A Review of Evidence for the Involvement of the Circadian Clock Genes into Malignant Transformation of Thyroid Tissue
Abstract
1. Introduction
2. Results
3. Discussion
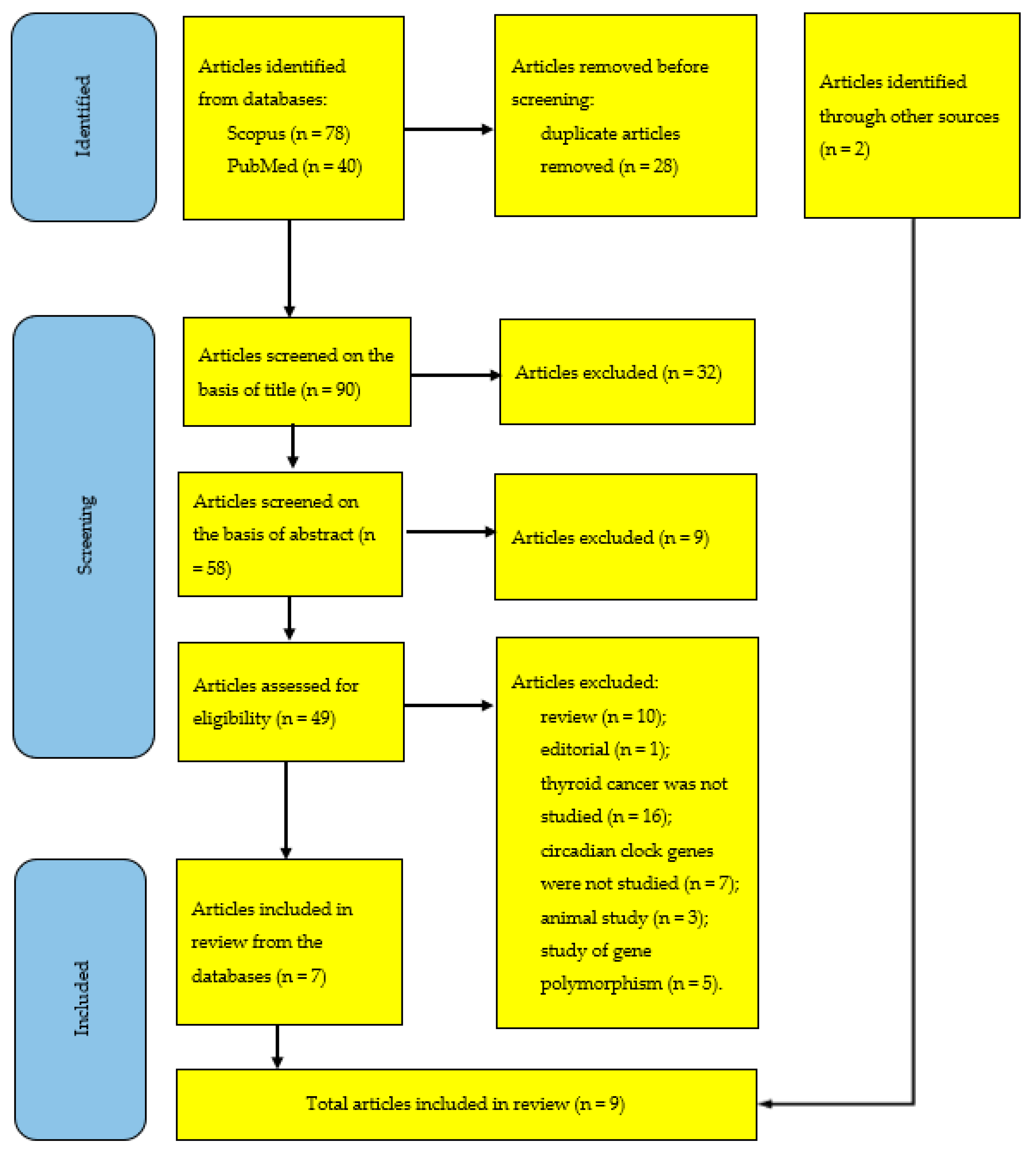
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mannic, T.; Meyer, P.; Triponez, F.; Pusztaszeri, M.; Le Martelot, G.; Mariani, O.; Schmitter, D.; Sage, D.; Philippe, J.; Dibner, C. Circadian clock characteristics are altered in human thyroid malignant nodules. J. Clin. Endocrinol. Metab. 2013, 98, 4446–4456. [Google Scholar] [CrossRef] [PubMed]
- Mond, M.; Alexiadis, M.; Eriksson, N.; Davis, M.J.; Muscat, G.E.; Fuller, P.J.; Gilfillan, C. Nuclear receptor expression in human differentiated thyroid tumors. Thyroid 2014, 24, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Chitikova, Z.; Pusztaszeri, M.; Makhlouf, A.M.; Berczy, M.; Delucinge-Vivier, C.; Triponez, F.; Meyer, P.; Philippe, J.; Dibner, C. Identification of new biomarkers for human papillary thyroid carcinoma employing NanoString analysis. Oncotarget 2015, 6, 10978–10993. [Google Scholar] [CrossRef] [PubMed]
- Makhlouf, A.M.; Chitikova, Z.; Pusztaszeri, M.; Berczy, M.; Delucinge-Vivier, C.; Triponez, F.; Meyer, P.; Philippe, J.; Dibner, C. Identification of CHEK1, SLC26A4, c-KIT, TPO and TG as new biomarkers for human follicular thyroid carcinoma. Oncotarget 2016, 7, 45776–45788. [Google Scholar] [CrossRef]
- Gallo, C.; Fragliasso, V.; Donati, B.; Torricelli, F.; Tameni, A.; Piana, S.; Ciarrocchi, A. The bHLH transcription factor DEC1 promotes thyroid cancer aggressiveness by the interplay with NOTCH1. Cell Death Dis. 2018, 9, 871. [Google Scholar] [CrossRef]
- Sadowski, S.M.; Petrenko, V.; Meyer, P.; Pusztaszeri, M.; Brulhart-Meynet, M.C.; Heddad Masson, M.; Triponez, F.; Philippe, J.; Dibner, C. Validation of molecular biomarkers for preoperative diagnostics of human papillary thyroid carcinoma in fine needle aspirates. Gland. Surg. 2019, 8 (Suppl. S2), S62–S76. [Google Scholar] [CrossRef]
- Lou, X.; Wang, H.; Tu, Y.; Tan, W.; Jiang, C.; Sun, J.; Bao, Z. Alterations of sleep quality and circadian rhythm genes expression in elderly thyroid nodule patients and risks associated with thyroid malignancy. Sci. Rep. 2021, 11, 13682. [Google Scholar] [CrossRef]
- Xu, T.; Jin, T.; Lu, X.; Pan, Z.; Tan, Z.; Zheng, C.; Liu, Y.; Hu, X.; Ba, L.; Ren, H.; et al. A signature of circadian rhythm genes in driving anaplastic thyroid carcinoma malignant progression. Cell. Signal. 2022, 95, 110332. [Google Scholar] [CrossRef]
- Mou, Y.K.; Ren, C.; Li, Y.M.; Yu, G.H.; Zheng, G.B.; Song, H.; Lu, C.X.; Tian, R.X.; Song, X.C. Correlation analysis of clock genes and MEN2 medullary thyroid carcinoma. Chin. J. Otorhinolaryngol. Head Neck Surg. 2022, 7, 1079–1086. (In Chinese) [Google Scholar] [CrossRef]
- Ikegami, K.; Refetoff, S.; Van Cauter, E.; Yoshimura, T. Interconnection between circadian clocks and thyroid function. Nat. Rev. Endocrinol. 2019, 15, 590–600. [Google Scholar] [CrossRef]
- Malaguarnera, R.; Ledda, C.; Filippello, A.; Frasca, F.; Francavilla, V.C.; Ramaci, T.; Parisi, M.C.; Rapisarda, V.; Piro, S. Thyroid cancer and circadian clock disruption. Cancers 2020, 12, 3109. [Google Scholar] [CrossRef]
- Putilov, A. Prospects of testing diurnal profiles of expressions of TSH-R and circadian clock genes in thyrocytes for identification of preoperative biomarkers for thyroid carcinoma. Int. J. Mol. Sci. 2022, 23, 12208. [Google Scholar] [CrossRef]
- Philippe, J.; Dibner, C. Thyroid circadian timing: Roles in physiology and thyroid malignancies. J. Biol. Rhythm. 2015, 30, 76–83. [Google Scholar] [CrossRef]
- Dibner, C.; Sadowski, S.M.; Triponez, F.; Philippe, J. The search for preoperative biomarkers for thyroid carcinoma: Application of the thyroid circadian clock properties. Biomark. Med. 2017, 11, 285–293. [Google Scholar] [CrossRef]
- Angelousi, A.; Kassi, E.; Nasiri-Ansari, N.; Weickert, M.O.; Randeva, H.; Kaltsas, G. Clock genes alterations and endocrine disorders. Eur. J. Clin. Investig. 2018, 48, e12927. [Google Scholar] [CrossRef]
- Angelousi, A.; Kassi, E.; Ansari-Nasiri, N.; Randeva, H.; Kaltsas, G.; Chrousos, G. Clock genes and cancer development in particular in endocrine tissues. Endocr.-Relat. Cancer 2019, 26, R305–R317. [Google Scholar] [CrossRef]
- Bellastella, G.; Maiorino, M.I.; Scappaticcio, L.; De Bellis, A.; Mercadante, S.; Esposito, K.; Bellastella, A. Chronothyroidology: Chronobiological aspects in thyroid function and diseases. Life 2021, 11, 426. [Google Scholar] [CrossRef]
- Miro, C.; Docimo, A.; Barrea, L.; Verde, L.; Cernea, S.; Sojat, A.S.; Marina, L.V.; Docimo, G.; Colao, A.; Dentice, M.; et al. “Time” for obesity-related cancer: The role of the circadian rhythm in cancer pathogenesis and treatment. Semin. Cancer Biol. 2023, 91, 99–109. [Google Scholar] [CrossRef]
- Valderrabano, P.; McIver, B. Evaluation and management of indeterminate thyroid nodules: The revolution of risk stratification beyond cytological diagnosis. Cancer Control. 2017, 24, 1073274817729231. [Google Scholar] [CrossRef]
- Durante, C.; Grani, G.; Lamartina, L.; Filetti, S.; Mandel, S.J.; Cooper, D.S. The diagnosis and management of thyroid nodules: A review. JAMA 2018, 319, 914–924. [Google Scholar] [CrossRef]
- Gharib, H. Fine-needle aspiration biopsy of thyroid nodules: Advantages, limitations, and effect. Mayo Clin. Proc. 1994, 69, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Chmielik, E.; Rusinek, D.; Oczko-Wojciechowska, M.; Jarzab, M.; Krajewska, J.; Czarniecka, A.; Jarzab, B. Heterogeneity of Thyroid Cancer. Pathobiology 2018, 85, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Baloch, Z.W.; Asa, S.L.; Barletta, J.A.; Ghossein, R.A.; Juhlin, C.C.; Jung, C.K.; LiVolsi, V.A.; Papotti, M.G.; Sobrinho-Simões, M.; Tallini, G.; et al. Overview of the 2022 WHO Classification of Thyroid Neoplasms. Endocr. Pathol. 2022, 33, 27–63. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimpasic, T.; Ghossein, R.; Carlson, D.L.; Chernichenko, N.; Nixon, I.; Palmer, F.L.; Lee, N.Y.; Shaha, A.R.; Patel, S.G.; Tuttle, R.M.; et al. Poorly differentiated thyroid carcinoma presenting with gross extrathyroidal extension: 1986–2009 Memorial Sloan-Kettering Cancer Center experience. Thyroid 2013, 23, 997–1002. [Google Scholar] [CrossRef]
- Elisei, R.; Molinaro, E.; Agate, L.; Bottici, V.; Masserini, L.; Ceccarelli, C.; Lippi, F.; Grasso, L.; Basolo, F.; Bevilacqua, G.; et al. Are the clinical and pathological features of differentiated thyroid carcinoma really changed over the last 35 years? Study on 4187 patients from a single Italian institution to answer this question. J. Clin. Endocrinol. Metab. 2010, 95, 1516–1527. [Google Scholar] [CrossRef]
- Luster, M.; Hänscheid, H.; Freudenberg, L.S.; Verburg, F.A. Radioiodine therapy of metastatic lesions of differentiated thyroid cancer. J. Endocrinol. Investig. 2012, 35 (Suppl. S6), 21–29. [Google Scholar]
- Haugen, B.R. 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: What is new and what has changed? Cancer 2017, 123, 372–381. [Google Scholar] [CrossRef]
- Chiacchio, S.; Lorenzoni, A.; Boni, G.; Rubello, D.; Elisei, R.; Mariani, G. Anaplastic thyroid cancer: Prevalence, diagnosis and treatment. Minerva Endocrinol. 2008, 33, 341–357. [Google Scholar]
- Walsh, P.S.; Wilde, J.I.; Tom, E.Y.; Reynolds, J.D.; Chen, D.C.; Chudova, D.I.; Pagan, M.; Pankratz, D.G.; Wong, M.; Veitch, J.; et al. Analytical performance verification of a molecular diagnostic for cytology-indeterminate thyroid nodules. J. Clin. Endocrinol. Metab. 2012, 97, E2297–E2306. [Google Scholar] [CrossRef]
- Rappa, G.; Puglisi, C.; Santos, M.F.; Forte, S.; Memeo, L.; Lorico, A. Extracellular vesicles from thyroid carcinoma: The new frontier of liquid biopsy. Int. J. Mol. Sci. 2019, 20, 1114. [Google Scholar] [CrossRef]
- Alexander, E.K.; Kennedy, G.C.; Baloch, Z.W.; Cibas, E.S.; Chudova, D.; Deggans, J.; Friedman, L.; Kloos, R.T.; LiVolsi, V.A.; Mandel, S.J.; et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N. Engl. J. Med. 2012, 367, 705–715. [Google Scholar] [CrossRef]
- Rossi, E.D.; Pantanowitz, L.; Faquin, W.C. The role of molecular testing for the indeterminate thyroid FNA. Genes 2019, 10, 736. [Google Scholar] [CrossRef]
- Muzza, M.; Colombo, C.; Pogliaghi, G.; Karapanou, O.; Fugazzola, L. Molecular markers for the classification of cytologically indeterminate thyroid nodules. J. Endocrinol. Investig. 2020, 43, 703–716. [Google Scholar] [CrossRef]
- Rao, S.N.; Bernet, V. Indeterminate thyroid nodules in the era of molecular genomics. Mol. Genet. Genom. Med. 2020, 8, e1288. [Google Scholar] [CrossRef]
- Karapanou, O. The role of molecular genetics in the presurgical management of thyroid nodules. Minerva Endocrinol. 2021, 46, 21–34. [Google Scholar] [CrossRef]
- Patel, S.G.; Carty, S.E.; Lee, A.J. Molecular testing for thyroid nodules including its interpretation and use in clinical practice. Ann. Surg. Oncol. 2021, 28, 8884–8891. [Google Scholar] [CrossRef]
- Rosbash, M. Circadian rhythms and the transcriptional feedback loop (Nobel lecture). Angew. Chem. Int. Ed. Engl. 2021, 60, 8650–8666. [Google Scholar] [CrossRef]
- Ye, R.; Selby, C.P.; Ozturk, N.; Annayev, Y.; Sancar, A. Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 2011, 286, 25891–25902. [Google Scholar] [CrossRef]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15, R271–R277. [Google Scholar] [CrossRef]
- Sato, F.; Kawamoto, T.; Fujimoto, K.; Noshiro, M.; Honda, K.K.; Honma, S.; Honma, K.; Kato, Y. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. Eur. J. Biochem. 2004, 271, 4409–4419. [Google Scholar] [CrossRef]
- Crumbley, C.; Wang, Y.; Kojetin, D.J.; Burris, T.P. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J. Biol. Chem. 2010, 285, 35386–35392. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.; Azzi, A. Peripheral circadian oscillators in mammals. Handb. Exp. Pharmacol. 2013, 217, 45–66. [Google Scholar] [CrossRef]
- Albrecht, U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Honma, S. The mammalian circadian system: A hierarchical multi-oscillator structure for generating circadian rhythm. J. Physiol. Sci. 2018, 68, 207–219. [Google Scholar] [CrossRef]
- Gamble, K.L.; Berry, R.; Frank, S.J.; Young, M.E. Circadian clock control of endocrine factors. Nat. Rev. Endocrinol. 2014, 10, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U.; Albrecht, U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [PubMed]
- Koronowski, K.B.; Sassone-Corsi, P. Communicating clocks shape circadian homeostasis. Science 2021, 371, eabd0951. [Google Scholar] [CrossRef]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef]
- Fu, L.; Kettner, N.M. The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 2013, 119, 221–282. [Google Scholar] [CrossRef]
- Li, H.X. The role of circadian clock genes in tumors. OncoTargets Ther. 2019, 12, 3645–3660. [Google Scholar] [CrossRef]
- Morgan, M.N.; Dvuchbabny, S.; Martinez, C.A.; Kerr, B.; Cistulli, P.A.; Cook, K.M. The cancer clock is (not) ticking: Links between circadian rhythms and cancer. Clocks Sleep 2019, 1, 435–458. [Google Scholar] [CrossRef]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between circadian clock and cancer: New frontiers for cancer treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef]
- Hernández-Rosas, F.; López-Rosas, C.A.; Saavedra-Vélez, M.V. Disruption of the molecular circadian clock and cancer: An epigenetic link. Biochem. Genet. 2020, 58, 189–209. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Umemura, Y.; Yagita, K. Circadian clock and cancer: From a viewpoint of cellular differentiation. Int. J. Urol. 2020, 27, 518–524. [Google Scholar] [CrossRef]
- Sancar, A.; Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science 2021, 371, eabb0738. [Google Scholar] [CrossRef]
- Yang, Y.; Lindsey-Boltz, L.A.; Vaughn, C.M.; Selby, C.P.; Cao, X.; Liu, Z.; Hsu, D.S.; Sancar, A. Circadian clock, carcinogenesis, chronochemotherapy connections. J. Biol. Chem. 2021, 297, 101068. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, C.; Cao, Y.; Li, J.; Bi, F. Major roles of the circadian clock in cancer. Cancer Biol. Med. 2023, 20, 1–24. [Google Scholar] [CrossRef]
- Sulli, G.; Rommel, A.; Wang, X.; Kolar, M.J.; Puca, F.; Saghatelian, A.; Plikus, M.V.; Verma, I.M.; Panda, S. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 2018, 553, 351–355. [Google Scholar] [CrossRef]
- Cha, H.K.; Chung, S.; Lim, H.Y.; Jung, J.W.; Son, G.H. Small molecule modulators of the circadian molecular clock with implications for neuropsychiatric diseases. Front. Mol. Neurosci. 2019, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Machekano, R.; McHenry, C.R. The utility of preoperative serum thyroid-stimulating hormone level for predicting malignant nodular thyroid disease. Am. J. Surg. 2010, 199, 294–297, discussion 298. [Google Scholar] [CrossRef] [PubMed]
- Fiore, E.; Rago, T.; Provenzale, M.A.; Scutari, M.; Ugolini, C.; Basolo, F.; Di Coscio, G.; Berti, P.; Grasso, L.; Elisei, R.; et al. Lower levels of TSH are associated with a lower risk of papillary thyroid cancer in patients with thyroid nodular disease: Thyroid autonomy may play a protective role. Endocr. Relat. Cancer 2009, 16, 1251–1260. [Google Scholar] [CrossRef]
- Kim, S.S.; Lee, B.J.; Lee, J.C.; Song, S.H.; Kim, B.H.; Son, S.M.; Kim, I.J.; Kim, Y.K.; Kang, Y.H. Preoperative serum thyroid stimulating hormone levels in well-differentiated thyroid carcinoma is a predictive factor for lateral lymph node metastasis as well as extrathyroidal extension in Korean patients: A single-center experience. Endocrine 2011, 39, 259–265. [Google Scholar] [CrossRef] [PubMed]
- McLeod, D.S.; Cooper, D.S.; Ladenson, P.W.; Ain, K.B.; Brierley, J.D.; Fein, H.G.; Haugen, B.R.; Jonklaas, J.; Magner, J.; Ross, D.S.; et al. Prognosis of differentiated thyroid cancer in relation to serum thyrotropin and thyroglobulin antibody status at time of diagnosis. Thyroid 2014, 24, 35–42. [Google Scholar] [CrossRef]
- Shi, R.L.; Liao, T.; Qu, N.; Liang, F.; Chen, J.Y.; Ji, Q.H. The usefulness of preoperative thyroid-stimulating hormone for predicting differentiated thyroid microcarcinoma. Otolaryngol. Head Neck Surg. 2016, 154, 256–262. [Google Scholar] [CrossRef]
- Su, A.; Zhao, W.; Wu, W.; Wei, T.; Ruan, M.; Li, Z.; Zhu, J. The association of preoperative thyroid-stimulating hormone level and the risk of differentiated thyroid cancer in patients with thyroid nodules: A systematic review and meta-analysis. Am. J. Surg. 2020, 220, 634–641. [Google Scholar] [CrossRef]
- Kim, H.; Jung, J.; Cho, Y.S.; Choi, J.Y.; Park, H.; Lee, Y.B.; Kim, S.W.; Chung, J.H.; Kim, T.H. Pattern analysis for prognosis of differentiated thyroid cancer according to preoperative serum thyrotropin levels. Sci. Rep. 2021, 11, 22322. [Google Scholar] [CrossRef]
- Baser, H.; Topaloglu, O.; Tam, A.A.; Evranos, B.; Alkan, A.; Sungu, N.; Dumlu, E.G.; Ersoy, R.; Cakir, B. Higher TSH can be used as an additional risk factor in prediction of malignancy in euthyroid thyroid nodules evaluated by cytology based on Bethesda system. Endocrine 2016, 53, 520–529. [Google Scholar] [CrossRef]
- Kaliszewski, K.; Diakowska, D.; Rzeszutko, M.; Nowak, Ł.; Wojtczak, B.; Sutkowski, K.; Ludwig, M.; Ludwig, B.; Mikuła, A.; Greniuk, M.; et al. Assessment of preoperative TSH serum level and thyroid cancer occurrence in patients with AUS/FLUS thyroid nodule diagnosis. Biomedicines 2022, 10, 1916. [Google Scholar] [CrossRef]
- Golbert, L.; de Cristo, A.P.; Faccin, C.S.; Farenzena, M.; Folgierini, H.; Graudenz, M.S.; Maia, A.L. Serum TSH levels as a predictor of malignancy in thyroid nodules: A prospective study. PLoS ONE 2017, 12, e0188123. [Google Scholar] [CrossRef]
- Lucke, C.; Hehrmann, R.; von Mayersbach, K.; von zur Mühlen, A. Studies on circadian variations of plasma TSH, thyroxine and triiodothyronine in man. Acta Endocrinol. 1977, 86, 81–88. [Google Scholar] [CrossRef]
- Brabant, G.; Prank, K.; Ranft, U.; Schuermeyer, T.; Wagner, T.O.; Hauser, H.; Kummer, B.; Feistner, H.; Hesch, R.D.; von zur Mühlen, A. Physiological regulation of circadian and pulsatile thyrotropin secretion in normal man and woman. J. Clin. Endocrinol. Metab. 1990, 70, 403–409. [Google Scholar] [CrossRef]
- Roelfsema, F.; Veldhuis, J.D. Thyrotropin secretion patterns in health and disease. Endocr. Rev. 2013, 34, 619–657. [Google Scholar] [CrossRef]
- Danilenko, K.V.; Putilov, A.A. Diurnal and seasonal variations in cortisol, prolactin, TSH and thyroid hormones in women with and without seasonal affective disorder. J. Interdisc. Cycle Res. 1993, 24, 185–196. [Google Scholar] [CrossRef]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef]
- Morris, C.J.; Aeschbach, D.; Scheer, F.A. Circadian system, sleep and endocrinology. Mol. Cell. Endocrinol. 2012, 349, 91–104. [Google Scholar] [CrossRef]
- Custro, N.; Scafidi, V.; Notarbartolo, A. Alterations in circadian rhythm of serum thyrotropin in critically ill patients. Acta Endocrinol. 1992, 127, 18–22. [Google Scholar] [CrossRef]
- Chiamolera, M.I.; Wondisford, F.E. Minireview: Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology 2009, 150, 1091–1096. [Google Scholar] [CrossRef]
- García-Jiménez, C.; Santisteban, P. TSH signalling and cancer. Arq. Bras. Endocrinol. Metabol. 2007, 51, 654–671. [Google Scholar] [CrossRef]
- Spitzweg, C.; Joba, W.; Heufelder, A.E. Expression of thyroid-related genes in human thymus. Thyroid 1999, 9, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Bidart, J.M.; Caillou, B.; Mahé, C.; Lacroix, L.; Filetti, S.; Schlumberger, M. Expression of the Na+/I− symporter gene in human thyroid tumors: A comparison study with other thyroid-specific genes. J. Clin. Endocrinol. Metab. 1999, 84, 3228–3234. [Google Scholar] [CrossRef]
- Tanaka, K.; Sonoo, H.; Yamamoto, Y.; Udagawa, K.; Kunisue, H.; Arime, I.; Yamamoto, S.; Kurebayashi, J.; Shimozuma, K. Changes of expression level of the differentiation markers in papillary thyroid carcinoma under thyrotropin suppression therapy in vivo immunohistochemical detection of thyroglobulin, thyroid peroxidase, and thyrotropin receptor. J. Surg. Oncol. 2000, 75, 108–116. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, M.; Sponziello, M.; Puppin, C.; Celano, M.; Maggisano, V.; Baldan, F.; Biffoni, M.; Bulotta, S.; Durante, C.; Filetti, S.; et al. Different expression of TSH receptor and NIS genes in thyroid cancer: Role of epigenetics. J. Mol. Endocrinol. 2014, 52, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Jiang, N.S.; Gorman, C.A.; Lee, C.Y. Thyrotropin receptors in normal and pathological human thyroid tissues. J. Clin. Endocrinol. Metab. 1978, 47, 870–876. [Google Scholar] [CrossRef]
- Ohta, K.; Endo, T.; Onaya, T. The mRNA levels of thyrotropin receptor, thyroglobulin and thyroid peroxidase in neoplastic human thyroid tissues. Biochem. Biophys. Res. Commun. 1991, 174, 1148–1153. [Google Scholar] [CrossRef]
- Elisei, R.; Pinchera, A.; Romei, C.; Gryczynska, M.; Pohl, V.; Maenhaut, C.; Fugazzola, L.; Pacini, F. Expression of thyrotropin receptor (TSH-R), thyroglobulin, thyroperoxidase, and calcitonin messenger ribonucleic acids in thyroid carcinomas: Evidence of TSH-R gene transcript in medullary histotype. J. Clin. Endocrinol. Metab. 1994, 78, 867–871. [Google Scholar] [CrossRef]
- Brabant, G.; Maenhaut, C.; Köhrle, J.; Scheumann, G.; Dralle, H.; Hoang-Vu, C.; Hesch, R.D.; von zur Mühlen, A.; Vassart, G.; Dumont, J.E. Human thyrotropin receptor gene: Expression in thyroid tumors and correlation to markers of thyroid differentiation and dedifferentiation. Mol. Cell. Endocrinol. 1991, 82, R7–R12. [Google Scholar] [CrossRef]
- Hoang-Vu, C.; Dralle, H.; Scheumann, G.; Maenhaut, C.; Horn, R.; von zur Mühlen, A.; Brabant, G. Gene expression of differentiation- and dedifferentiation markers in normal and malignant human thyroid tissues. Exp. Clin. Endocrinol. 1992, 100, 51–56. [Google Scholar] [CrossRef]
- Tanaka, K.; Inoue, H.; Miki, H.; Masuda, E.; Kitaichi, M.; Komaki, K.; Uyama, T.; Monden, Y. Relationship between prognostic score and thyrotropin receptor (TSH-R) in papillary thyroid carcinoma: Immunohistochemical detection of TSH-R. Br. J. Cancer 1997, 76, 594–599. [Google Scholar] [CrossRef]
- Zhou, C.; Li, J.; Wang, Y.; Xue, S.; Zhang, Y. Association of BRAF gene and TSHR with cervical lymph node metastasis of papillary thyroid microcarcinoma. Oncol. Lett. 2019, 17, 183–194. [Google Scholar] [CrossRef]
- Chinnappa, P.; Taguba, L.; Arciaga, R.; Faiman, C.; Siperstein, A.; Mehta, A.E.; Reddy, S.K.; Nasr, C.; Gupta, M.K. Detection of thyrotropin-receptor messenger ribonucleic acid (mRNA) and thyroglobulin mRNA transcripts in peripheral blood of patients with thyroid disease: Sensitive and specific markers for thyroid cancer. J. Clin. Endocrinol. Metab. 2004, 89, 3705–3709. [Google Scholar] [CrossRef]
- Chia, S.Y.; Milas, M.; Reddy, S.K.; Siperstein, A.; Skugor, M.; Brainard, J.; Gupta, M.K. Thyroid-stimulating hormone receptor messenger ribonucleic acid measurement in blood as a marker for circulating thyroid cancer cells and its role in the preoperative diagnosis of thyroid cancer. J. Clin. Endocrinol. Metab. 2007, 92, 468–475. [Google Scholar] [CrossRef]
- Haus, E.L.; Smolensky, M.H. Shift work and cancer risk: Potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med. Rev. 2013, 17, 273–284. [Google Scholar] [CrossRef]
- Rizza, S.; Neri, A.; Capanna, A.; Grecuccio, C.; Pietroiusti, A.; Magrini, A.; Federici, M.; Coppeta, L. Night shift working is associated with an increased risk of thyroid nodules. J. Occup. Environ. Med. 2020, 62, 1–3. [Google Scholar] [CrossRef]
- Luo, J.; Sands, M.; Wactawski-Wende, J.; Song, Y.; Margolis, K.L. Sleep disturbance and incidence of thyroid cancer in postmenopausal women the Women’s Health Initiative. Am. J. Epidemiol. 2013, 177, 42–49. [Google Scholar] [CrossRef]
- Jung, S.J.; Lee, J.; Choi, J.W.; Kim, S.; Shin, A.; Lee, Y.J. Association between sedative-hypnotic medication use and incidence of cancer in Korean Nation Health Insurance Service data. Sleep Med. 2019, 60, 159–164. [Google Scholar] [CrossRef]
- Zhang, D.; Jones, R.R.; James, P.; Kitahara, C.M.; Xiao, Q. Associations between artificial light at night and risk for thyroid cancer: A large US cohort study. Cancer 2021, 127, 1448–1458. [Google Scholar] [CrossRef]
- Peeri, N.C.; Tao, M.H.; Demissie, S.; Nguyen, U.D.T. Sleep duration, chronotype, and insomnia and the risk of lung cancer: United Kingdom Biobank Cohort. Cancer Epidemiol. Biomark. Prev. 2022, 31, 766–774. [Google Scholar] [CrossRef]
- Jensen, L.D.; Oliva, D.; Andersson, B.Å.; Lewin, F. A multidisciplinary perspective on the complex interactions between sleep, circadian, and metabolic disruption in cancer patients. Cancer Metastasis Rev. 2021, 40, 1055–1071. [Google Scholar] [CrossRef]
- Almaida-Pagan, P.F.; Torrente, M.; Campos, M.; Provencio, M.; Madrid, J.A.; Franco, F.; Morilla, B.R.; Cantos, B.; Sousa, P.A.; Madrid, M.J.M.; et al. Chronodisruption and Ambulatory Circadian Monitoring in Cancer Patients: Beyond the Body Clock. Curr. Oncol. Rep. 2022, 24, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Katchy, C.A.; Fu, L. Circadian gene variants in cancer. Ann. Med. 2014, 46, 208–220. [Google Scholar] [CrossRef]
- Morales-Santana, S.; Morell, S.; Leon, J.; Carazo-Gallego, A.; Jimenez-Lopez, J.C.; Morell, M. Overview of the polymorphisms of circadian genes associated with endocrine cancer. Front. Endocrinol. 2019, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, C.; Li, H.; Zhang, C.; Wu, H.; Huang, F. Effect of the interaction between cadmium exposure and CLOCK gene polymorphisms on thyroid cancer: A case-control study in China. Biol. Trace Elem. Res. 2020, 196, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Shilts, J.; Chen, G.; Hughey, J.J. Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ 2018, 6, e4327. [Google Scholar] [CrossRef]
- Pogliaghi, G. Liquid biopsy in thyroid cancer: From circulating biomarkers to a new prospective of tumor monitoring and therapy. Minerva Endocrinol. 2021, 46, 45–61. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, M.J.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
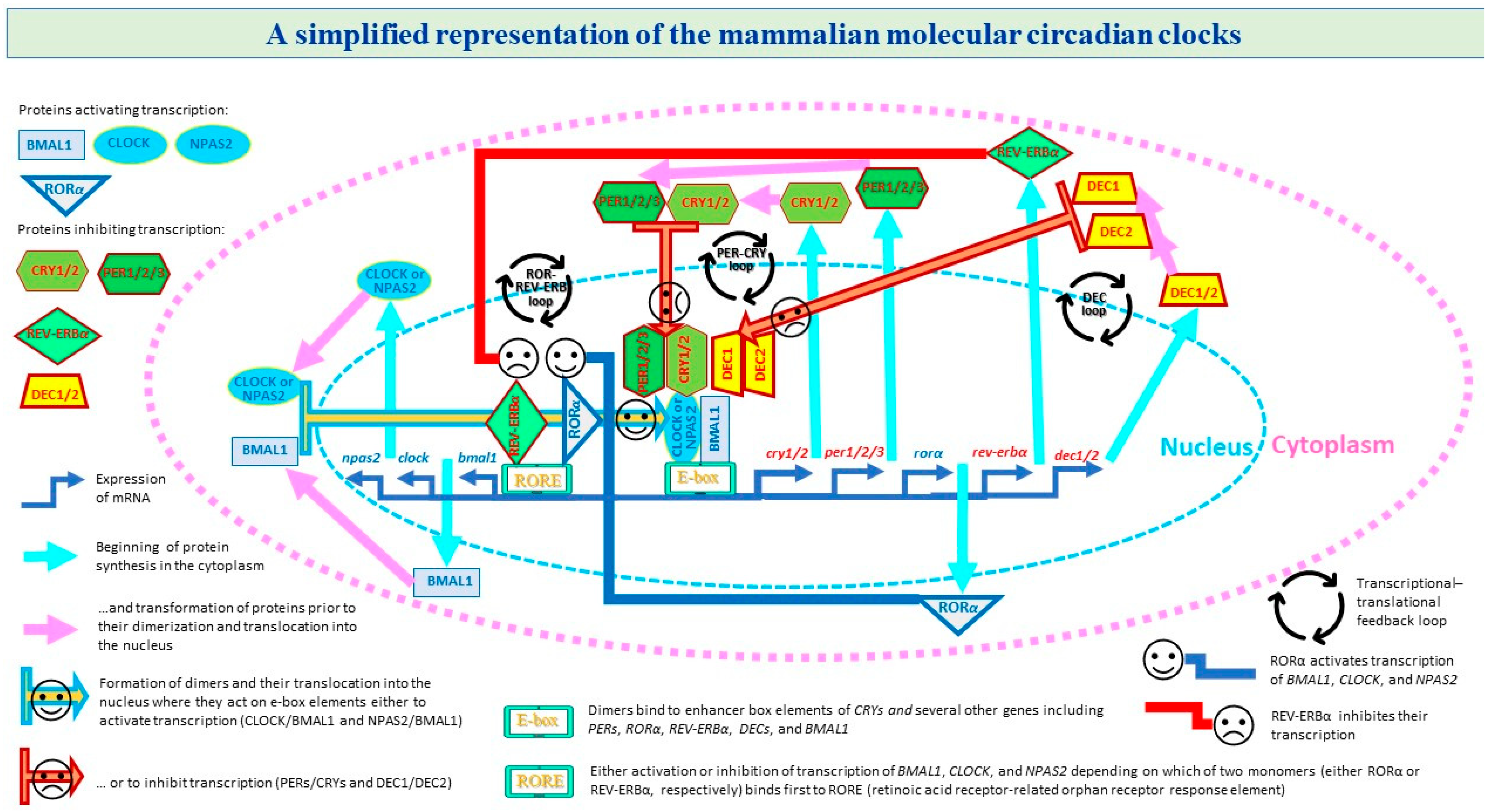
| Feedback Loop | Name | Circadian Clock Transcript | ||||
|---|---|---|---|---|---|---|
| Gene | Protein | Upregulated | Downregulated | Nonsignificant | No Data | |
| Monomers or dimers are promoters | ||||||
| All three | BMAL1 or ARNTL | BMAL1 or ARNTL | FTC and PTC [1] PTC [3] PTC [6] TC [7] MTC [9] | [2,4,5,8] | ||
| All three | CLOCK | CLOCK | TC [7] MTC [9] | PTC [3] | [1,2,4,5,6,8] | |
| All three | NPAS2 | NPAS2 | ATC [8] | [1,2,3,4,6,7,9] | ||
| ROR-REV-ERB | RORα | Nuclear receptor ROR-alpha | PTC [2] | PTC [3] | [1,4,5,6,7,8,9] | |
| Monomers or dimers are suppressors | ||||||
| ROR-REV-ERB | REV-ERBα or NR1D1 | REV-ERBα | PTC [2] | FTC, PTC [1] PTC [3] | [4,5,6,7,8,9] | |
| PER-CRY | CRY1 | CRY1 | FTC, PTC [1] PTC [3] TC [7] | [2,4,5,6,8,9] | ||
| PER-CRY | CRY2 | CRY2 | FTC and PTC [1] PDTC [4] TC [7] | PTC [3] | [2,5,6,8,9] | |
| PER-CRY | PER1 | PER1 | FTC, PTC [1] PTC [3] TC [7] | [2,4,5,6,8,9] | ||
| PER-CRY | PER2 | PER2 | TC [7] | FTC and PDTC [4] | FTC, PTC [1] PTC [3] | [2,5,6,8,9] |
| PER-CRY | PER3 | PER3 | FTC, PTC [1] PTC [3] | [2,4,5,6,7,8,9] | ||
| DEC | DEC1 | DEC1 | PTC [5] | [1,2,3,4,6,7,8,9] | ||
| DEC | DEC2 | DEC2 | PTC [5] | [1,2,3,4,6,7,8,9] | ||
| Name | Circadian Rhythmicity of Transcript | Compared to | ||
|---|---|---|---|---|
| Gene | Protein | Altered Phase | Altered Amplitude | Other TC Type |
| BMAL1 or ARNTL | BMAL1 or ARNTL | PDTC | PTC or HB | |
| REV-ERBα or NR1D1 | REV-ERBα | PDTC | PTC or HB | |
| CRY1 | CRY1 | PDTC | PTC or HB | |
| CRY2 | CRY2 | PDTC | PTC or HB | |
| PER1 | PER1 | PDTC | PTC or HB | |
| PER2 | PER2 | PDTC | PTC or HB | |
| PER3 | PER3 | PDTC | PTC or HB | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Putilov, A.A.; Budkevich, E.V.; Budkevich, R.O. A Review of Evidence for the Involvement of the Circadian Clock Genes into Malignant Transformation of Thyroid Tissue. Clocks & Sleep 2023, 5, 384-398. https://doi.org/10.3390/clockssleep5030029
Putilov AA, Budkevich EV, Budkevich RO. A Review of Evidence for the Involvement of the Circadian Clock Genes into Malignant Transformation of Thyroid Tissue. Clocks & Sleep. 2023; 5(3):384-398. https://doi.org/10.3390/clockssleep5030029
Chicago/Turabian StylePutilov, Arcady A., Elena V. Budkevich, and Roman O. Budkevich. 2023. "A Review of Evidence for the Involvement of the Circadian Clock Genes into Malignant Transformation of Thyroid Tissue" Clocks & Sleep 5, no. 3: 384-398. https://doi.org/10.3390/clockssleep5030029
APA StylePutilov, A. A., Budkevich, E. V., & Budkevich, R. O. (2023). A Review of Evidence for the Involvement of the Circadian Clock Genes into Malignant Transformation of Thyroid Tissue. Clocks & Sleep, 5(3), 384-398. https://doi.org/10.3390/clockssleep5030029







