Power Analysis for Human Melatonin Suppression Experiments
Abstract
1. Introduction
2. Results
2.1. Overview
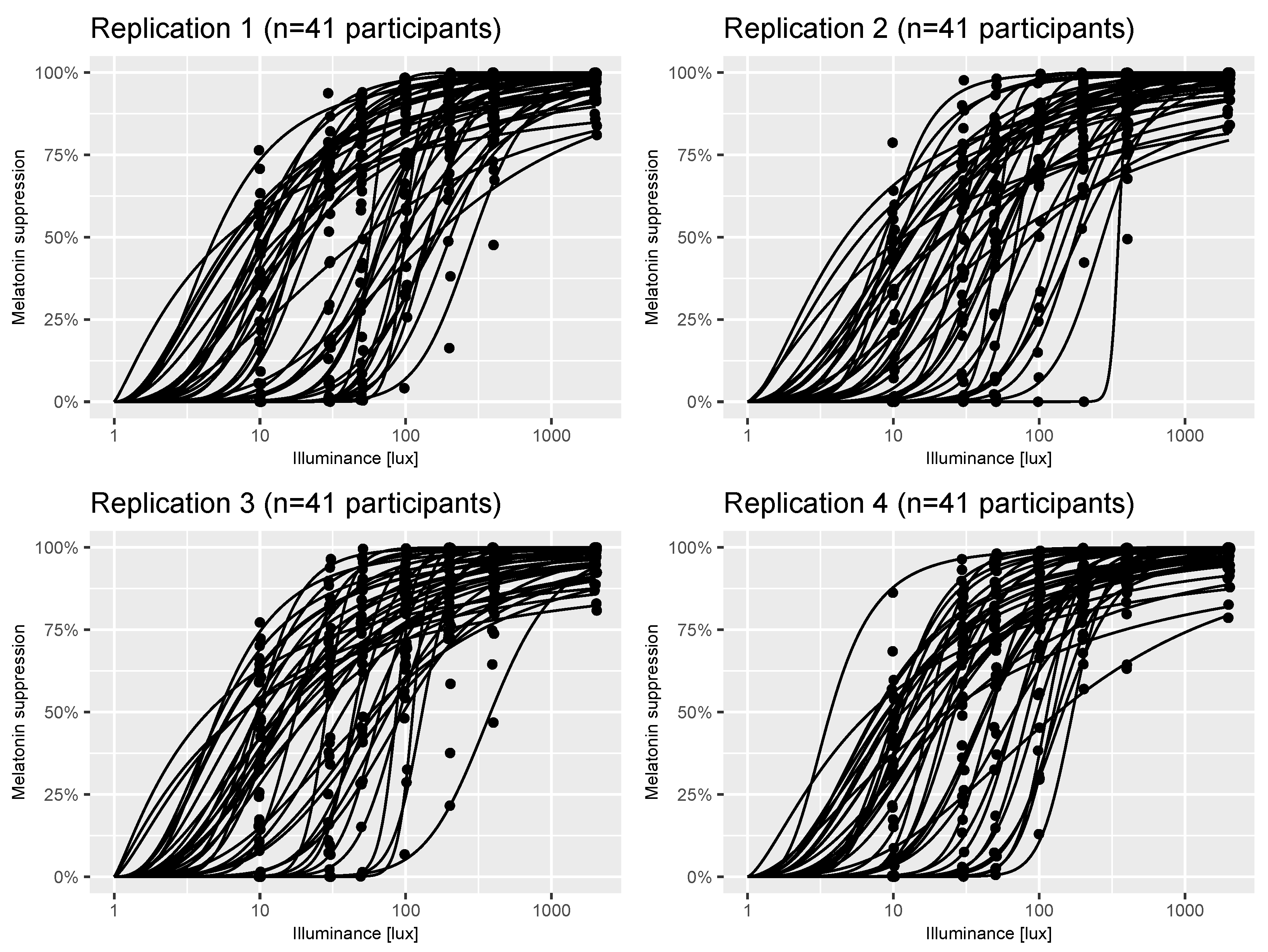
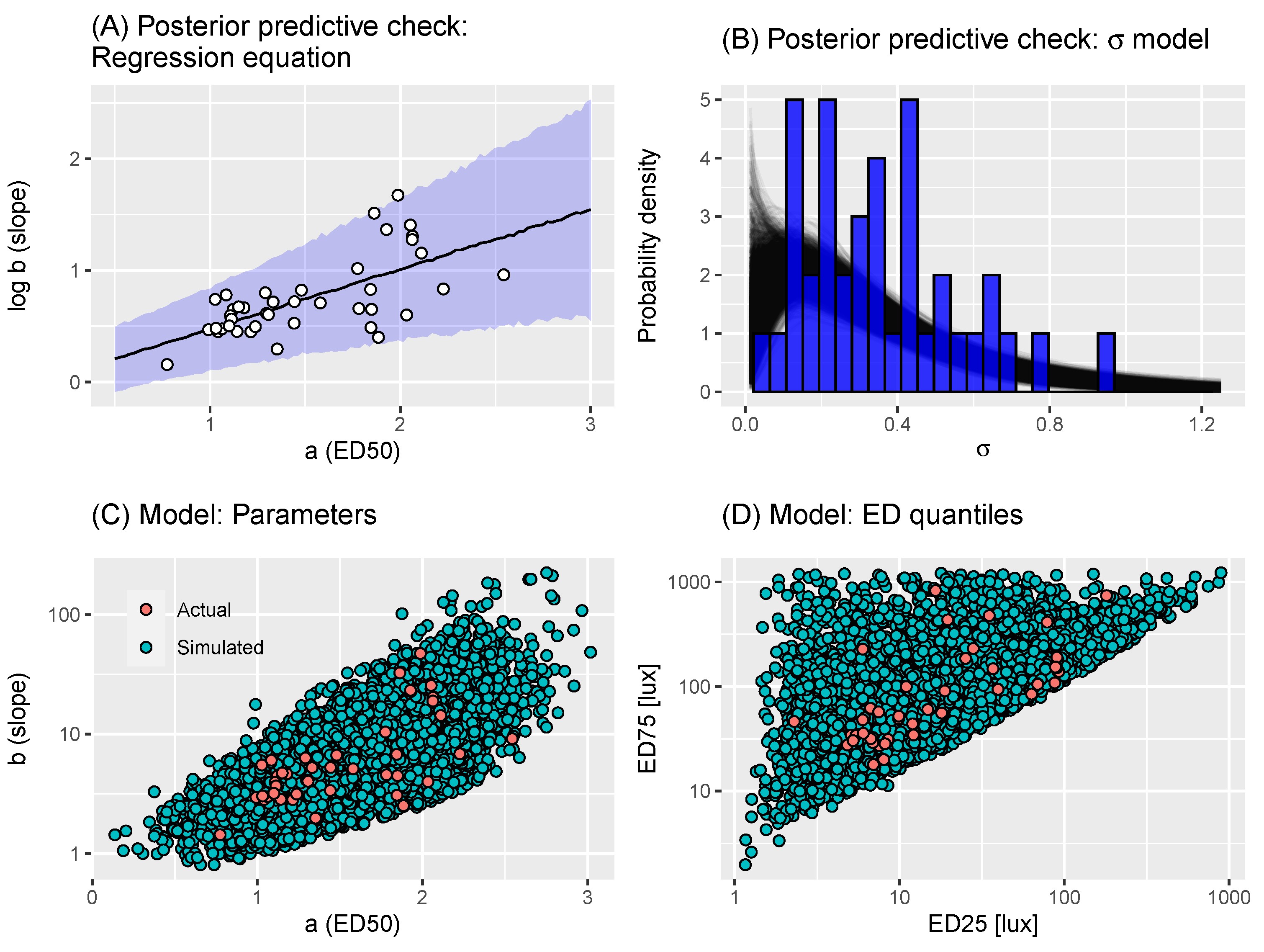
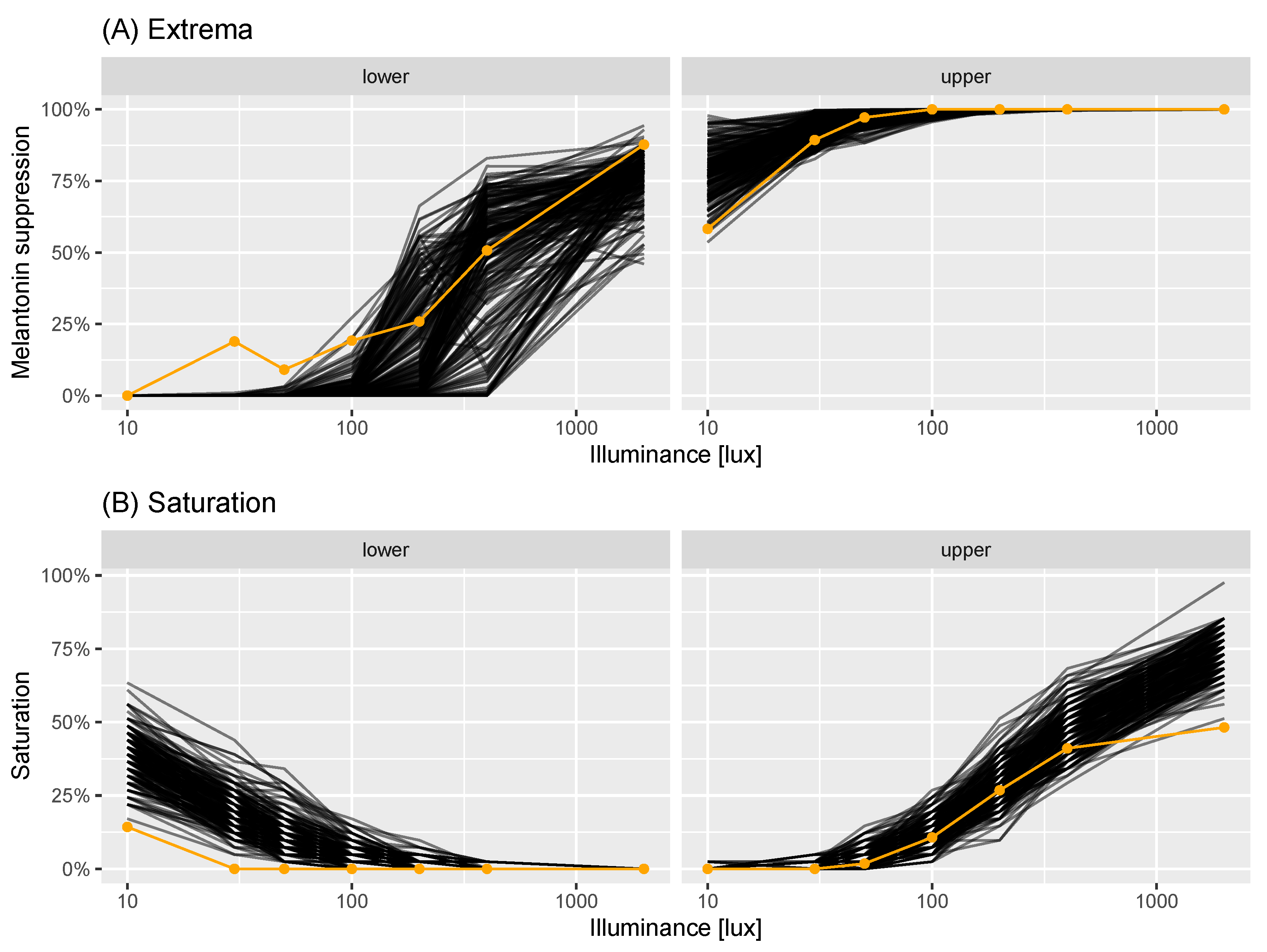
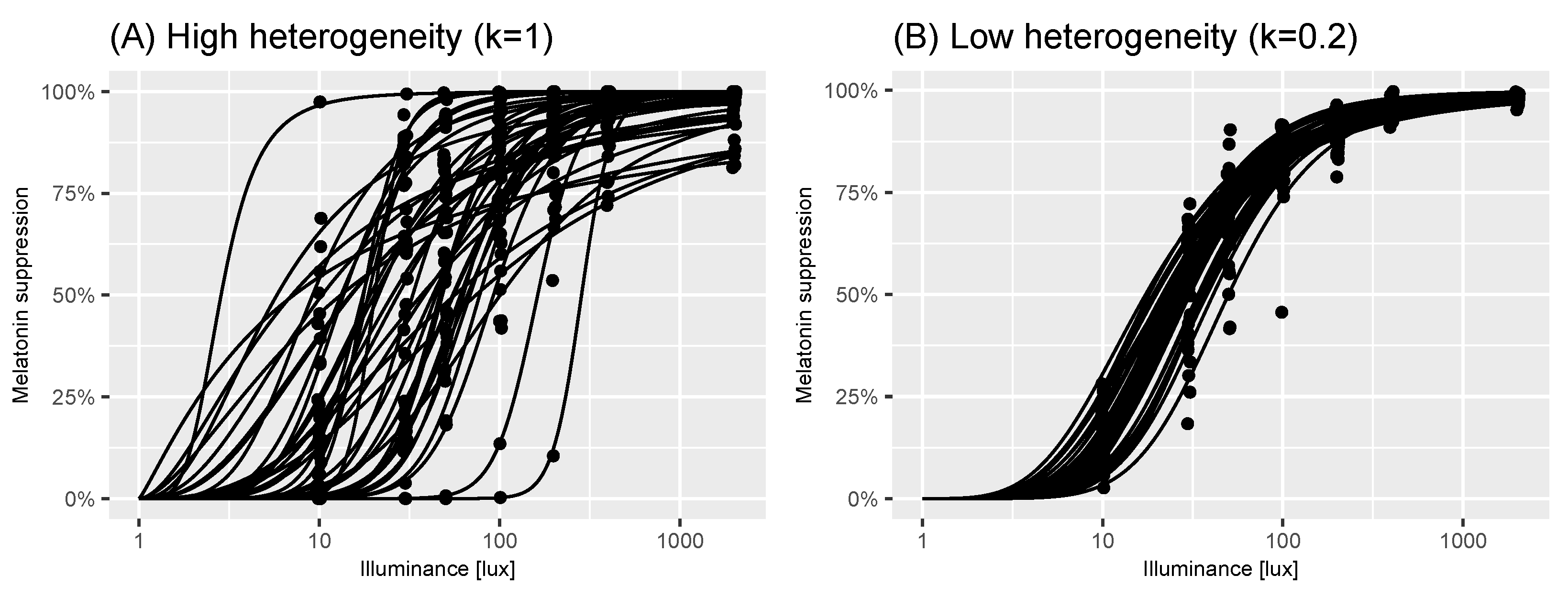
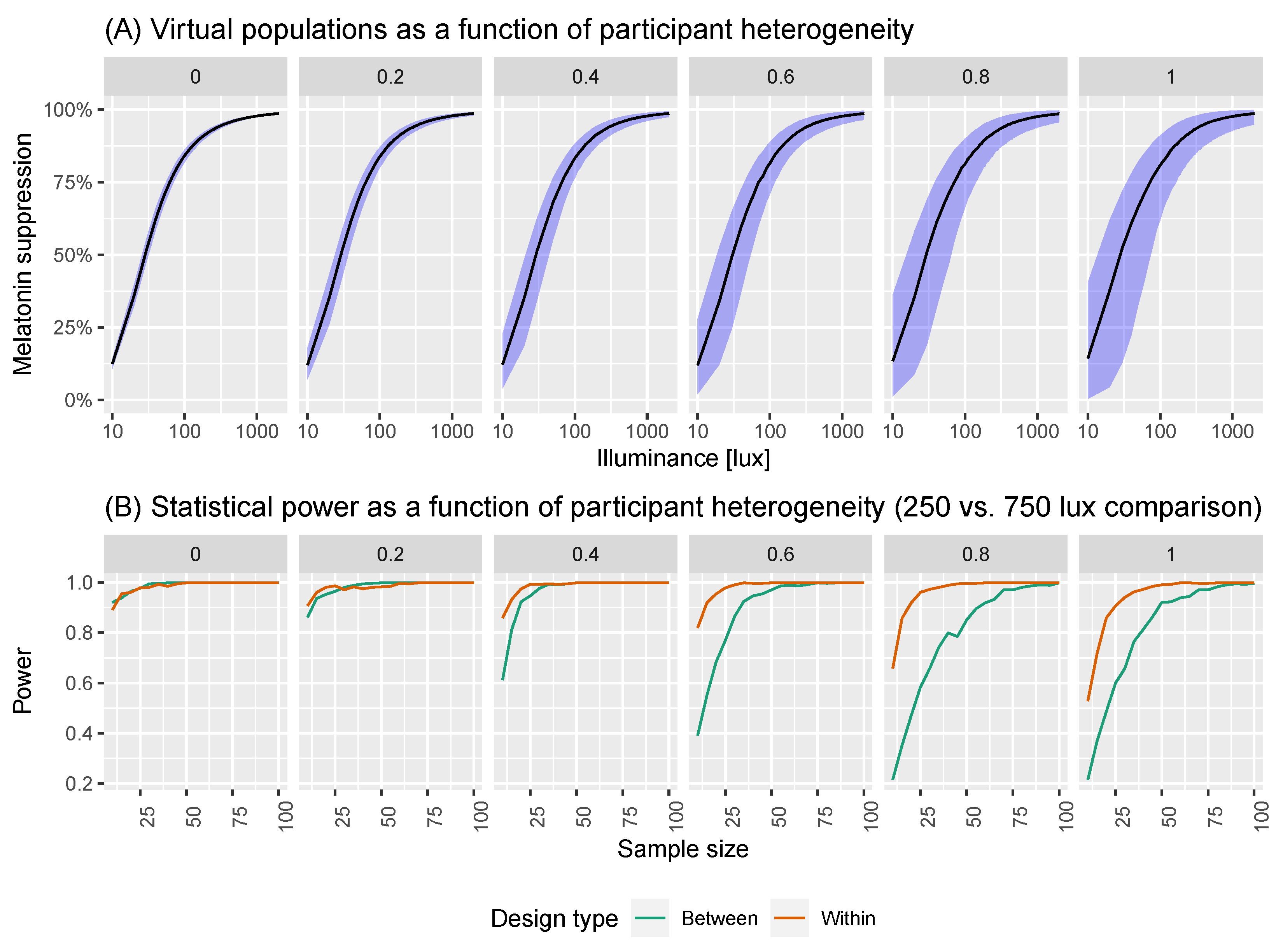
2.2. A Model of Virtual Experiments
| Algorithm 1 Virtual individual generation. Takes as input posterior draws of in Equation (2) |
|
| Algorithm 2 Virtual individual generation: reduced individual variance. Takes as input posterior draws of in Equation (2) and |
|
2.3. Power Calculations
2.4. Extension to Melanopic EDI
3. Discussion
3.1. Limitations
3.2. Future Directions
4. Conclusions
5. Methods
5.1. Sample Characteristics
“A total of 61 participants were enrolled, of whom 3 were excluded based on actigraphy, and 2 did not complete the study beyond the baseline DLMO. Overall, 56 healthy young Caucasian adults (29 women, 27 men; 20.8 ± 2.6 y of age) completed the study. Participants were free from any medical or psychological conditions, had a BMI of 18–30 kg/m2, and were not taking any medications at the time of the study. Participants had not recently traveled across time zones (1 mo per time zone, up to 3 mo) or engaged in shiftwork in the previous 12 mo. Women were naturally cycling (i.e., free from hormonal contraception) and had a regular menstrual cycle of 21–36 d in duration. Participants were healthy sleepers, reporting no subjective problems or previous diagnoses, and having a regular bedtime before 1 AM. A score of 10 or greater on the Epworth Sleepiness Scale was exclusionary, and participants were predominantly intermediate chronotypes (MEQ score of 52.7 ± 9.2). Participants had an average bedtime and waketime of 23:04 (SD = 44 min) and 07:04 (SD = 44 min), respectively. DLMO occurred on average at 21:05 (SD = 70 min), 2.22 h before bedtime. A total of nine participants wore prescription glasses at each test session.”
5.2. Reproducibility of Results
5.3. Shiny App
5.4. Code and Data Availability
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blume, C.; Garbazza, C.; Spitschan, M. Effects of light on human circadian rhythms, sleep and mood. Somnologie 2019, 23, 147–156. [Google Scholar] [CrossRef]
- Vetter, C.; Pattison, P.M.; Houser, K.; Herf, M.; Phillips, A.J.K.; Wright, K.P.; Skene, D.J.; Brainard, G.C.; Boivin, D.B.; Glickman, G. A review of human physiological responses to light: Implications for the development of integrative lighting solutions. Leukos 2021, 18, 387–414. [Google Scholar] [CrossRef]
- Schlangen, L.J.M.; Price, L.L.A. The lighting environment, its metrology, and non-visual responses. Front. Neurol. 2021, 12, 624861. [Google Scholar] [CrossRef]
- Provencio, I.; Jiang, G.; De Grip, W.J.; Hayes, W.P.; Rollag, M.D. Melanopsin: An opsin in melanophores, brain, and eye. Proc. Natl. Acad. Sci. USA 1998, 95, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Provencio, I.; Rodriguez, I.R.; Jiang, G.; Hayes, W.P.; Moreira, E.F.; Rollag, M.D. A novel human opsin in the inner retina. J. Neurosci. 2000, 20, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Do, M.T.H. Melanopsin and the intrinsically photosensitive retinal ganglion cells: Biophysics to behavior. Neuron 2019, 104, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Spitschan, M. Melanopsin contributions to non-visual and visual function. Curr. Opin. Behav. Sci. 2019, 30, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Brainard, G.C.; Hanifin, J.P.; Greeson, J.M.; Byrne, B.; Glickman, G.; Gerner, E.; Rollag, M.D. Action spectrum for melatonin regulation in humans: Evidence for a novel circadian photoreceptor. J. Neurosci. 2001, 21, 6405–6412. [Google Scholar] [CrossRef] [PubMed]
- Thapan, K.; Arendt, J.; Skene, D.J. An action spectrum for melatonin suppression: Evidence for a novel non-rod, non-cone photoreceptor system in humans. J. Physiol. 2001, 535, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Nowozin, C.; Wahnschaffe, A.; Rodenbeck, A.; de Zeeuw, J.; Hadel, S.; Kozakov, R.; Schopp, H.; Munch, M.; Kunz, D. Applying melanopic lux to measure biological light effects on melatonin suppression and subjective sleepiness. Curr. Alzheimer Res. 2017, 14, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Prayag, A.S.; Najjar, R.P.; Gronfier, C. Melatonin suppression is exquisitely sensitive to light and primarily driven by melanopsin in humans. J. Pineal Res. 2019, 66, e12562. [Google Scholar] [CrossRef]
- Brown, T.M. Melanopic illuminance defines the magnitude of human circadian light responses under a wide range of conditions. J. Pineal Res. 2020, 69, e12655. [Google Scholar] [CrossRef]
- Gimenez, M.C.; Stefani, O.; Cajochen, C.; Lang, D.; Deuring, G.; Schlangen, L.J.M. Predicting melatonin suppression by light in humans: Unifying photoreceptor-based equivalent daylight illuminances, spectral composition, timing and duration of light exposure. J. Pineal Res. 2022, 72, e12786. [Google Scholar] [CrossRef]
- Cajochen, C.; Munch, M.; Kobialka, S.; Krauchi, K.; Steiner, R.; Oelhafen, P.; Orgul, S.; Wirz-Justice, A. High sensitivity of human melatonin, alertness, thermoregulation, and heart rate to short wavelength light. J. Clin. Endocrinol. Metab. 2005, 90, 1311–1316. [Google Scholar] [CrossRef]
- Nagare, R.; Rea, M.S.; Plitnick, B.; Figueiro, M.G. Nocturnal melatonin suppression by adolescents and adults for different levels, spectra, and durations of light exposure. J. Biol. Rhythm. 2019, 34, 178–194. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.E.; Hazelhoff, E.M.; Martial, F.P.; Cajochen, C.; Lucas, R.J. Exploiting metamerism to regulate the impact of a visual display on alertness and melatonin suppression independent of visual appearance. Sleep 2018, 41, zsy100. [Google Scholar] [CrossRef]
- Spitschan, M.; Lazar, R.; Yetik, E.; Cajochen, C. No evidence for an S cone contribution to acute neuroendocrine and alerting responses to light. Curr. Biol. 2019, 29, R1297–R1298. [Google Scholar] [CrossRef] [PubMed]
- Kennaway, D.J. A critical review of melatonin assays: Past and present. J. Pineal Res. 2019, 67, e12572. [Google Scholar] [CrossRef] [PubMed]
- Kennaway, D.J. Measuring melatonin by immunoassay. J. Pineal Res. 2020, 69, e12657. [Google Scholar] [CrossRef] [PubMed]
- Chellappa, S.L. Individual differences in light sensitivity affect sleep and circadian rhythms. Sleep 2021, 44, zsaa214. [Google Scholar] [CrossRef]
- Spitschan, M.; Santhi, N. Individual differences and diversity in human physiological responses to light. EBioMedicine 2021, 75, 103640. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Vidafar, P.; Burns, A.C.; McGlashan, E.M.; Anderson, C.; Rajaratnam, S.M.; Lockley, S.W.; Cain, S.W. High sensitivity and interindividual variability in the response of the human circadian system to evening light. Proc. Natl. Acad. Sci. USA 2019, 116, 12019–12024. [Google Scholar] [CrossRef]
- Spitschan, M.; Lambert, B. melluxdrc: Power Analysis for Human Melatonin Suppression Experiments (Main Package). 2021. Available online: https://github.com/mellux-project/melluxdrc (accessed on 1 February 2024).
- Spitschan, M.; Lambert, B. mellux-app: Power Analysis for Human Melatonin Suppression Experiments (Shiny app). 2021. Available online: https://github.com/mellux-project/mellux-app (accessed on 1 February 2024).
- Wand, M. KernSmooth: Functions for Kernel Smoothing Supporting Wand & Jones (1995), R package version 2.23-15; 2015; Available online: https://rdrr.io/cran/KernSmooth/ (accessed on 1 February 2024).
- Carpenter, B.; Gelman, A.; Hoffman, M.D.; Lee, D.; Goodrich, B.; Betancourt, M.; Brubaker, M.; Guo, J.; Li, P.; Riddell, A. Stan: A probabilistic programming language. J. Stat. Softw. 2017, 76, 1–32. [Google Scholar] [CrossRef]
- Vehtari, A.; Gelman, A.; Simpson, D.; Carpenter, B.; Bürkner, P.C. Rank-normalization, folding, and localization: An improved for assessing convergence of MCMC. arXiv 2019, arXiv:1903.08008. [Google Scholar] [CrossRef]
- Lambert, B. A Student’s Guide to Bayesian Statistics; Sage: Newcastle upon Tyne, UK, 2018. [Google Scholar]
- Spitschan, M.; Lambert, B. mellux-model_fitting: Power Analysis for Human Melatonin Suppression Experiments (Model Fitting Code). 2021. Available online: https://www.biorxiv.org/content/10.1101/2023.01.30.526303v1 (accessed on 1 February 2024).
- Lambert, B. An Introduction to Inverse Transform Sampling. Youtube. 2018. Available online: https://www.youtube.com/watch?v=rnBbYsysPaU (accessed on 1 February 2024).
- CIE S 026:2018; CIE System for Metrology of Optical Radiation for ipRGC-Influenced Responses to Light. CIE: Vienna, Austria, 2018.
- Spitschan, M.; Mead, J.; Roos, C.; Lowis, C.; Griffiths, B.; Mucur, P.; Herf, M.; Nam, S.; Veitch, J.A. luox: Validated reference open-access and open-source web platform for calculating and sharing physiologically relevant quantities for light and lighting. Wellcome Open Res 2022, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Spitschan, M.; Lambert, B. mellux-Simulations: Power Analysis for Human Melatonin Suppression Experiments (Simulations). 2021. Available online: https://github.com/mellux-project/mellux-simulations (accessed on 1 February 2024).




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spitschan, M.; Vidafar, P.; Cain, S.W.; Phillips, A.J.K.; Lambert, B.C. Power Analysis for Human Melatonin Suppression Experiments. Clocks & Sleep 2024, 6, 114-128. https://doi.org/10.3390/clockssleep6010009
Spitschan M, Vidafar P, Cain SW, Phillips AJK, Lambert BC. Power Analysis for Human Melatonin Suppression Experiments. Clocks & Sleep. 2024; 6(1):114-128. https://doi.org/10.3390/clockssleep6010009
Chicago/Turabian StyleSpitschan, Manuel, Parisa Vidafar, Sean W. Cain, Andrew J. K. Phillips, and Ben C. Lambert. 2024. "Power Analysis for Human Melatonin Suppression Experiments" Clocks & Sleep 6, no. 1: 114-128. https://doi.org/10.3390/clockssleep6010009
APA StyleSpitschan, M., Vidafar, P., Cain, S. W., Phillips, A. J. K., & Lambert, B. C. (2024). Power Analysis for Human Melatonin Suppression Experiments. Clocks & Sleep, 6(1), 114-128. https://doi.org/10.3390/clockssleep6010009









