomol. Ther. 2022, 30, 48–54. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-Chain Fatty Acids and Their Association with Signalling Pathways in Inflammation, Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.-M.; Zhao, H.-L.; Guo, G.-J.; Xu, J.; Zhou, Y.-L.; Huang, H.-L.; Nie, Y.-Q. Characterization of short-chain fatty acids in patients with ulcerative colitis: A meta-analysis. BMC Gastroenterol. 2022, 22, 117. [Google Scholar] [CrossRef]
- Carroll, I.M.; Maharshak, N. Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J. Gastroenterol. 2013, 19, 7531–7543. [Google Scholar] [CrossRef]
| Symptom | I + D (n = 60) | Control (n = 20) | p |
|---|---|---|---|
| Pain or discomfort in upper abdomen | 2 [1–3] | 1 [1–1] | <0.001 |
| Heartburn | 2 [1–3] | 1 [1–1] | <0.001 |
| Acid reflux | 1 [1–3] | 1 [1–1] | 0.002 |
| Hunger pangs | 1 [1–3] | 1 [1–1] | 0.002 |
| Nausea | 2 [1–3] | 1 [1–1] | <0.001 |
| Rumbling | 4 [3–5] | 1 [1–1] | <0.001 |
| Stomach felt bloating | 3 [3–5] | 1 [1–1] | <0.001 |
| Burping | 2 [2–4] | 1 [1–1] | <0.001 |
| Passing gas or flatus | 3 [2–4] | 1 [1–1] | <0.001 |
| Constipation | 1 [1–1] | 1 [1–1] | 0.116 |
| Increased stool frequency | 3 [2–4] | 1 [1–1] | <0.001 |
| Loose stool | 4 [3–5] | 1 [1–1] | <0.001 |
| Hard stool | 1 [1–2] | 1 [1–1] | 0.056 |
| Urgent need to have a bowel movement | 1 [1–2] | 1 [1–1] | <0.001 |
| Sensation of incomplete bowel emptying | 2 [2–3] | 1 [1–1] | <0.001 |
| Taxon/Index | Taxon Rank | I + D (n = 60) | Control (n = 20) | p |
|---|---|---|---|---|
| Shannon index | - | 4.52 [4.08–4.83] | 4.54 [4.43–4.79] | 0.562 |
| Chao1 index | - | 567 [456–758] | 600 [477–690] | 1.000 |
| ACE index | - | 554 [455–746] | 589 [469–680] | 0.918 |
| Firmicutes | phylum | 83,560.2 | 70,600.2 | 0.075 |
| Bacteroidota | phylum | 13,721.4 | 27,140.8 | 0.075 |
| Actinobacteriota | phylum | 3733.0 | 4357.4 | 0.411 |
| Proteobacteria | phylum | 2410.6 | 2739.5 | 0.570 |
| Verrucomicrobiota | phylum | 628.7 | 138.8 | 0.340 |
| Desulfobacterota | phylum | 98.8 | 137.6 | 0.189 |
| Clostridia | class | 66,610.1 | 64,506.2 | 0.632 |
| Bacilli | class | 14,653.1 | 3373.5 | 0.102 |
| Bacteroidia | class | 13,719.2 | 27,140.7 | 0.115 |
| Actinobacteria | class | 3147.1 | 3317.0 | 0.434 |
| Gammaproteobacteria | class | 2339.4 | 2712.1 | 0.678 |
| Negativicutes | class | 2157.1 | 2683.9 | 0.737 |
| Verrucomicrobiae | class | 628.7 | 138.6 | 0.434 |
| Coriobacteriia | class | 583.6 | 1040.1 | 0.853 |
| Taxon | Taxon Rank | LogFC | I + D (n = 60) | Control (n = 20) | p |
|---|---|---|---|---|---|
| Holdemanella | genus | 9.01 | 6183.8 | 0.0 | <0.001 |
| Erysipelotrichaceae | family | 5.02 | 7294.1 | 212.9 | 0.035 |
| Prevotellaceae | family | 2.88 | 1956.4 | 254.5 | 0.036 |
| Erysipelotrichales | order | 2.70 | 9965.0 | 1527.8 | 0.001 |
| Agathobacter | genus | 2.28 | 7555.1 | 1547.1 | 0.003 |
| Slackia | genus | 2.23 | 44.4 | 0.0 | 0.009 |
| Lactococcus | genus | 1.84 | 40.9 | 2.7 | 0.011 |
| Pseudomonadales | order | 1.73 | 144.8 | 35.3 | 0.006 |
| Pseudomonadaceae | family | 1.69 | 139.5 | 35.1 | 0.013 |
| Pseudomonas | genus | 1.67 | 137.1 | 34.9 | 0.009 |
| Stenotrophomonas | genus | 1.60 | 76.4 | 17.1 | 0.003 |
| Xanthomonadaceae | family | 1.60 | 77.1 | 17.4 | 0.003 |
| Xanthomonadales | order | 1.60 | 77.1 | 17.4 | 0.001 |
| Rhizobiaceae | family | 1.10 | 28.4 | 6.8 | 0.036 |
| Erysipelatoclostridiaceae | family | 1.02 | 2669.3 | 1314.8 | 0.039 |
| Lachnospiraceae | family | 0.33 | 32,140.1 | 25,607.6 | 0.035 |
| Lachnospirales | order | 0.33 | 32,154.7 | 25,619.3 | 0.02 |
| Flavonifractor | genus | −1.07 | 68.5 | 156.4 | 0.039 |
| Bacteroidaceae | family | −1.15 | 9830.4 | 21,868.1 | 0.036 |
| Tannerellaceae | family | −1.33 | 590.3 | 1501.8 | 0.036 |
| Rikenellaceae | family | −1.42 | 893.9 | 2404.9 | 0.047 |
| Bilophila | genus | −1.46 | 42.3 | 137.4 | 0.009 |
| Merdibacter | genus | −1.49 | 6.2 | 39.3 | 0.003 |
| Peptostreptococcaceae | family | −1.51 | 332.4 | 971.8 | 0.044 |
| Parabacteroides | genus | −1.53 | 504.6 | 1475.0 | 0.018 |
| Eisenbergiella | genus | −1.96 | 6.3 | 59.4 | 0.003 |
| Hungatella | genus | −2.08 | 16.5 | 108.7 | 0.009 |
| Burkholderiaceae | family | −2.14 | 11.6 | 92.1 | <0.001 |
| Ralstonia | genus | −2.17 | 11.2 | 92.1 | <0.001 |
| Frisingicoccus | genus | −3.99 | 0.2 | 182.1 | 0.003 |
| Parameter | Group | Mean Value | SD | p |
|---|---|---|---|---|
| Absolute total level of SCFAs | I + D | 3.19 | 1.69 | 0.003 |
| Control | 1.81 | 1.29 | ||
| Absolute level of acetic acid (C2) | I + D | 1.58 | 0.78 | 0.003 |
| Control | 0.93 | 0.64 | ||
| Absolute level of propionic acid (C3) | I + D | 0.75 | 0.49 | 0.002 |
| Control | 0.37 | 0.3 | ||
| Absolute level of butyric acid (C4) | I + D | 0.62 | 0.40 | <0.001 |
| Control | 0.24 | 0.21 | ||
| Absolute level of isoacids | I + D | 0.19 | 0.10 | 0.028 |
| Control | 0.14 | 0.1 | ||
| IsoCn/Cn | I + D | 0.57 | 1.4 | 0.013 |
| Control | 0.52 | 0.23 | ||
| Anaerobic index | I + D | −0.79 | 0.33 | 0.046 |
| Control | −0.69 | 0.19 | ||
| Relative level of acetic acid (C2) | I + D | 0.55 | 0.07 | 0.111 |
| Control | 0.56 | 0.12 | ||
| Relative level of propionic acid (C3) | I + D | 0.24 | 0.05 | 0.463 |
| Control | 0.23 | 0.04 | ||
| Relative level of butyric acid (C4) | I + D | 0.21 | 0.06 | 0.178 |
| Control | 0.17 | 0.07 | ||
| Relative level of isoacids | I + D | 0.07 | 0.035 | 0.107 |
| Control | 0.09 | 0.03 |
| Gut Microbiota Taxon | Acetic Acid (C2) | Propionic Acid (C3) | Butyric Acid (C4) | Isoacids | Total Acid Content | Anaerobic Index |
|---|---|---|---|---|---|---|
| Acidaminococcaceae | −0.55; <0.001 | 0.31; 0.04 | ||||
| Actinomycetaceae | 0.30; 0.05 | 0.39; 0.008 | 0.30; 0.04 | −0.45; 0.002 | ||
| Aerococcaceae | 0.42; 0.004 | 0.35; 0.02 | 0.55; <0.001 | 0.46; 0.001 | ||
| Akkermansiaceae | 0.38; 0.01 | 0.32; 0.04 | ||||
| Anaerovoracaceae | −0.61; <0.001 | −0.63; <0.001 | −0.71; <0.001 | −0.65; <0.001 | 0.51; <0.001 | |
| Atopobiaceae | 0.39; 0.008 | |||||
| Bacteroidaceae | 0.39; 0.008 | |||||
| Barnesiellaceae | −0.35; 0.02 | −0.49; <0.001 | −0.36; 0.02 | 0.60; <0.001 | ||
| Bifidobacteriaceae | −0.32; 0.03 | |||||
| Burkholderiaceae | −0.32; 0.03 | −0.38; 0.01 | −0.31; 0.04 | |||
| Butyricicoccaceae | 0.37; 0.01 | |||||
| Carnobacteriaceae | 0.33; 0.03 | 0.52; <0.001 | 0.47; 0.001 | 0.47; 0.001 | −0.65; <0.001 | |
| Chitinophagaceae | −0.31; 0.04 | 0.32; 0.03 | ||||
| Christensenellaceae | −0.34; 0.02 | −0.47; 0.001 | −0.54; <0.001 | −0.46; 0.001 | 0.57; <0.001 | |
| Clostridiaceae | −0.31; 0.04 | −0.45; 0.002 | −0.34; 0.02 | 0.35; 0.02 | ||
| Comamonadaceae | −0.41; 0.005 | −0.30; 0.05 | 0.42; 0.004 | |||
| Defluviitaleaceae | −0.40; 0.007 | |||||
| Enterococcaceae | 0.51; <0.001 | 0.32; 0.03 | −0.30; 0.04 | |||
| Erysipelotrichaceae | −0.37; 0.01 | |||||
| Fusobacteriaceae | 0.29; 0.05 | 0.31; 0.04 | ||||
| Gemellaceae | 0.46; 0.002 | 0.55; <0.001 | 0.63; <0.001 | 0.57; <0.001 | −0.64; <0.001 | |
| Lachnospiraceae | 0.32; 0.03 | 0.50; <0.001 | 0.32; 0.04 | −0.55; <0.001 | ||
| Lactobacillaceae | −0.35; 0.02 | −0.30; 0.04 | ||||
| Leuconostocaceae | −0.48; 0.0008 | 0.36; 0.02 | ||||
| Marinifilaceae | −0.37; 0.01 | −0.41; 0.005 | −0.39; 0.008 | −0.33; 0.03 | 0.45; 0.002 | |
| Methanobacteriaceae | −0.48; 0.001 | −0.42; 0.004 | −0.45; 0.002 | −0.40; 0.006 | ||
| Micrococcaceae | 0.34; 0.02 | 0.38; 0.01 | 0.31; 0.04 | −0.63; <0.001 | ||
| Monoglobaceae | 0.32; 0.03 | |||||
| Mycobacteriaceae | −0.32; 0.03 | −0.39; 0.008 | −0.34; 0.02 | −0.33; 0.03 | 0.37; 0.01 | |
| Oscillospiraceae | −0.33; 0.03 | −0.45; 0.002 | −0.31; 0.04 | 0.34; 0.02 | ||
| Oxalobacteraceae | −0.31; 0.04 | −0.35; 0.02 | −0.45; 0.002 | −0.34; 0.02 | 0.43; 0.004 | |
| Pasteurellaceae | 0.34; 0.02 | 0.57; <0.001 | 0.36; 0.01 | −0.65; <0.001 | ||
| Peptococcaceae | −0.33; 0.03 | −0.33; 0.02 | ||||
| Peptostreptococcaceae | −0.30; 0.05 | −0.45; 0.002 | −0.33; 0.03 | 0.32; 0.03 | ||
| Puniceicoccaceae | −0.32; 0.03 | −0.37; 0.01 | −0.38; 0.01 | −0.32; 0.03 | 0.40; 0.007 | |
| Rikenellaceae | −0.41; 0.005 | −0.40; 0.007 | −0.36; 0.01 | 0.56; <0.001 | ||
| Saccharimonadaceae | −0.56; <0.001 | |||||
| Selenomonadaceae | 0.33; 0.02 | 0.41; 0.005 | −0.30; 0.05 | −0.50; <0.001 | ||
| Solimonadaceae | −0.38; 0.009 | −0.40; 0.007 | −0.34; 0.02 | −0.36; 0.01 | ||
| Solirubrobacteraceae | −0.30; 0.05 | −0.34; 0.02 | −0.32; 0.03 | −0.31; 0.04 | 0.33; 0.03 | |
| Streptococcaceae | 0.37; 0.01 | 0.42; 0.004 | 0.33; 0.03 | −0.74; <0.001 | ||
| Sutterellaceae | 0.33; 0.03 | |||||
| Tannerellaceae | −0.31; 0.04 | −0.44; 0.002 | −0.32; 0.03 | 0.55; <0.001 | ||
| Vibrionaceae | −0.31; 0.04 | |||||
| Weeksellaceae | −0.30; 0.05 | −0.34; 0.02 | −0.32; 0.03 | −0.31; 0.04 | 0.33; 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovaleva, A.; Poluektova, E.; Maslennikov, R.; Zolnikova, O.; Shifrin, O.; Kudryavtseva, A.; Krasnov, G.; Fedorova, M.; Karchevskaya, A.; Ivashkin, V. Structure and Metabolic Activity of the Gut Microbiota in Diarrhea-Predominant Irritable Bowel Syndrome Combined with Functional Dyspepsia. Gastrointest. Disord. 2023, 5, 296-309. https://doi.org/10.3390/gidisord5030024
Kovaleva A, Poluektova E, Maslennikov R, Zolnikova O, Shifrin O, Kudryavtseva A, Krasnov G, Fedorova M, Karchevskaya A, Ivashkin V. Structure and Metabolic Activity of the Gut Microbiota in Diarrhea-Predominant Irritable Bowel Syndrome Combined with Functional Dyspepsia. Gastrointestinal Disorders. 2023; 5(3):296-309. https://doi.org/10.3390/gidisord5030024
Chicago/Turabian StyleKovaleva, Aleksandra, Elena Poluektova, Roman Maslennikov, Oxana Zolnikova, Oleg Shifrin, Anna Kudryavtseva, George Krasnov, Maria Fedorova, Anna Karchevskaya, and Vladimir Ivashkin. 2023. "Structure and Metabolic Activity of the Gut Microbiota in Diarrhea-Predominant Irritable Bowel Syndrome Combined with Functional Dyspepsia" Gastrointestinal Disorders 5, no. 3: 296-309. https://doi.org/10.3390/gidisord5030024
APA StyleKovaleva, A., Poluektova, E., Maslennikov, R., Zolnikova, O., Shifrin, O., Kudryavtseva, A., Krasnov, G., Fedorova, M., Karchevskaya, A., & Ivashkin, V. (2023). Structure and Metabolic Activity of the Gut Microbiota in Diarrhea-Predominant Irritable Bowel Syndrome Combined with Functional Dyspepsia. Gastrointestinal Disorders, 5(3), 296-309. https://doi.org/10.3390/gidisord5030024






