Abstract
Circulating tumour DNAs and non-coding RNAs present in body fluids have been under investigation as tools for cancer diagnosis, disease monitoring, and prognosis for many years. These so-called liquid biopsies offer the opportunity to obtain information about the molecular make-up of a cancer in a minimal invasive way and offer the possibility to implement theranostics for precision oncology. Furthermore, liquid biopsies could overcome the limitations of tissue biopsies in capturing the complexity of tumour heterogeneity within the primary cancer and among different metastatic sites. Liquid biopsies may also be implemented to detect early tumour formation or to monitor cancer relapse of response to therapy with greater sensitivity compared with the currently available protein-based blood biomarkers. Most colorectal cancers are often diagnosed at late stages and have a high mortality rate. Hence, biomolecules as nucleic acids present in liquid biopsies might have prognostic potential and could serve as predictive biomarkers for chemotherapeutic regimens. This review will focus on the role of circulating tumour DNAs and non-coding RNAs as diagnostic, prognostic, and predictive biomarkers in the context of colorectal cancer.
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy with 1.3 million new cases worldwide and 700,000 cancer-related deaths per year [1]. During the last years, the survival rate of CRC patients has improved significantly, owing to different treatment strategies including surgery, chemotherapy, and radiotherapy. CRC development is a multifactorial disease caused by a combination of poor lifestyle habits such as obesity, smoking, alcohol, and red meat consumption combined with genetic susceptibility and inflammatory conditions of the digestive tract [2]. Around 85% of CRC cases are sporadic, with age as the most critical risk factor, whereas nearly 10% of CRC cases are associated with familiar conditions [3] and hereditary syndromes such as Lynch syndrome (2–5%), familial adenomatous polyposis (FAP) (<1%), and MYH gene-associated polyposis (<1%) [4].
The vast majority of CRCs are adenocarcinomas arising from epithelial cells of the colorectal mucosa [5]. Normally, CRC develops through the gradual accumulation of genetic and epigenetic changes and a stepwise progression from benign adenoma to malignant adenocarcinoma. This neoplastic transformation is thought to occur in a timeframe of 10 years [6]. The development of distant metastasis from primary CRC may happen as early as one year from the emergence of primary CRC [7]. In general, a time window exists for intervention before metastatic disease progression. Thus, CRC screening and early diagnosis might offer an option to improve CRC outcomes [6].
Nevertheless, early diagnosis of CRC still represents a major challenge and the currently available screening methods are invasive (video colonoscopy), require radiation exposure (virtual colonoscopy), are unpleasant for the patient (stool-based analysis) or expensive, and are not ubiquitously accepted (capsule colonoscopy), making the global take-up rate low [8]. Furthermore, the currently used blood biomarkers—carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9)—have limited utility owing to low sensitivity and specificity, especially in early stages of the disease, thus these protein-based assays are used by clinicians mostly for disease monitoring post-surgery [9]. The new serum methylated Septin-9 assay also showed unexpectedly low sensitivity in stage I CRC and advanced adenomas [10], highlighting the importance of novel and more accurate tests and methodologies in this space. The importance of improvements in early detection is underlined by the fact that diagnosis at early stages results in a five-year survival rate of 90% compared with a survival rate of 13% for CRC diagnosed in stage IV [11]. Another critical need in managing CRC patients is to detect the development of acquired resistance to anti-cancer treatment early and to improve the prognosis in advanced stages of CRC using targeted therapies. Identification of biomarkers for early detection of CRC as well as biomarkers for therapy success or for detection of acquired resistance is urgently needed, albeit it is mandatory that they should have high specificity and sensitivity. To reach this goal, molecular biomarkers such as circulating tumour DNA and non-coding RNAs have been widely investigated and liquid biopsies might represent the new era for the discovery of biomarkers [12,13,14].
Herein, we will discuss the challenges of liquid biopsies in comparison with tissue biopsies and summarise the current status of molecular biomarkers with a special focus on circulating tumour DNA, microRNAs, and long non-coding RNAs for diagnosis, disease monitoring, and prognosis of CRC patients.
2. Current Status of Liquid Biopsy in Clinical Setting versus Traditional Biopsy
The concept of liquid biopsy concept was initially described in 1948 [15], but only several decades later it became possible to use liquid biopsies for clinical purposes (diagnosis, prognostication, and prediction) (Figure 1).
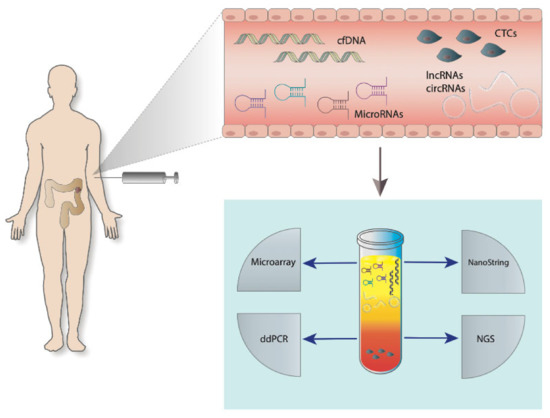
Figure 1.
Liquid biopsies in the management of colorectal cancer (CRC). Disease specific circulating free DNA (cfDNA), circular RNAs (circRNAs), long-non coding RNA (lncRNAs), microRNAs, and circulating tumour cells (CTCs) are present in the blood of a colon cancer patient. Analysis of nucleic-acids isolated from a blood sample can be performed by different molecular biological methods such as microarrays, NanoString technology, digital droplet PCR (ddPCR), or next-generation sequencing (NGS).
The major application of liquid biopsies in recent years has been focussed on the detection of circulating tumour DNA and non-coding RNAs (circular RNAs, microRNAs, and long non-coding RNAs), the latter present as free floating and proteins packed forms or encapsulated in exosomes [12,16]. Another application of liquid biopsy that has been widely investigated is the detection of circulating tumour cells (CTCs) [17], albeit this application requires a higher volume of sample body fluids, and it is considered more expensive. Furthermore, the outcomes seem to be highly dependent on tumour stage as the fraction of CTCs appears to correlate with the tumour stage and increases in advanced and metastatic CRC [18]. Thus, in general, liquid biopsies might represent an alternative or complementary method to implement early diagnosis and provide more specific prognosis and therapeutic prediction to tissue biopsies. The most significant differences including advantages and disadvantages between liquid and tissue biopsies are summarized in Table 1.

Table 1.
Overview about advantages and disadvantages of the two biopsy forms.
One of the most relevant advantages emerging from many reports is that liquid biopsies can effectively capture tumour heterogeneity, thus appearing as essential tools to monitor treatment responses as well as genomic variations [19]. This overcomes a disadvantage of tissue biopsies and enables implementation of theranostics in the clinical settings, resulting in better treatments for cancer patients [20,21]. However, limitations in liquid biopsies still do exist; for example, the lack of unified methods and standardization [22], the final diagnosis of cancers still relies on specialised pathological assessment, and it is only possible with morphological and immune-colorimetric evaluation of validated markers, thus liquid biopsy cannot be used [23].
In the following, we will discuss the different molecular tumour markers present in liquid biopsies that can be used as biomarkers for early diagnosis, disease monitoring, and therapy.
3. Biomolecules
3.1. Circulating Tumour DNAs (ctDNAs)
Under physiological as well as pathological conditions (like cancer), cells are constantly cleared by apoptosis and necrosis. One consequence is the permanent release of DNA fragments into the circulation [24,25]. The amount of circulating tumour DNA (ctDNA) varies between less than 0.1% to over 10% of the total circulating cell-free DNA in cancer patients, depending on cellular turnover, cancer stage, and response to treatment [26,27].
3.2. Circulating Tumour Cells (CTCs)
Besides ctDNAs, intact tumour cells are also released into the circulation by cancer areas from primary tumours as well as metastatic deposits [28]. The concentration of CTCs in blood is extremely low and, especially at early stages, often impossible to detect [29,30]. Isolation and detection of CTCs is still a challenge and special flow through specific devices is in use for capturing the circulating tumour cells with antibodies directed against tumour cell specific antigens [30]. Because of the fact that the amount of CTCs in the circulation increases in parallel with the tumour burden, this could become an accurate method for monitoring cancer growth in real time [31,32,33].
3.3. Non-Coding RNAs
Non-coding RNAs represent a large family of RNAs that are not coding for known proteins and form a significant proportion of the human genome [34,35].
After being considered as “junk” for a long time, it is nowadays well established that non-coding RNAs play important roles in maintaining cells’ homeostasis and functions. Often, non-coding RNAs are expressed in a cell- and tissue-specific pattern, and are specifically deregulated in cancer [35].
Non-coding RNAs are a heterogeneous group of RNA molecules in terms of length, structure, and biological function. They are divided into two categories according to their length: (a) non-coding RNAs with less than 200 nucleotides are summarized as small non-coding RNAs and include micro RNA, vault RNAs, small interfering RNA, piwi-interacting RNA, small nucleolar RNA, small nuclear RNA, transfer RNAs, ribosomal RNAs, endogenous small interfering RNAs, sno-derived RNAs, transcription initiation RNAs, microRNA-offset-RNAs, circular RNAs, extracellular RNAs, and small Cajal body RNA; (b) non-coding RNAs with more than 200 nucleotides are called long non-coding RNAs [35,36,37,38]. A summary of functionally validated non-coding RNAs can be found in the “ncRNA database”.
Long non-coding RNAs and miRNAs have been established to play a central role in gene expression control, either as transcriptional or post-transcriptional regulators [39]. In cancer microRNAs as well as long non-coding RNAs, influence all hallmarks of cancer by limiting the expression of tumour suppressors and increasing the activation of oncogenic pathways [35,40]. Both groups of non-coding RNAs also emerge as important mediators in drug-sensitivity and drug-resistance mechanisms [35,41,42].
3.4. Micro RNAs (miRNAs)
MiRNAs are composed of 18–24 nucleotides and they function as regulators of gene expression at the translational level [43]. In most cases, miRNAs act as negative regulators of gene expression upon binding to the 3’-untranslated region (3’-UTR) of the target mRNA, resulting in down-regulating the protein synthesis rate [44]. Nevertheless, some miRNAs bind to the 5’-untraslated region (5’-UTR) or to the coding sequence of their target mRNAs [45].
MiRNA expression patterns are tissue-specific and altered miRNAs’ expression, which results in deregulated proteins’ expression, is observed in the context of virtually any cancer [35,41,46,47,48,49,50,51]. Deregulated expression of miRNAs is caused by diverse mechanisms such as promoter methylation, mutations, amplifications, or deletions involving miRNA coding regions, transcriptional regulation by proteins (e.g., p53 and c-myc), or long-non coding RNAs, as well as down-regulation by mutations in genes coding for miRNA biogenesis-related enzymes (i.e., Drosha, Exportin 5, Dicer, Argonaute 2) [44,52,53,54].
In addition to acting within cells, circulating cell-free miRNAs have been detected in body fluids [55,56]. MiRNAs are either released by passive leakage from lytic cells or actively secreted via extracellular vesicles (e.g., exosomes), as well as bound to high-density lipoprotein (HDL) [57,58]. Most of the extracellular miRNAs are exported in conjunction with RNA-binding proteins, such as AGO2 and NPM1, and thus protected against degradation [59,60,61].
3.5. Circular RNAs (circRNAs)
CircRNAs were first discovered as viroids in RNA viruses [62], but further studies have clearly shown that circRNAs are also present in eukaryotes, and they have been recently identified as a further class of non-coding RNAs [63,64,65]. Compared with linear RNAs, circRNAs are extremely resistant to RNA exonucleases as their chemical structure consisting of covalent closed-loop structures missing 3′-polyA tails and 5′-caps provides protection to degradation [66]. Therefore, circRNAs are extremely stable in body fluids and can be isolated from liquid biopsies [67,68,69]. Nowadays, it is widely accepted that circRNAs are important regulators that influence physiological and pathological processes [70,71,72,73,74] by regulating splicing mechanisms and acting as miRNA sponges [64]. Besides transcriptional regulation, circRNAs can also regulate protein functions by forming complexes with proteins and alter proteins’ functions [75,76].
3.6. Long Non-Coding RNAs (lncRNAs)
LncRNAs are RNA species spanning more than 200 nucleotides in length and represent important regulators during development and pathological processes [77,78,79,80,81]. Under physiological conditions, most lncRNAs are expressed at low levels as they are transcribed from promoters with low CpG dinucleotide contents [82], and an aberrant expression of lncRNAs is observed in several diseases owing to modulation of the chromatin state [83]. LncRNAs can be localized in the nucleus or cytoplasm [84,85] and, depending on the subcellular localization, they can directly regulate gene expression by the following: (a) influencing chromatin modification by binding to chromatin regulatory proteins; (b) regulating splicing and stability of mRNAs; as well as (c) indirectly in transcriptional and post-transcriptional gene expression mechanisms, by interacting with other RNAs and proteins [86,87,88].
A strong interaction between lncRNAs and miRNAs exists in RNA regulation processes [35,89,90,91]. LncRNAs can act as miRNA decoys by sequestering miRNAs or lncRNAs can compete with miRNAs for specific binding sites in the non-coding regions of mRNAs and preventing the transcriptional repression caused by miRNA [35,89,91]. Furthermore, some lncRNAs can be processed into miRNAs [35,89,90,91].
Altered lncRNA expression has been observed in cancer and changes in lncRNAs expression can be detected in body fluids like urine and blood [92,93,94,95]. LncRNAs are released either encapsulated in exosomes or inside apoptotic bodies and they are bound to RNA-binding proteins [96,97]; therefore, they are resistant to RNase degradation [93,98].
4. Techniques and Methods for Analysing Nucleic-Acids in Liquid Biopsy
4.1. Microarray
This method has been used in many studies for screening purposes, in the evaluation of known nucleic-acid sequences panels. Several commercially available microarrays for detecting the expression of ctDNA, circRNAs, miRNAs, and lncRNAs have often been used for genome-wide analysis expression of differences between normal and pathological samples [99,100,101,102,103,104]. However, all these microarrays have the limitation that an amplification step is always necessary before chip-based measurement takes place.
4.2. Nanostring nCounter
NanoString nCounter technology on the contrary is another high-throughput screening method for nucleic acid sequences (genes, miRNAs, DNA) that directly detects the gene target expression levels without any enzymatic amplification reaction. NanoString nCounter allows counting molecules in a given sample directly using barcoded target 5′-end sequence-specific probes for capturing and purification as well as a barcoded 3′-end target sequence-specific fluorescent-labelled probe for detection [105,106,107]. Many reports have shown in the last years how nanostring technology can be used in order to detect, for instance, miRNAs panels in plasma/serum of patients for diagnostic or disease recurrence control in CRC and other cancers [108,109]. The limitation of this technology is the requirement of validation of the hits identified with orthogonal technologies like qPCR, ddPCR, immunohistochemistry, and florescence in-situ hybridization [110,111,112].
4.3. Digital-Droplet PCR (ddPCR)
A new generation of PCR called ddPCR has emerged in the last years allowing absolute quantification of nucleic acid molecules encapsulated in discrete, volumetrically defined, water-in-oil droplet partitions. The nucleic acid template sample is randomly distributed into these partitions, such that some droplets contain no nucleic acid template and others contain one or more copies of the template defined by a Poisson distribution [113,114,115]. As an end-point measurement, following PCR amplification in single droplets, ddPCR requires neither parallel amplification of any housekeeping gene for normalization nor the need of standard curves’ generation for detection and quantification [113,116,117,118,119,120]. On the basis of its high sensitivity, ddPCR is often used for identification of rare or low-level mutations in cancer [25,119,121,122] and as an independent method for verification of next-generation sequencing [123]. The detection is limited to only a few hotspots per reaction, and this is a bottleneck of ddPCR technology.
4.4. Next Generation Sequencing (NGS)
Several different sequencing platforms are currently available for NGS (e.g., Ion Torrent’s PGM, Pacific Biosciences’ RS, and Illumina platforms) and the preparation of sequence libraries is specific to each platform [124,125]. RNA must be reverse-transcribed into cDNA [126] and then cDNA is used in the following steps, which are also mandatory for using DNA as starting material. The pre-processing can be subdivided into several steps. Extracted genomic DNA is randomly fragmented into a library of small DNA sequences (fragment size is platform-specific and varies between 100 bp and 20 kbp) [127,128]. Specific adapters (still platform-specific) are ligated to the fragments at the 3′ and 5′ends. Depending on the sequencing platform, the libraries must be pre-amplified prior to sequencing or can be used directly without any amplification step [124,129,130]. In last years, NGS has become the gold standard for sequencing of gene-panels given its high-throughput and sensitivity. The ability of sample pooling has also allowed a reduction of costs, making it highly attractive for diagnostic purposes. This has increased its application for the analysis of gene panels for cancer genome profiling or whole genome or exome sequencing in liquid biopsy.
5. Liquid Biopsy and Biomarkers
Several studies have tried to identify molecular biomarkers for diagnostic and prognostic purposes using primary lesions as starting material and correlate this information with clinical assessment and disease stage [131,132]. Most of the studies involved the investigation of classical mutation pattern in CRC, as described by Vogelstein, thus with the use of markers like APC (Adenomatous polyposis coli; mutation rate 80%), KRAS (Kirsten rat sarcoma; mutation rate 40%), BRAF (v-raf murine sarcoma viral oncogene homolog B1; mutation rate is between 8 and 10%), and TP53 (tumor protein 53; mutation rate 60%) [133]. Therefore, mutations in APC, BRAF, KRAS, and TP53 have been the focus of investigations and their potential as prognostic and predictive factors in early as well as advanced cancers has been evaluated in order to potentially identify subgroups of patients with common characteristics [134,135]. Early studies focused only on protein biomarkers mainly based on immunostaining of known deregulated proteins using cancer tissues [136], but recently, genetic DNA-based biomarkers have been introduced into the diagnostic settings [137]. Nevertheless, more complex cancer heterogeneities become evident, thus this approach might be hampered by the lack of necessary sensitivity in capturing this complexity. Another issue that needs to be addressed is given by the scenario where the assessment of primary cancer does not necessarily reflect the same mutational burden observed in metastatic deposits [7,138].
In current clinical settings, several blood-based tests are used for CRC detection, monitoring recurrence, and prognosis. These tests detect the tumour markers carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9), or septin-9 (SEPT9) [139,140,141,142]. Nevertheless, it is well established that they all have several weaknesses in regard to specificity and sensitivity. CEA, for instance, is elevated in patients with different gastrointestinal cancers [143] and is also not suited for early detection of CRC owing to a low sensitivity (5–10%) [144]. However, over-expression of CEA is in use for monitoring CRC and as a prognostic factor [145]. CA19-9 is not specific for a tumour type, cannot be detected in early state of cancer disease, and is also less sensitive than CEA [143,146] in diagnostic setting. Another biomarker test is based on SEPT9 methylated DNA in serum. Despite having only 70% sensitivity, but 90% specificity for CRC [147,148], it is currently the most sensitive and specific diagnostic tool for early detection of CRC [149].
Beside these proteins used in diagnostic setting, some other proteins carried by extracellular vesicles (e.g., exosomes) have recently gained attention and seem to be directly involved in the carcinogenesis of CRC [150,151,152,153,154]. Even if this field is in its early infancy, it is worth mentioning that Frizzled-10 protein (FZD-10), a receptor for the Wnt pathway, is present in exosomes isolated from colorectal cancer cell lines. SiRNA-based experiments demonstrated a role of FZD-10 containing exosome for cancer cell viability and establishment of metastatic sites [152]. FZD-10 is also present at a higher level in extracellular vesicle of CRC patients compared with healthy volunteers. In addition, the level of FZD-10 protein in extracellular vesicles seems to depend on the stage of CRC disease and FZD-10 expression returned to the one observed in the control group after successful treatment of the cancer patients. Therefore, FZD-10 is currently discussed as a new prognostic and diagnostic cancer marker [153,155]. Nevertheless, FZD-10 in extracellular vesicles is not CRC-specific and is also found to be elevated in patients with cholangiocarcinoma, hepatocarcinoma, and gastric cancer [152,153].
To overcome the limitations of protein markers in diagnostic settings, further DNA- and RNA-based biomarkers, present in liquid biopsies, have been evaluated as potentially new prognostic and predictive markers [137,156,157,158,159,160]. Using minimal invasive approaches could significantly improve the diagnostic and offer new therapeutic possibilities for CRC patients by improving cancer monitoring, as outlined in the following sections.
5.1. Circulating Tumour DNAs (ctDNAs)
The promising value of ctDNAs for stratification of cancer patients was underlined by the FDA approval for an EGFR (epidermal growth factor receptor) mutation test based on liquid biopsy in 2016 [161]. This test is used to select non-small cell lung cancer patients who could benefit from EGFR inhibitor therapy [161]. Similarly to this test, detecting KRAS-mutations in CRC patients using liquid biopsies as starting material could be a potential tool to select patients to tailored therapies [162]. This is supported by the fact that around 40% of all CRC patients have KRAS-mutations, and thus will not benefit from an anti-EGFR antibody therapy, which is often used for CRC treatment [163]. Unfortunately, this test is not yet approved for clinical use, although recently, the FDA approved a similar test for the detection of SEPT9 promoter region methylation in serum samples from CRC patients [149]. The methylation of SEPT9 promoter is accepted as a specific biomarker and is in clinical use for early detection of CRC [147,148]. Compared with the CEA biomarker, which is in clinical use, ctDNA analysis has been proven to be a more accurate predictor of CRC relapse, albeit only in a research setting [164]. So, not surprisingly, several studies proved that the overall amount of all circulating DNA (ctDNA and cell-free DNA) in CRC patients correlates with disease-free survival and overall survival. This correlation was found in all CRC tumour stages and was irrespective of the use of adjuvant chemotherapy [165,166,167,168,169,170,171,172,173]. In addition, ctDNA reflects tumour heterogeneity with an overall accuracy of 97% when compared with corresponding tissue biopsies, but it seems to be even superior to tissue biopsies in some cases [174].
Few reports examined ctDNA in CRC patients with primary or acquired resistance to EGFR blockade and emergence of mutated KRAS clones in blood was observed during treatment [175]. Recently, sequential ctDNA profiling was used in a phase II clinical study to monitor acquired resistance to cetuximab in RAS wild-type CRC patients. This study demonstrated the potential of liquid biopsies in combination with a mathematical framework model of tumor evolution for an individualized forecast of cancer progression, thus providing a novel opportunity for adaptive personalized therapies based on ctDNA analysis [122]. Other reports demonstrated that ctDNA analysis is able to monitor the emergence of resistant KRAS mutated clones for up to 10 months before radiographic confirmation of disease progression [176,177]. In another study in a phase II trial, the amount of RAS mutated ctDNA was determined in metastatic CRC patients under regorafenib therapy. The decrease of RAS mutant in plasma within 8 weeks of treatment was associated with better progression-free survival and overall survival. Combination of ctDNA analysis with radiological assessment was able to predict duration of anti-angiogenic response to regorafenib, thus suggesting that this result could be used to improve patient management in the future. This study adds further importance to the need for ctDNA markers owing to a lack of predictive biomarkers and potential toxicities related to regorafenib treatment [178].
Selection of CRC patients who could benefit from a HER-2-targeted antibody-therapy was addressed in another study, where a HER-2 high copy number variation was determined using cfDNA from blood with an elevated accuracy (97%) in 28 out of 29 patients. While these findings will need to be verified in larger study cohorts, they still provide a strong indication that copy number variation analysis based on ctDNA could predict the benefit from HER-2-targeted therapy [179].
Furthermore, detection of ctDNA following surgery might be an indicator of minimal residual disease, thus anticipating clinical and radiological assessment of recurrence [171,180]. In a large prospective study based on 230 CRC patients with resected stage II cancer, ctDNA from mutated genes was detected post-surgery in 14 patients. At follow-up, radiological recurrence was verified in 11 (79%) of these patients. From patients without any detectable post-operative ctDNA, disease recurrence was identified in 9% of them [170]. Similar results were obtained in a prospective study with 130 stage I-III CRC patients. Plasma was collected pre- and post-surgery and ctDNAs were analysed by NGS. Longitudinal ctDNA analysis identified 14 of 16 (87.5%) CRC patients that relapsed. In this study, all patients who have had ctDNA in their blood after adjuvant chemotherapy relapsed. Thus, ctDNA analyses predicted recurrence up to 16.5 months before radiographic confirmation of disease progression. Furthermore, in 82% of ctDNA-positive relapse samples, new actionable mutations have been identified that would enable further personalized treatment of the patients [181].
5.2. Circulating Tumour Cells (CTCs)
Detection of CTCs in body fluids of colorectal cancer patients still represents a big challenge and an intense area of work as CTCs are only present in body fluids at a very low level [182]. This limitation has been exploited and resulted in a FDA-approved microfluidic device [183] for capturing and concentrate circulating tumour cells. The process is based on magnetic particles coated with antibodies targeting the epithelial cell adhesion molecule (EpCAM), antigen that is over-expressed in different tumours including CRC [184]. According to several studies, the number of circulating tumour cells has the potential to monitor therapy success and indicate potential resistance against a given therapy [185,186,187].
An increasing amount of circulating tumour cells in patients’ blood is associated with poor prognosis in CRC and, for instance, it has been suggested that if three or more circulating tumour cells are present in 7.5 mL of blood, patients have a reduced overall survival [188,189]. Moreover, circulating tumour cells are an important source for molecular characterization of chemotherapy resistant clones [185,186,187,190,191,192,193,194], and thus represent an important tool for personalized treatment approaches.
5.3. MicroRNAs (miRNAs)
During recent years, miRNAs have been evaluated and proposed as promising new biomarkers for diagnostic purposes, for monitoring therapy success, and for detection of drug resistance towards treatment [195]. A significant number of studies are related to miRNA expression in tissue, but in the following, only studies based on liquid biopsies will be summarized.
CRC patients are characterised by a high-level expression of miR-92 and miR-29a in plasma. These miRNAs could be suitable biomarkers for screening tests and might represent a diagnostic tool for early detection of CRC [196,197,198]. A signature of significantly up-regulated miRNAs (miR-17, miR-18a, miR-18b, miR-181a, miR-125a, miR-320c) in plasma of CRC patients has been published. In this study, miRNA screening was performed comparing a pool sample of plasma from 30 CRC patients against a pool of plasma from 10 healthy individuals. Differentially expressed miRNAs were then verified in a larger training cohort of CRC patients by real-time PCR analysis. Finally, the diagnostic potential of this miRNA signature was verified in an independent validation cohort. The specificity of this miRNA signature for CRC was guaranteed by including healthy volunteers in both cohorts [199]. Some miRNAs of this signature were already proposed earlier in another study [200], where over-expression of miR-125a and miR-320c has been observed in CRC; therefore, co-evaluation of these miRNAs has been suggested as a possible biomarker combination for early-stage detection of CRC. For CRC diagnostic purposes, addition of miR-125a to CEA resulted in a better diagnostic model compared with CEA alone [200]. In another study, miR-96 and miR-149 were found to be down-regulated in plasma from CRC patients compared with a healthy control group. The CRC specificity of the deregulated miRNAs expression was underlined by the fact that expression returned to the one observed in the control group after surgical intervention within two months [201]. Very recently, miR-23a and miR-301a were found to be up-regulated in CRC patients compared with healthy individuals. Moreover, in this case, both miRNAs seem to be CRC-specific as they are significantly down-regulated after surgical intervention [202]. Furthermore, miRNAs’ expression could be used to discriminate between metastatic and non-metastatic CRC patients. A high expression of miR-200c in serum from CRC patients can be used as a predictive biomarker for the presence of local and distant metastasis [203]. Using miR-320d as a biomarker, patients with metastatic cancer have been detected with a sensitivity of 62% and a specificity of 65%. Combined analysis with CEA, this increased the sensitivity and specificity to 63% and 91%, respectively [204]. All these examples highlight the potential of miRNAs as diagnostic markers for difficult to diagnose early stages of CRC, as well as in metastatic disease settings. Furthermore, miRNAs can be used in addition to the currently used biomarker CEA and by combining them, and the specificity as well as the sensitivity of CRC diagnosis is significant increased.
Other studies addressed the role of miRNAs as prognostic markers. In different studies, up-regulated expression of miR-21, miR-193a, miR-203, miR-1290, miR-1229, and miR-17/92 cluster has been connected to poor prognosis of CRC patients owing to metastasis, post-treatment relapse, and poor overall survival [203,205,206,207,208,209,210,211,212,213], and it would be of interest if a combined miRNA signature could represent a robust prognostic tool. On the contrary, two down-regulated miRNAs (miR-4772-3p and miR-6869) have also been identified as prognostic CRC biomarkers. The reduced expression of both miRNAs was correlated with a high risk of tumour recurrence and death of patients [214,215]. Patients with a low serum level of miR-6869 had a poor three-year survival rate [215], whereas miR-4772 was a better predictor of recurrence than tumour location and lymph node metastasis [214]. Another report showed that a significant up-regulation of tumour-derived miR-27a and miR-130a in plasma of CRC patients correlates with tumour grade and stage as well as poor overall survival [216]. Furthermore, both miRNAs might be used for identification of CRC by comparison with healthy individuals; miR-27a had a sensitivity of 82% and a specificity of 91%, while miR-130a had a sensitivity of 70% and specificity of 100% [216]. In regard to disease-free survival and overall survival of CRC patients, the serum level of miR-548c and miR-6803 appears to be predictive. An increase in miR-6803 and reduced level of miR-548c are poor prognostic markers, especially in later stages of the disease and in cases where liver metastasis is present [217,218].
Serum of patients with resistance to 5-fluoruracil and oxaliplatin treatment is characterised by an up-regulation of miR-21, miR-96, miR-1229, and miR-1246 compared with respective chemo-sensitive CRC patients [219]. CRC patients receiving mFOLFOX6 (5-fluorouracil, leucovorin, and oxaliplatin)-based chemotherapy have an increased blood level of miR-125b when they become resistant to this therapy, resulting in shorter progression-free survival [220]. In contrast, patients with partial response to mFOLFOX6 treatment have a significantly lower miR-125b level post-treatment in comparison with pre-treatment and patients with stable disease had no change in miR-125 expression. These differences between the patients’ groups can be detected within one-month post-treatment [220]. Therefore, miRNAs might represent an accurate tool for prediction of therapeutic success and resistance.
5.4. Circular RNAs (circRNAs)
The evaluation of circRNAs in the context of liquid biopsies is still in its infancy and is mostly restricted to the observation of different expression levels between pathological and physiological conditions without any molecular analysis of the mechanisms. CircRNA microarray assay, based on plasma samples from four CRC patients and healthy controls, revealed circ-0082182, circ-0000370, and circ-0035445 as up-regulated in CRC and as possible diagnostic markers [221]. Although this finding has been confirmed by qPCR, it is desirable to analyse a larger patient cohort before this panel could become a diagnostic tool and a non-invasive biomarker panel for early CRC. Three other circRNAs (circ-CCDC66, circ-ABCC1, and circ-STIL) have been found to be down-regulated in plasma of CRC patients compared with healthy controls [222]. The predictive power of these three circRNAs appears to be higher compared with the currently used protein biomarker CEA and CA19-9 [222]. In serum of CRC patients, a 14-fold increased level of circ-0004771 was present compared with patients with benign intestinal diseases and healthy volunteers. The sensitivity and specificity of these levels in differentiating CRC patients from healthy donors were 80.91% and 82.86%, respectively. The CRC specificity of circ-004771 is further underlined by the fact that the level significantly decreased post-operatively and upon CRC treatment [223]. Another study on serum found an increased expression of circ-0001649 in patients with CRC after surgery [224]. Evidence from in vitro assays shows that over-expression of circ-0001649 inhibited proliferation, migration, and cell invasion [225], but the type of action of circ-0001649 and circ-0004771 in CRC is still unknown. Another report has instead evidenced that up-regulation of circ_0007142 could be used as an indication for lymphatic metastasis and differentiation of CRC [226].
5.5. Long Non-Coding RNAs (lncRNAs)
So far, research on lncRNAs has been less extensive compared with studies involving miRNAs. Nevertheless, it is well established that lncRNAs could be useful biomarkers for CRC [227]. Three lncRNAs (HOTAIR, CRNDE, and RPPH1) were found to be up-regulated in blood with prognostic and diagnostic benefit for CRC patients [228,229,230]. Of special interest is lncRNA HOTAIR, as its expression level correlates directly with tumour stage and overall survival of cancer patients [228,231]. Therefore, detection of lncRNA HOTAIR in blood of CRC patients could serve as an additional prognostic marker [228], but some caution is necessary because an increased amount of lncRNA HOTAIR is also observed during the course of other diseases, for example, gastric and pancreatic cancer, thus its specificity still needs to be investigated [232,233]. LncRNA CRNDE is higher expressed in serum from CRC patients compared with patients with benign colorectal disease or healthy controls [229]. LncRNA CRNDE expression might be connected to lymph node metastasis and is indicative of a low overall survival in CRC. In addition, the prognostic value of lncRNA CRNDE appears to be better than CEA, with a sensitivity of 70% versus 37% and a specificity of 94% versus 89% [229]. LncRNA RPPH1 expression can also be used as a prognostic factor. Expression of lncRNA RPPH1 is significantly elevated in plasma of CRC patients in advanced stages, and decreases post-surgery [230]. A high level of lncRNA RPPH1 in CRC is an indicator of poor overall survival and poor disease-free survival, and this lncRNA could be a better prognostic marker than CEA and CA19-9 [230].
LncRNA UCA1 has been shown to be down-regulated in serum of CRC patients compared with healthy volunteers [234]. In this study, identification of cancer patients from the healthy control group was possible with 100% sensitivity and 43% specificity [234].
The aforementioned studies demonstrate clearly the potential prognostic and diagnostic use of lncRNA isolated from liquid biopsies, and that they could potentially be more robust and reliable predictors compared with the currently used biomarkers CEA and CA19-9.
6. Conclusions and Perspectives
Early detection and non-invasive management of CRC are clear areas of unmet clinical need. The potential benefit of an early detection of CRC is underlined by the fact that the five-year survival rate is around 90% for patients diagnosed at early stage compared with 13% for patients diagnosed at stage IV [11]. Liquid biopsies offer a minimal invasive method for gaining access to CTCs and ctDNAs as well as lncRNAs, circRNAs, and miRNAs. All of these molecules can be used as prognostic and diagnostic markers, often with better specificity and sensitivity when used alone, compared with CEA and CA19-9, or they might also add value to the protein biomarkers currently in use. Before introducing these new nucleic acids-based biomarkers into the clinic, some standardizations of the methods and platforms are necessary [22]. In particular, the down-stream processes, such as the isolation process of material from the liquid biopsies and the sensitivity of detection methods (such as ddPCR or NGS), must be addressed and solved before liquid biopsies can be used as an important tool in clinical routine for (early) diagnosis and prognosis of CRC patients. Nevertheless, it is already proven that liquid biopsies and especially non-coding RNAs (lncRNAs and miRNAs) will be a great support in precision oncology and personalised medicine in the near future. In general, liquid biopsy has the potential to bring molecular diagnostic tests and targeted therapies together in a highly synergistic way. Liquid biopsies open the door for longitudinal analyses and offer the ability to routinely test patients frequently over prolonged periods of time. Compared with tissue biopsy, it facilitates routine screening and monitoring of patients throughout the whole treatment; therapy response and advent of resistance as well as changes in the molecular basis of the disease can be monitored in real time. The liquid biopsy approach has reduced workflow and turnaround time when compared with tissue biopsy. Last, but not least, liquid biopsy has the potential to support the analysis of a greater number of validated biomarkers, which will give much greater depth of support to clinical decision making. Up to now, liquid biopsies are mostly associated with oncology, but liquid biopsies have the potential to also become a useful tool for analyzing molecular variations in the context of other complex multifactorial diseases.
Author Contributions
Conceptualization, J.C.H., A.L., and N.V.; writing—original draft preparation, J.C.H., M.G., M.R., and A.L.; writing—review and editing, J.C.H., N.V., M.B.M., and A.F.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
N.V. received speaker honorarium from the companies Bayer, Eli-Lilly, Pfizer, and Merck. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Lieberman, D. Progress and challenges in colorectal cancer screening and surveillance. Gastroenterology 2010, 138, 2115–2126. [Google Scholar] [CrossRef]
- Pinol, V.; Castells, A.; Andreu, M.; Castellvi-Bel, S.; Alenda, C.; Llor, X.; Xicola, R.M.; Rodriguez-Moranta, F.; Paya, A.; Jover, R.; et al. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA 2005, 293, 1986–1994. [Google Scholar] [CrossRef]
- Farrington, S.M.; Tenesa, A.; Barnetson, R.; Wiltshire, A.; Prendergast, J.; Porteous, M.; Campbell, H.; Dunlop, M.G. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am. J. Hum. Genet. 2005, 77, 112–119. [Google Scholar] [CrossRef]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [CrossRef]
- Binefa, G.; Rodriguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014, 20, 6786–6808. [Google Scholar] [CrossRef]
- Lote, H.; Spiteri, I.; Ermini, L.; Vatsiou, A.; Roy, A.; McDonald, A.; Maka, N.; Balsitis, M.; Bose, N.; Simbolo, M.; et al. Carbon dating cancer: Defining the chronology of metastatic progression in colorectal cancer. Ann. Oncol. 2017, 28, 1243–1249. [Google Scholar] [CrossRef]
- Issa, I.A.; Noureddine, M. Colorectal cancer screening: An updated review of the available options. World J. Gastroenterol. 2017, 23, 5086–5096. [Google Scholar] [CrossRef]
- Herszenyi, L.; Farinati, F.; Cardin, R.; Istvan, G.; Molnar, L.D.; Hritz, I.; De Paoli, M.; Plebani, M.; Tulassay, Z. Tumor marker utility and prognostic relevance of cathepsin B, cathepsin L, urokinase-type plasminogen activator, plasminogen activator inhibitor type-1, CEA and CA 19-9 in colorectal cancer. BMC Cancer 2008, 8, 194. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castanos-Velez, E.; Blumenstein, B.A.; Rosch, T.; Osborn, N.; et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef]
- Toiyama, Y.; Okugawa, Y.; Fleshman, J.; Richard Boland, C.; Goel, A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 274–282. [Google Scholar] [CrossRef]
- Jiang, N.; Pan, J.; Fang, S.; Zhou, C.; Han, Y.; Chen, J.; Meng, X.; Jin, X.; Gong, Z. Liquid biopsy: Circulating exosomal long noncoding RNAs in cancer. Clin. Chim. Acta 2019, 495, 331–337. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’Homme. CR Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Marrugo-Ramirez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Soda, N.; Rehm, B.H.A.; Sonar, P.; Nguyen, N.T.; Shiddiky, M.J.A. Advanced liquid biopsy technologies for circulating biomarker detection. J. Mater. Chem. B 2019, 7, 6670–6704. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pelle, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10. [Google Scholar] [CrossRef]
- Shin, S.H.; Bode, A.M.; Dong, Z. Precision medicine: The foundation of future cancer therapeutics. NPJ Precis. Oncol. 2017, 1, 12. [Google Scholar] [CrossRef]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef]
- Mino-Kenudson, M. Cons: Can liquid biopsy replace tissue biopsy?-the US experience. Transl. Lung Cancer Res. 2016, 5, 424–427. [Google Scholar] [CrossRef]
- Stroun, M.; Maurice, P.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Lefort, F.; Rossier, A.; Chen, X.Q.; Anker, P. The origin and mechanism of circulating DNA. Ann. N.Y. Acad. Sci. 2000, 906, 161–168. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Parkinson, D.R.; Dracopoli, N.; Petty, B.G.; Compton, C.; Cristofanilli, M.; Deisseroth, A.; Hayes, D.F.; Kapke, G.; Kumar, P.; Lee, J.; et al. Considerations in the development of circulating tumor cell technology for clinical use. J. Transl. Med. 2012, 10, 138. [Google Scholar] [CrossRef]
- Young, R.; Pailler, E.; Billiot, F.; Drusch, F.; Barthelemy, A.; Oulhen, M.; Besse, B.; Soria, J.C.; Farace, F.; Vielh, P. Circulating tumor cells in lung cancer. Acta Cytol. 2012, 56, 655–660. [Google Scholar] [CrossRef]
- Van der Toom, E.E.; Verdone, J.E.; Gorin, M.A.; Pienta, K.J. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget 2016, 7, 62754–62766. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Sefrioui, D.; Blanchard, F.; Toure, E.; Basile, P.; Beaussire, L.; Dolfus, C.; Perdrix, A.; Paresy, M.; Antonietti, M.; Iwanicki-Caron, I.; et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer 2017, 117, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Diamantopoulos, M.A.; Tsiakanikas, P.; Scorilas, A. Non-coding RNAs: The riddle of the transcriptome and their perspectives in cancer. Ann. Transl. Med. 2018, 6, 241. [Google Scholar] [CrossRef] [PubMed]
- Hahne, J.C.; Valeri, N. Non-Coding RNAs and Resistance to Anticancer Drugs in Gastrointestinal Tumors. Front. Oncol. 2018, 8, 226. [Google Scholar] [CrossRef]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef]
- Brosnan, C.A.; Voinnet, O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009, 21, 416–425. [Google Scholar] [CrossRef]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [Google Scholar] [CrossRef]
- Finotti, A.; Fabbri, E.; Lampronti, I.; Gasparello, J.; Borgatti, M.; Gambari, R. MicroRNAs and Long Non-coding RNAs in Genetic Diseases. Mol. Diagn. Ther. 2019, 23, 155–171. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Ghidini, M.; Hahne, J.C.; Frizziero, M.; Tomasello, G.; Trevisani, F.; Lampis, A.; Passalacqua, R.; Valeri, N. MicroRNAs as Mediators of Resistance Mechanisms to Small-Molecule Tyrosine Kinase Inhibitors in Solid Tumours. Target. Oncol. 2018, 13, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer—A brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef]
- Balatti, V.; Pekarky, Y.; Croce, C.M. Role of microRNA in chronic lymphocytic leukemia onset and progression. J. Hematol. Oncol. 2015, 8, 12. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Garibaldi, F.; Falcone, E.; Trisciuoglio, D.; Colombo, T.; Lisek, K.; Walerych, D.; Del Sal, G.; Paci, P.; Bossi, G.; Piaggio, G.; et al. Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene 2016, 35, 3760–3770. [Google Scholar] [CrossRef]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Nat. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Nat. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Nat. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Nat. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.; Baird, A.M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Sun, L.F.; Zhang, B.; Chen, X.J.; Wang, X.Y.; Zhang, B.W.; Ji, Y.Y.; Wu, K.C.; Wu, J.; Jin, Z.B. Circular RNAs in human and vertebrate neural retinas. RNA Biol. 2019, 16, 821–829. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Zhao, S.Y.; Wang, J.; Ouyang, S.B.; Huang, Z.K.; Liao, L. Salivary Circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as Novel Biomarkers for the Diagnosis of Oral Squamous Cell Carcinoma. Cell Physiol. Biochem. 2018, 47, 2511–2521. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Cui, Y. SomamiR 2.0: A database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016, 44, D1005–D1010. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Zheng, Q.; Bao, C.; He, J.; Chen, B.; Lyu, D.; Zheng, B.; Xu, Y.; Long, Z.; et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017, 388, 208–219. [Google Scholar] [CrossRef]
- Drula, R.; Braicu, C.; Harangus, A.; Nabavi, S.M.; Trif, M.; Slaby, O.; Ionescu, C.; Irimie, A.; Berindan-Neagoe, I. Critical function of circular RNAs in lung cancer. Wiley Interdiscip. Rev. RNA 2020, e1592. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Yang, J.; Li, X.; Liu, D.; Fu, L.; Wang, X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol. Cancer 2020, 19, 58. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Liu, P.; Xu, Z.; Li, N.; Yi, Z.; Tie, X.; Zhang, Y.; Gao, L. CircRNA_001569 promotes cell proliferation through absorbing miR-145 in gastric cancer. J. Biochem. 2019, 165, 27–36. [Google Scholar] [CrossRef]
- Lai, Z.; Yang, Y.; Yan, Y.; Li, T.; Li, Y.; Wang, Z.; Shen, Z.; Ye, Y.; Jiang, K.; Wang, S. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle 2017, 16, 2301–2311. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Herriges, M.J.; Swarr, D.T.; Morley, M.P.; Rathi, K.S.; Peng, T.; Stewart, K.M.; Morrisey, E.E. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014, 28, 1363–1379. [Google Scholar] [CrossRef]
- Li, G.; Zhang, H.; Wan, X.; Yang, X.; Zhu, C.; Wang, A.; He, L.; Miao, R.; Chen, S.; Zhao, H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed. Res. Int. 2014, 2014, 780521. [Google Scholar] [CrossRef]
- Ounzain, S.; Pezzuto, I.; Micheletti, R.; Burdet, F.; Sheta, R.; Nemir, M.; Gonzales, C.; Sarre, A.; Alexanian, M.; Blow, M.J.; et al. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell Cardiol. 2014, 76, 55–70. [Google Scholar] [CrossRef]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 2, e01749. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA expression. Cell Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Int. Rev. RNA 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.Y.; Hao, Q.Y.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Chang, C.P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Lopez-Urrutia, E.; Bustamante Montes, L.P.; Ladron de Guevara Cervantes, D.; Perez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Martignano, F.; Rossi, L.; Maugeri, A.; Galla, V.; Conteduca, V.; De Giorgi, U.; Casadio, V.; Schepisi, G. Urinary RNA-based biomarkers for prostate cancer detection. Clin. Chim. Acta 2017, 473, 96–105. [Google Scholar] [CrossRef]
- Terracciano, D.; Ferro, M.; Terreri, S.; Lucarelli, G.; D’Elia, C.; Musi, G.; de Cobelli, O.; Mirone, V.; Cimmino, A. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: New architects in cancer prognostic biomarkers. Transl. Res. 2017, 184, 108–117. [Google Scholar] [CrossRef]
- Zhou, X.Y.; Yin, C.Q.; Dang, Y.N.; Ye, F.; Zhang, G.X. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shao, Y.F.; Zhang, X.J.; Zheng, T.; Miao, M.; Qin, L.J.; Wang, B.J.; Ye, G.L.; Xiao, B.X.; Guo, J.M. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015, 36, 2007–2012. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Thum, T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating Long Noncoding RNA, LIPCAR, Predicts Survival in Patients With Heart Failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef]
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2019, 20, 1420–1433. [Google Scholar] [CrossRef]
- Slattery, M.L.; Herrick, J.S.; Mullany, L.E.; Valeri, N.; Stevens, J.; Caan, B.J.; Samowitz, W.; Wolff, R.K. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int. J. Cancer 2015, 137, 428–438. [Google Scholar] [CrossRef]
- Braconi, C.; Valeri, N.; Kogure, T.; Gasparini, P.; Huang, N.; Nuovo, G.J.; Terracciano, L.; Croce, C.M.; Patel, T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc. Nat. Acad. Sci. USA 2011, 108, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Wernert, N.; Kaminski, A.; Haddouti el, M.; Hahne, J.C. Tumor-stroma interactions of metastatic prostate cancer cell lines: Analyses using microarrays. Methods Mol. Biol. 2007, 382, 223–237. [Google Scholar] [CrossRef]
- Liu, C.G.; Calin, G.A.; Volinia, S.; Croce, C.M. MicroRNA expression profiling using microarrays. Nat. Protoc. 2008, 3, 563–578. [Google Scholar] [CrossRef]
- Li, M.H.; Fu, S.B.; Xiao, H.S. Genome-wide analysis of microRNA and mRNA expression signatures in cancer. Acta Pharmacol. Sin. 2015, 36, 1200–1211. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef]
- Malkov, V.A.; Serikawa, K.A.; Balantac, N.; Watters, J.; Geiss, G.; Mashadi-Hossein, A.; Fare, T. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res. Notes 2009, 2, 80. [Google Scholar] [CrossRef]
- Valeri, N.; Braconi, C.; Gasparini, P.; Murgia, C.; Lampis, A.; Paulus-Hock, V.; Hart, J.R.; Ueno, L.; Grivennikov, S.I.; Lovat, F.; et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014, 25, 469–483. [Google Scholar] [CrossRef]
- Fassan, M.; Realdon, S.; Cascione, L.; Hahne, J.C.; Munari, G.; Guzzardo, V.; Arcidiacono, D.; Lampis, A.; Brignola, S.; Dal Santo, L.; et al. Circulating microRNA expression profiling revealed miR-92a-3p as a novel biomarker of Barrett’s carcinogenesis. Pathol. Res. Pract. 2020, 216, 152907. [Google Scholar] [CrossRef]
- Zhang, J.; Raju, G.S.; Chang, D.W.; Lin, S.H.; Chen, Z.; Wu, X. Global and targeted circulating microRNA profiling of colorectal adenoma and colorectal cancer. Cancer 2018, 124, 785–796. [Google Scholar] [CrossRef]
- Hyeon, J.; Cho, S.Y.; Hong, M.E.; Kang, S.Y.; Do, I.; Im, Y.H.; Cho, E.Y. NanoString nCounter(R) Approach in Breast Cancer: A Comparative Analysis with Quantitative Real-Time Polymerase Chain Reaction, In Situ Hybridization, and Immunohistochemistry. J. Breast Cancer 2017, 20, 286–296. [Google Scholar] [CrossRef]
- Eastel, J.M.; Lam, K.W.; Lee, N.L.; Lok, W.Y.; Tsang, A.H.F.; Pei, X.M.; Chan, A.K.C.; Cho, W.C.S.; Wong, S.C.C. Application of NanoString technologies in companion diagnostic development. Expert Rev. Mol. Diagn. 2019, 19, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, G.S.; Feldman, T.; Rimsza, L.M.; Westin, J.R.; Witzig, T.E.; Zinzani, P.L. Integrating precision medicine through evaluation of cell of origin in treatment planning for diffuse large B-cell lymphoma. Blood Cancer J. 2019, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Nat. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Bhat, S.; Herrmann, J.; Armishaw, P.; Corbisier, P.; Emslie, K.R. Single molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal. Bioanal. Chem. 2009, 394, 457–467. [Google Scholar] [CrossRef]
- Bhat, S.; Curach, N.; Mostyn, T.; Bains, G.S.; Griffiths, K.R.; Emslie, K.R. Comparison of methods for accurate quantification of DNA mass concentration with traceability to the international system of units. Anal. Chem. 2010, 82, 7185–7192. [Google Scholar] [CrossRef]
- Dong, L.; Meng, Y.; Sui, Z.; Wang, J.; Wu, L.; Fu, B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci. Rep. 2015, 5, 13174. [Google Scholar] [CrossRef]
- Sanders, R.; Huggett, J.F.; Bushell, C.A.; Cowen, S.; Scott, D.J.; Foy, C.A. Evaluation of digital PCR for absolute DNA quantification. Anal. Chem. 2011, 83, 6474–6484. [Google Scholar] [CrossRef]
- Lo, Y.M.; Lun, F.M.; Chan, K.C.; Tsui, N.B.; Chong, K.C.; Lau, T.K.; Leung, T.Y.; Zee, B.C.; Cantor, C.R.; Chiu, R.W. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Nat. Acad. Sci. USA 2007, 104, 13116–13121. [Google Scholar] [CrossRef]
- Sclafani, F.; Chau, I.; Cunningham, D.; Hahne, J.C.; Vlachogiannis, G.; Eltahir, Z.; Lampis, A.; Braconi, C.; Kalaitzaki, E.; De Castro, D.G.; et al. KRAS and BRAF mutations in circulating tumour DNA from locally advanced rectal cancer. Sci. Rep. 2018, 8, 1445. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.H.; Cunningham, D.; Werner, B.; Vlachogiannis, G.; Spiteri, I.; Heide, T.; Mateos, J.F.; Vatsiou, A.; Lampis, A.; Damavandi, M.D.; et al. Longitudinal Liquid Biopsy and Mathematical Modeling of Clonal Evolution Forecast Time to Treatment Failure in the PROSPECT-C Phase II Colorectal Cancer Clinical Trial. Cancer Discov. 2018, 8, 1270–1285. [Google Scholar] [CrossRef] [PubMed]
- White, R.A., 3rd; Blainey, P.C.; Fan, H.C.; Quake, S.R. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics 2009, 10, 116. [Google Scholar] [CrossRef]
- Buermans, H.P.; den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Dyer, N.P.; Shahrezaei, V.; Hebenstreit, D. LiBiNorm: An htseq-count analogue with improved normalisation of Smart-seq2 data and library preparation diagnostics. PeerJ 2019, 7, e6222. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef]
- Head, S.R.; Komori, H.K.; LaMere, S.A.; Whisenant, T.; Van Nieuwerburgh, F.; Salomon, D.R.; Ordoukhanian, P. Library construction for next-generation sequencing: Overviews and challenges. Biotechniques 2014, 56, 61–77. [Google Scholar] [CrossRef]
- Dabney, J.; Meyer, M. Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques 2012, 52, 87–94. [Google Scholar] [CrossRef]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Nat. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef]
- Roth, A.D.; Tejpar, S.; Delorenzi, M.; Yan, P.; Fiocca, R.; Klingbiel, D.; Dietrich, D.; Biesmans, B.; Bodoky, G.; Barone, C.; et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010, 28, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, G.; Southward, K.; Handley, K.; Magill, L.; Beaumont, C.; Stahlschmidt, J.; Richman, S.; Chambers, P.; Seymour, M.; Kerr, D.; et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J. Clin. Oncol. 2011, 29, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Farina-Sarasqueta, A.; van Lijnschoten, G.; Moerland, E.; Creemers, G.J.; Lemmens, V.E.; Rutten, H.J.; van den Brule, A.J. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann. Oncol. 2010, 21, 2396–2402. [Google Scholar] [CrossRef] [PubMed]
- Jass, J.R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007, 50, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Shi, Q.; Smyrk, T.C.; Thibodeau, S.N.; Dienstmann, R.; Guinney, J.; Bot, B.M.; Tejpar, S.; Delorenzi, M.; Goldberg, R.M.; et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015, 148, 88–99. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, D.; Ralton, L.D.; O’Shea, A.; Murray, G.I. The proteomics of colorectal cancer: Identification of a protein signature associated with prognosis. PLoS ONE 2011, 6, e27718. [Google Scholar] [CrossRef]
- De Rosa, M.; Rega, D.; Costabile, V.; Duraturo, F.; Niglio, A.; Izzo, P.; Pace, U.; Delrio, P. The biological complexity of colorectal cancer: Insights into biomarkers for early detection and personalized care. Ther. Adv. Gastroenterol. 2016, 9, 861–886. [Google Scholar] [CrossRef]
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef]
- DeVos, T.; Tetzner, R.; Model, F.; Weiss, G.; Schuster, M.; Distler, J.; Steiger, K.V.; Grutzmann, R.; Pilarsky, C.; Habermann, J.K.; et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin. Chem. 2009, 55, 1337–1346. [Google Scholar] [CrossRef]
- Vukobrat-Bijedic, Z.; Husic-Selimovic, A.; Sofic, A.; Bijedic, N.; Bjelogrlic, I.; Gogov, B.; Mehmedovic, A. Cancer Antigens (CEA and CA 19-9) as Markers of Advanced Stage of Colorectal Carcinoma. Med. Arch. 2013, 67, 397–401. [Google Scholar] [CrossRef]
- Nicholson, B.D.; Shinkins, B.; Pathiraja, I.; Roberts, N.W.; James, T.J.; Mallett, S.; Perera, R.; Primrose, J.N.; Mant, D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Svobodova, S.; Topolcan, O.; Holubec, L., Jr.; Levy, M.; Pecen, L.; Svacina, S. Parameters of biological activity in colorectal cancer. Anticancer Res. 2011, 31, 373–378. [Google Scholar] [PubMed]
- Polat, E.; Duman, U.; Duman, M.; Atici, A.E.; Reyhan, E.; Dalgic, T.; Bostanci, E.B.; Yol, S. Diagnostic value of preoperative serum carcinoembryonic antigen and carbohydrate antigen 19-9 in colorectal cancer. Curr. Oncol. 2014, 21, e1–e7. [Google Scholar] [CrossRef]
- Duffy, M.J. Carcinoembryonic antigen as a marker for colorectal cancer: Is it clinically useful? Clin. Chem. 2001, 47, 624–630. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C.; Asco. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef] [PubMed]
- Bagaria, B.; Sood, S.; Sharma, R.; Lalwani, S. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol. Med. 2013, 10, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Molnar, B.; Toth, K.; Bartak, B.K.; Tulassay, Z. Plasma methylated septin 9: A colorectal cancer screening marker. Expert Rev. Mol. Diagn. 2015, 15, 171–184. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, P.M.; Liu, R.B. Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J. Gastrointest. Oncol. 2018, 10, 15–22. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef]
- Lugini, L.; Valtieri, M.; Federici, C.; Cecchetti, S.; Meschini, S.; Condello, M.; Signore, M.; Fais, S. Exosomes from human colorectal cancer induce a tumor-like behavior in colonic mesenchymal stromal cells. Oncotarget 2016, 7, 50086–50098. [Google Scholar] [CrossRef] [PubMed]
- Scavo, M.P.; Depalo, N.; Rizzi, F.; Ingrosso, C.; Fanizza, E.; Chieti, A.; Messa, C.; Denora, N.; Laquintana, V.; Striccoli, M.; et al. FZD10 Carried by Exosomes Sustains Cancer Cell Proliferation. Cells 2019, 8, 777. [Google Scholar] [CrossRef] [PubMed]
- Principia, S.M.; Antonio, C.; Nicoletta, D.; Elisabetta, F.; Bianco, M.G.; Nunzio, D.; Valentino, L.; Maria Lucia, C.; Dionigi, L.; Claudio, L.; et al. Frizzled-10 Extracellular Vesicles Plasma Concentration Is Associated with Tumoral Progression in Patients with Colorectal and Gastric Cancer. J. Oncol. 2019, 2019, 2715968. [Google Scholar] [CrossRef] [PubMed]
- Sundararajan, V.; Sarkar, F.H.; Ramasamy, T.S. Correction to: The versatile role of exosomes in cancer progression: Diagnostic and therapeutic implications. Cell Oncol. 2018, 41, 463. [Google Scholar] [CrossRef]
- Scavo, M.P.; Fucci, L.; Caldarola, L.; Mangia, A.; Azzariti, A.; Simone, G.; Gasparini, G.; Krol, S. Frizzled-10 and cancer progression: Is it a new prognostic marker? Oncotarget 2018, 9, 824–830. [Google Scholar] [CrossRef]
- Hollis, M.; Nair, K.; Vyas, A.; Chaturvedi, L.S.; Gambhir, S.; Vyas, D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J. Gastroenterol. 2015, 21, 8284–8292. [Google Scholar] [CrossRef]
- Weng, M.; Wu, D.; Yang, C.; Peng, H.; Wang, G.; Wang, T.; Li, X. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl. Res. 2017, 181, 108–120. [Google Scholar] [CrossRef]
- Zhang, J.X.; Song, W.; Chen, Z.H.; Wei, J.H.; Liao, Y.J.; Lei, J.; Hu, M.; Chen, G.Z.; Liao, B.; Lu, J.; et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: A microRNA expression analysis. Lancet Oncol. 2013, 14, 1295–1306. [Google Scholar] [CrossRef]
- Calapre, L.; Warburton, L.; Millward, M.; Ziman, M.; Gray, E.S. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett. 2017, 404, 62–69. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabieres, C. Liquid biopsy in 2016: Circulating tumour cells and cell-free DNA in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 73–74. [Google Scholar] [CrossRef]
- Kwapisz, D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 2017, 5, 46. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Tsai, H.L.; Jiang, J.K.; Sung, Y.C.; Huang, C.W.; Yeh, Y.M.; Chen, L.T.; Wang, J.Y. Emergence of RAS mutations in patients with metastatic colorectal cancer receiving cetuximab-based treatment: A study protocol. BMC Cancer 2019, 19, 640. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Scholer, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef] [PubMed]
- El Messaoudi, S.; Mouliere, F.; Du Manoir, S.; Bascoul-Mollevi, C.; Gillet, B.; Nouaille, M.; Fiess, C.; Crapez, E.; Bibeau, F.; Theillet, C.; et al. Circulating DNA as a Strong Multimarker Prognostic Tool for Metastatic Colorectal Cancer Patient Management Care. Clin. Cancer Res. 2016, 22, 3067–3077. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, J.; Lenz, H.J.; Siena, S.; Sobrero, A.; Falcone, A.; Ychou, M.; Humblet, Y.; Bouche, O.; Mineur, L.; Barone, C.; et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015, 16, 937–948. [Google Scholar] [CrossRef]
- Basnet, S.; Zhang, Z.Y.; Liao, W.Q.; Li, S.H.; Li, P.S.; Ge, H.Y. The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis. J. Cancer 2016, 7, 1105–1113. [Google Scholar] [CrossRef]
- Parseghian, C.M.; Loree, J.M.; Morris, V.K.; Liu, X.; Clifton, K.K.; Napolitano, S.; Henry, J.T.; Pereira, A.A.; Vilar, E.; Johnson, B.; et al. Anti-EGFR-resistant clones decay exponentially after progression: Implications for anti-EGFR re-challenge. Ann. Oncol. 2019, 30, 243–249. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra392. [Google Scholar] [CrossRef]
- Wang, J.Y.; Hsieh, J.S.; Chang, M.Y.; Huang, T.J.; Chen, F.M.; Cheng, T.L.; Alexandersen, K.; Huang, Y.S.; Tzou, W.S.; Lin, S.R. Molecular detection of APC, K- ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J. Surg. 2004, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.L.; Lim, J.S.; Sinha, A.; Gopinathan, A.; Lim, R.; Tan, C.S.; Soh, T.; Venkatesh, S.; Titin, C.; Sapari, N.S.; et al. Tumour pharmacodynamics and circulating cell free DNA in patients with refractory colorectal carcinoma treated with regorafenib. J. Transl. Med. 2015, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Spindler, K.G. Methodological, biological and clinical aspects of circulating free DNA in metastatic colorectal cancer. Acta Oncol. 2017, 56, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Kidess, E.; Jeffrey, S.S. Circulating tumor cells versus tumor-derived cell-free DNA: Rivals or partners in cancer care in the era of single-cell analysis? Genome Med. 2013, 5, 70. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef]
- Misale, S.; Arena, S.; Lamba, S.; Siravegna, G.; Lallo, A.; Hobor, S.; Russo, M.; Buscarino, M.; Lazzari, L.; Sartore-Bianchi, A.; et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci. Transl. Med. 2014, 6, 224ra226. [Google Scholar] [CrossRef]
- Khan, K.; Rata, M.; Cunningham, D.; Koh, D.M.; Tunariu, N.; Hahne, J.C.; Vlachogiannis, G.; Hedayat, S.; Marchetti, S.; Lampis, A.; et al. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut 2018, 67, 1484–1492. [Google Scholar] [CrossRef]
- Siravegna, G.; Sartore-Bianchi, A.; Nagy, R.J.; Raghav, K.; Odegaard, J.I.; Lanman, R.B.; Trusolino, L.; Marsoni, S.; Siena, S.; Bardelli, A. Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-targeted Therapy in Metastatic Colorectal Cancer. Clin. Cancer Res. 2019, 25, 3046–3053. [Google Scholar] [CrossRef]
- Ryan, B.M.; Lefort, F.; McManus, R.; Daly, J.; Keeling, P.W.; Weir, D.G.; Kelleher, D. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: Strong prognostic indicator in postoperative follow up. Gut 2003, 52, 101–108. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Nat. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed]
- Su, D.W.; Nieva, J. Biophysical technologies for understanding circulating tumor cell biology and metastasis. Transl. Lung Cancer Res. 2017, 6, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Dizdar, L.; Fluegen, G.; van Dalum, G.; Honisch, E.; Neves, R.P.; Niederacher, D.; Neubauer, H.; Fehm, T.; Rehders, A.; Krieg, A.; et al. Detection of circulating tumor cells in colorectal cancer patients using the GILUPI CellCollector: Results from a prospective, single-center study. Mol. Oncol. 2019, 13, 1548–1558. [Google Scholar] [CrossRef] [PubMed]
- Bork, U.; Rahbari, N.N.; Scholch, S.; Reissfelder, C.; Kahlert, C.; Buchler, M.W.; Weitz, J.; Koch, M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: A prospective study. Br. J. Cancer 2015, 112, 1306–1313. [Google Scholar] [CrossRef]
- Gazzaniga, P.; Gianni, W.; Raimondi, C.; Gradilone, A.; Lo Russo, G.; Longo, F.; Gandini, O.; Tomao, S.; Frati, L. Circulating tumor cells in high-risk nonmetastatic colorectal cancer. Tumor Biol. 2013, 34, 2507–2509. [Google Scholar] [CrossRef]
- Tsai, W.S.; Chen, J.S.; Shao, H.J.; Wu, J.C.; Lai, J.M.; Lu, S.H.; Hung, T.F.; Chiu, Y.C.; You, J.F.; Hsieh, P.S.; et al. Circulating Tumor Cell Count Correlates with Colorectal Neoplasm Progression and Is a Prognostic Marker for Distant Metastasis in Non-Metastatic Patients. Sci. Rep. 2016, 6, 24517. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Bartkowiak, K.; Pantel, K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol. Oncol. 2016, 10, 443–449. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wu, C.H.; Lu, C.Y.; Hsieh, J.S.; Wu, D.C.; Huang, S.Y.; Lin, S.R. Molecular detection of circulating tumor cells in the peripheral blood of patients with colorectal cancer using RT-PCR: Significance of the prediction of postoperative metastasis. World J. Surg. 2006, 30, 1007–1013. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.A.; Mitchell, E.; Miller, M.C.; et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009, 20, 1223–1229. [Google Scholar] [CrossRef]
- Gorges, T.M.; Penkalla, N.; Schalk, T.; Joosse, S.A.; Riethdorf, S.; Tucholski, J.; Lucke, K.; Wikman, H.; Jackson, S.; Brychta, N.; et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin. Cancer Res. 2016, 22, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Renehan, A.G.; Backen, A.; Gollins, S.; Chau, I.; Hasan, J.; Valle, J.W.; Morris, K.; Beech, J.; Ashcroft, L.; et al. Circulating Tumor Cell Enumeration in a Phase II Trial of a Four-Drug Regimen in Advanced Colorectal Cancer. Clin. Colorectal Cancer 2015, 14, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Romiti, A.; Raffa, S.; Di Rocco, R.; Roberto, M.; Milano, A.; Zullo, A.; Leone, L.; Ranieri, D.; Mazzetta, F.; Medda, E.; et al. Circulating tumor cells count predicts survival in colorectal cancer patients. J. Gastrointestin. Liver Dis. 2014, 23, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Scherag, F.D.; Niestroj-Pahl, R.; Krusekopf, S.; Lucke, K.; Brandstetter, T.; Ruhe, J. Highly Selective Capture Surfaces on Medical Wires for Fishing Tumor Cells in Whole Blood. Anal. Chem. 2017, 89, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018, 10, 59. [Google Scholar] [CrossRef]
- Ju, J. miRNAs as biomarkers in colorectal cancer diagnosis and prognosis. Bioanalysis 2010, 2, 901–906. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Ng, E.K.; Chong, W.W.; Jin, H.; Lam, E.K.; Shin, V.Y.; Yu, J.; Poon, T.C.; Ng, S.S.; Sung, J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, M.; Shan, X.; Zhou, X.; Wang, T.; Zhang, J.; Tao, J.; Cheng, W.; Chen, G.; Li, J.; et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019, 687, 246–254. [Google Scholar] [CrossRef]
- Wang, J.; Yan, F.; Zhao, Q.; Zhan, F.; Wang, R.; Wang, L.; Zhang, Y.; Huang, X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 2017, 7, 4150. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Guo, X.; Zhou, L.; Jia, Z.; Peng, Z.; Tang, Y.; Liu, W.; Zhu, B.; Wang, L.; et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J. Cell Mol. Med. 2017, 21, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Karimi, N.; Ali Hosseinpour Feizi, M.; Safaralizadeh, R.; Hashemzadeh, S.; Baradaran, B.; Shokouhi, B.; Teimourian, S. Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J. Chin. Med. Assoc. 2019, 82, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Hur, K.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 2014, 259, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhao, Y.; Song, X.; Song, X.; Niu, L.; Xie, L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 2019, 33, e23004. [Google Scholar] [CrossRef]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N.; et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598–78613. [Google Scholar] [CrossRef]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Nat. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef]
- Imaoka, H.; Toiyama, Y.; Fujikawa, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; Kato, T.; et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 2016, 27, 1879–1886. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017, 66, 654–665. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Hu, X.; Mu, J.; Samykutty, A.; Zhuang, X.; Deng, Z.; Kumar, A.; Zhang, L.; Merchant, M.L.; et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat. Commun. 2017, 8, 14448. [Google Scholar] [CrossRef]
- Hu, H.Y.; Yu, C.H.; Zhang, H.H.; Zhang, S.Z.; Yu, W.Y.; Yang, Y.; Chen, Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int. J. Biol. Macromol. 2019, 132, 470–477. [Google Scholar] [CrossRef]
- Kral, J.; Korenkova, V.; Novosadova, V.; Langerova, L.; Schneiderova, M.; Liska, V.; Levy, M.; Veskrnova, V.; Spicak, J.; Opattova, A.; et al. Expression profile of miR-17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis 2018, 39, 1359–1367. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, M.; Iinuma, H.; Yagi, T.; Matsuda, K.; Hashiguchi, Y. Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology 2017, 92, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Jiang, W.; Zhou, L.; Chen, Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Eng, C.; Shen, J.; Lu, Y.; Takata, Y.; Mehdizadeh, A.; Chang, G.J.; Rodriguez-Bigas, M.A.; Li, Y.; Chang, P.; et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 2016, 7, 76250–76260. [Google Scholar] [CrossRef]
- Yan, S.; Liu, G.; Jin, C.; Wang, Z.; Duan, Q.; Xu, J.; Xu, D. MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-kappaB signaling pathway in colorectal cancer. J. Cell Physiol. 2018, 233, 6660–6668. [Google Scholar] [CrossRef]
- Liu, X.; Pan, B.; Sun, L.; Chen, X.; Zeng, K.; Hu, X.; Xu, T.; Xu, M.; Wang, S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol. Biomarkers Prev. 2018, 27, 746–754. [Google Scholar] [CrossRef]
- Peng, Z.Y.; Gu, R.H.; Yan, B. Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J. Cell Biochem. 2018. [Google Scholar] [CrossRef]
- Yan, S.; Jiang, Y.; Liang, C.; Cheng, M.; Jin, C.; Duan, Q.; Xu, D.; Yang, L.; Zhang, X.; Ren, B.; et al. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J. Cell Biochem. 2018, 119, 4113–4119. [Google Scholar] [CrossRef]
- Jin, G.; Liu, Y.; Zhang, J.; Bian, Z.; Yao, S.; Fei, B.; Zhou, L.; Yin, Y.; Huang, Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 2019, 84, 315–325. [Google Scholar] [CrossRef]
- Yagi, T.; Iinuma, H.; Hayama, T.; Matsuda, K.; Nozawa, K.; Tsukamoto, M.; Shimada, R.; Akahane, T.; Tsuchiya, T.; Ozawa, T.; et al. Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol. Clin. Oncol. 2019, 11, 416–424. [Google Scholar] [CrossRef]
- Ye, D.X.; Wang, S.S.; Huang, Y.; Chi, P. A 3-circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int. 2019, 19, 276. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cai, D.; Li, W.; Yu, T.; Mao, H.; Jiang, S.; Xiao, B. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin. Biochem. 2019, 74, 60–68. [Google Scholar] [CrossRef]
- Pan, B.; Qin, J.; Liu, X.; He, B.; Wang, X.; Pan, Y.; Sun, H.; Xu, T.; Xu, M.; Chen, X.; et al. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front. Genet. 2019, 10, 1096. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Qiu, C.; Wang, M.; Mao, N.; Wu, S.; Dai, Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem. Biophys. Res. Commun. 2018, 497, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qiu, S.; Luo, P.; Zhou, H.; Jing, W.; Liang, C.; Tu, J. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018, 22, 135–142. [Google Scholar] [CrossRef]
- Zhu, C.L.; Sha, X.; Wang, Y.; Li, J.; Zhang, M.Y.; Guo, Z.Y.; Sun, S.A.; He, J.D. Circular RNA hsa_circ_0007142 Is Upregulated and Targets miR-103a-2-5p in Colorectal Cancer. J. Oncol. 2019, 2019, 9836819. [Google Scholar] [CrossRef]
- Xie, X.; Tang, B.; Xiao, Y.F.; Xie, R.; Li, B.S.; Dong, H.; Zhou, J.Y.; Yang, S.M. Long non-coding RNAs in colorectal cancer. Oncotarget 2016, 7, 5226–5239. [Google Scholar] [CrossRef]
- Svoboda, M.; Slyskova, J.; Schneiderova, M.; Makovicky, P.; Bielik, L.; Levy, M.; Lipska, L.; Hemmelova, B.; Kala, Z.; Protivankova, M.; et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014, 35, 1510–1515. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Gao, S.; Jing, F.; Yang, Y.; Du, L.; Zheng, G.; Li, P.; Li, C.; Wang, C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 2016, 7, 85551–85563. [Google Scholar] [CrossRef]
- Liang, Z.X.; Liu, H.S.; Wang, F.W.; Xiong, L.; Zhou, C.; Hu, T.; He, X.W.; Wu, X.J.; Xie, D.; Wu, X.R.; et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019, 10, 829. [Google Scholar] [CrossRef]
- Oehme, F.; Krahl, S.; Gyorffy, B.; Muessle, B.; Rao, V.; Greif, H.; Ziegler, N.; Lin, K.; Thepkaysone, M.L.; Polster, H.; et al. Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019, 16, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.J.; Xie, S.L.; Li, Q.; Ma, J.; Wang, G.Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 2011, 39, 2119–2128. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.H.; Wang, X.L.; Tang, H.M.; Jiang, T.; Chen, J.; Lu, S.; Qiu, G.Q.; Peng, Z.H.; Yan, D.W. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol. Rep. 2014, 32, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, C.; Brex, D.; Caponnetto, A.; Cirnigliaro, M.; Scalia, M.; Magnano, A.; Caltabiano, R.; Barbagallo, D.; Biondi, A.; Cappellani, A.; et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol. Ther. Nucleic Acids 2018, 12, 229–241. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).