2.1. Model Selection Estimators in Low Dimensions
This subsection explains why “sensible model selection estimators, including variable selection estimators,” produce fitted values (predictions) similar to that of the full OLS model when
n is much larger than
p. The result in Equation (
4) that the residuals from the model selection model and the full OLS model are highly correlated was a property of OLS and Mallow’s
criterion, not of any underlying model, but linearity forces the fitted values to be highly correlated. Hence the result works if OLS is consistent and the population model is linear, so for weighted least squares, AR(p) time series, serially correlated errors, et cetera. In particular, the cases do not need to be iid from some distribution. Since the correlation gets arbitrarily close to 1, the model selection estimator and full OLS estimator are estimating the same population parameter
, but it is possible that the model selection estimator picks the full OLS model with probability going to one.
Consider the OLS regression of
Y on a constant and
where, for example,
,
, or
. Let
I index the variables in the model so
means that
was selected. The full model
uses all
p predictors and the constant with
. Let
r be the residuals from the full OLS model and let
be the residuals from model
I that uses
. Suppose model
I uses
k predictors including a constant with
. Ref. [
5] proved that the model
I with
k predictors that minimizes [
6]
maximizes cor(
, that
and under linearity, cor(
forces
Thus
implies that
Let the model
minimize the
criterion among the models considered with
. Then
, and if PLS or PCR is selected using model selection (on models
with
corresponding to the
j-component regression) with the
criterion, and
, then cor(
Hence the correlation of ESP(I) and ESP(F) will typically also be high. (For PCR, the following variant should work better, as follows: take
and
as the
with the highest squared correlation with
Y,
as the
with the second highest squared correlation, etc.).
Machine learning methods for the multiple linear regression model can be incorporated as follows. Let k be the number of predictors selected by lasso, including a constant. Standardize the predictors to have unit sample variance and run the method. Let model I contain the variables corresponding to the predictors variables that have the largest . Fit the OLS model I to these predictors and a constant. If , use model I; otherwise, use the full OLS model. Many variants are possible. In low dimensions, comparisons between methods like lasso, PCR, PLS, and envelopes might use prediction intervals, the amount of dimension reduction, and standard errors if available.
If the above procedure is used, then model selection estimators, such as
, produce predictions that are similar to those of the OLS full model if
. Other model selection criterion, such as
k-fold cross validation, tend to behave like
in low dimensions, but getting bounds like Equation (
4) may be difficult. Empirically, variable selection estimators and model selection estimators often do not select the full model. Equation (
4) suggests that “weak” predictors will often be omitted, as long as cor(
stays high. (If the predictors are not orthogonal, “weak” might mean the predictor is not very useful given that the other predictors are in the model).
It is common in the model selection literature to assume, for the full model, that there is a model
S such that
for
, and
for
. Then model
I underfits unless
. If
, then an “important” predictor has been left out of the model. Then under the model
,
will not converge to 1 as
, and for large enough
n,
. Thus
as
. Hence
as
where
corresponds to the set of predictors selected by a variable selection method such as forward selection or lasso variable selection. Thus the probability that the model selection estimator underfits goes to zero as
if
p is fixed, the full model is one of the models considered, and the
criterion is used, as noted by [
7].
For real data, an important question in variable selection is whether
is a reasonable assumption. If
X has full rank
, then having
equal to zero for 20 decimal places may not be reasonable. See, for example, [
8,
9,
10]. Then the probability that the variable selection estimator chooses the full model goes to one if the probability of underfitting goes to 0 as
. If
is
, use zero padding to form the
vector
from
by adding 0s corresponding to the omitted variables.
For example, if
and
, then
and
This population model has
active predictors. Then the
possible subsets of
are
,
,
,
,
,
,
, and
. There are
subsets
and
such that
. Let
and
If
, then the observed variable selection estimator
As a statistic,
with probabilities
for
where there are
J subsets, e.g.,
. Theory for the variable selection estimator
is complicated. See [
7] for models such as multiple linear regression, GLMs, and [
11] Cox proportional hazards regression.
2.2. Sparse Fitted Models
A fitted or population regression model is sparse if a of the predictors are active (have nonzero or ) where with . Otherwise, the model is nonsparse. A high-dimensional population full regression model is abundant or dense if the regression information is spread out among the p predictors (nearly all of the predictors are active). Hence an abundant model is a nonsparse model. Under the above definitions, most classical low-dimensional models use sparse fitted models, and statisticians have over one hundred years of experience with such models.
The literature for high-dimensional sparse regression models often assumes that (i)
, that (ii)
where
I uses
k predictors including a constant, and that (iii)
. When these assumptions hold, the population model is sparse, the fitted model is sparse, and Equation (
3) becomes
, which can be small. Getting rid of assumption (i) and the assumption that
greatly increases the applicability of variable selection estimators, such as forward selection, lasso, and the elastic net, for high-dimensional data, even if
is huge. As argued in the following paragraphs, the sparse fitted model often fits the data well, and often
is a good estimator of
.
A sparse fitted model transforms a high-dimensional problem into a low-dimensional problem, and the sparse fitted model can be checked with the goodness of fit diagnostics available for that low-dimensional model. If the predictors used by the sparse fitted regression model are , and if the regression model depends on only through the sufficient predictor , then a useful diagnostic is the response plot of versus the response Y on the vertical axis. If there is goodness of fit, then tends to estimate regardless of whether the population model is sparse or nonsparse. Data splitting may be needed for valid inference such as hypothesis testing.
Suppose the cases are iid for . Then are iid, resulting in a valid sparse fitted model regardless of whether the population model is sparse or nonsparse. This null model omits all of the predictors. For high-dimensional data, a reasonable goal is to find a model that greatly outperforms the null model.
The sparse fitted model using
is often useful when there are one or more strong predictors. The following [
12] theorem gives two more situations where a sparse fitted model can greatly outperform the null model. The population models in Theorem 1 can be sparse or nonsparse. The high-dimensional multiple linear regression literature often assumes that the cases are iid from a multivariate normal distribution, and that the population model is sparse. Let
. For multiple linear regression, note that
unless
.
Theorem 1. Suppose the cases are iid from some distribution.
(a) If the joint distribution of is multivariate normal,then follows a multiple linear regression model, but so does , where , , and (b) If the response Y is binary, then binomial( where . Hence every linear combination of the predictors satisfies a binary regression model.
2.3. PCA-PLS
Another technique is to use PCA for dimension reduction. Let
be the PCA linear combinations (
) ordered with respect to the largest eigenvalues. Then use
in the regression or classification model where
k is chosen in some manner. This method can be used for models with
m response variables
. See, for example, [
13,
14,
15,
16].
Consider a low or high-dimensional regression or classification method with a univariate response variable
Y. Let
be the linear combinations ordered with respect to the highest squared correlations
where the sample correlation
. From a model selection viewpoint, using
should work much better than using
. Also, the PLS components
should be used instead of the PCA
, since the PLS components are chosen to be fairly highly correlated with
Y. See Equation (2). Ref. [
17] (pp. 71–72) shows that an equivalent way to compute the
k-component PLS estimator is to maximize
under some constraints. If the predictors are standardized to have unit sample variance, then this method becomes a correlation vector optimization problem. Ref. [
18] use the PLS components as predictors for nonlinear regression, but the above model selection viewpoint is new.
From canonical correlation analysis (CCA), if
are iid, then
This optimization problem is equivalent to maximizing
which has a maximum at
See [
19] (pp. 168, 282). Hence PLS is a lot like CCA for
but with more constraints, and PLS can be computed in high dimensions. From the dimension reduction literature, if
Y depends on
x only through
, then under the assumption of “linearly related predictors,”
estimates
for some constant
c which is often nonzero. See, for example, [
20] (p. 432).
The above results suggest computing lasso for multiple linear regression, find the number of predictors
k chosen by lasso, and take
k linear combinations
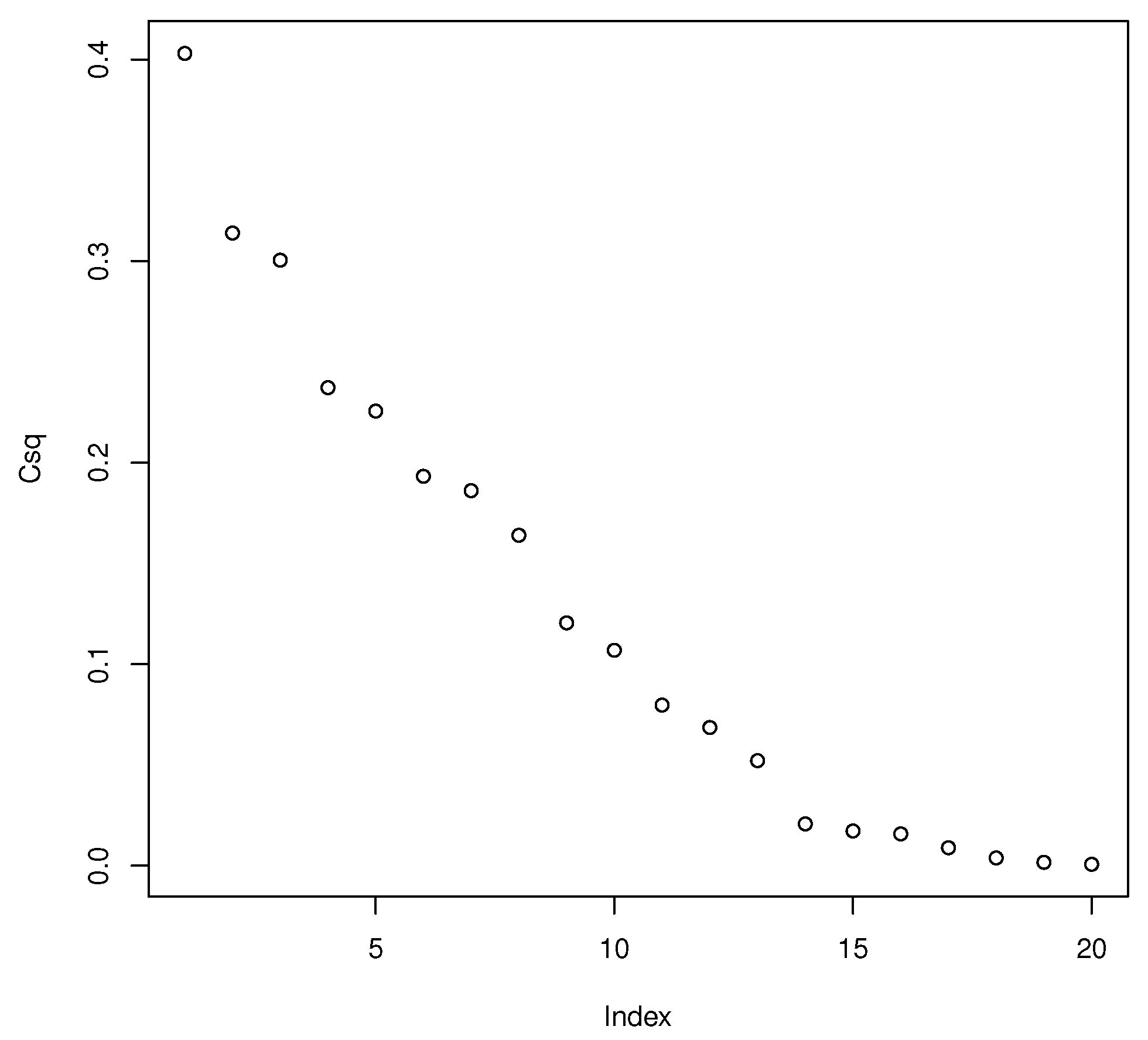
. An SC scree plot of
i versus
behaves like a scree plot of
i versus the eigenvalues. Hence quantities like
are of interest for
, and scree plot techniques could be adapted to choose
k. Many other possibilities exist, and there are many possibilities for models with
m response variables
. See [
18] for some ideas.
Another useful technique is to eliminate weak predictors before finding
. By Equation (
3),
may not be close to
in high dimensions, e.g.,
. For example, the sample eigenvectors
tend to be poor estimators of the population eigenvectors
of
. An exception is when the correlation Cor
for
where
is close to 1. See [
21]. One possibility is to take the
j predictors
with the highest squared correlations with
Y. The SC scree plot is useful. Then do lasso (meant for the multiple linear regression model) to further reduce the number of
. Here
j should be proportional to
n, for example,
, where
is an interesting choice. When
n is small, spurious correlations can be a problem as follows: if the actual correlation is near 0, the sample size
n may need to be large before the sample correlation
is near 0. For more on the importance of eliminating weak predictors and high-dimensional variable selection, see, for example, [
22,
23,
24,
25,
26].
2.4. Stack Low-Dimensional Estimators into a Vector
Another technique is to stack low-dimensional estimators into a vector. For example , and elements from an estimated covariance matrix such as . Using can give information about a multivariate regression. Let and denote the model that uses , and let I denote the model that uses . Then some of these estimators satisfy and , where the estimator is easy to compute in high dimensions but is singular if , while the estimator is easy to compute in high dimensions and is nonsingular if with . Often, the theory for F uses z while the theory for I uses . (Values of J much larger than 10 may be needed if some of the are skewed).
Then the following simple testing method reduces a possibly high-dimensional problem to a low-dimensional problem. Consider testing versus where A is a constant matrix and the hypothesis test is equivalent to testing versus where B is a constant matrix. For example, tests such as or are often of interest.
The marginal maximum likelihood estimator (MMLE) and one component partial least squares (OPLS) estimators stack low-dimensional estimators into a vector. In low dimensions, the OLS estimators are
and
For a multiple linear regression model with iid cases,
is a consistent estimator of
under mild regularity conditions, while
is a consistent estimator of
.
Refs. [
27,
28] showed that
estimates
where
and
for
. If
, then
. Let
Testing
versus
is equivalent to testing
versus
where
A is a
constant matrix and
.
The marginal maximum likelihood estimator (marginal least squares estimator) is due to [
22,
23]. This estimator computes the marginal regression of
Y on
resulting in the estimator
for
. Then
For multiple linear regression, the marginal estimators are the simple linear regression (SLR) estimators, and
Hence
If the
are the predictors standardized to have unit sample variances, then
where
denotes that
Y was regressed on
w, and
I is the
identity matrix. Hence the SC scree plot is closely related to the MMLE for multiple linear regression with standardized predictors.
Consider a subset of
k distinct elements from
. Stack the elements into a vector, and let each vector have the same ordering. For example, the largest subset of distinct elements corresponds to
For random variables
, use notation such as
the sample mean of
,
, and
. Let
For general vectors of elements, the ordering of the vectors will all be the same and be denoted vectors such as
and
.
Ref. [
29] proved that
if
is a
vector. The theorem may be a special case of the [
30] theory for the multivariate linear regression estimator when there are no predictors. Ref. [
31] also gave similar large sample theory, for example, for
, but the proof in [
29] and the estimator
are much simpler. Also see [
32].
The following [
12] large sample theory for
is also a special case. Let
and let
and
be defined below where
Then the low-order moments are needed for
to be a consistent estimator of
.
Theorem 2. Assume the cases are iid. Assume exist for and Let and . Let with sample mean . Let . Then (a) (b) Let and Then .
(c) Let A be a full rank constant matrix with , assume is true, and assume . Then This method of hypothesis testing does not depend on whether the population model is sparse or abundant, and it does need data splitting for valid inference. Data splitting with sparse fitted models can also be used for high-dimensional hypothesis testing. See, for example, [
12]. Ref. [
29] also provides the theory for the OPLS estimator and MMLE for multiple linear regression where heterogeneity is possible and where the predictors may have been standardized to have unit sample variances.
The MMLE for multiple linear regression is often used for variable selection if the predictors have been standardized to have unit sample variances. Then the
k predictors with the largest
correspond to the
k predictors with the largest squared correlations with
Y. Hence this method can be used to select
k in
Section 2.4.
2.5. Alternative Dispersion Estimators
Let be a symmetric positive semi-definite matrix such as , or . When is singular or ill conditioned, some common techniques are to replace with a symmetric positive definite matrix such as , where the constant , or . Regularized estimators are also used.
For
, a simple way to regularize a
correlation matrix
is to use
where
and
for
. Note that each correlation
is divided by the same factor
. If
is the
ith eigenvalue of
R, then
is the
ith eigenvalue of
. The eigenvectors of
R and
are the same since if
, then
Note that
, where
. See [
33,
34].
Following [
35], the condition number of a symmetric positive definite
matrix
A is
, where
are the eigenvalues of
A. Note that
A well conditioned matrix has condition number
for some number
c such as 50 or 500. Hence
is nonsingular for
and well conditioned if
or
if
. Taking
suggests using
The matrix can be further regularized by setting
if
, where
should be less than 0.5. Denote the resulting matrix by
. We suggest using
. Note that
. Using
is known as
thresholding. We recommend computing
and
for
50, 100, 200, 300, 400, and 500. Compute
R if it is nonsingular. Note that a regularized covariance matrix can be found using
where
and
.
A common type of regularization of a covariance matrix S is to use where the th element of and . The corresponding correlation matrix is the identity matrix, and Mahalanobis distances using the identity matrix correspond to Euclidean distances. These estimators tend to use too much regularization, and underfit. Note that as , , and has . Note that corresponds to using in Equation (9).
For the population correlation matrix and the population precision matrix , the literature often claims that most of the population correlations , so that the population matrix is sparse, and that is a good estimator of the population matrix. Assume that estimates a population dispersion matrix D. Note that this assumption always holds when . Note that estimates since estimates where for . However, by Equation (3), the estimator tends to not be good in high dimensions.
Consider testing
versus
, where a
statistic
satisfies
If
and
is true, then
as
. Then a Wald-type test rejects
if
where
if
, a chi-quare distribution with
g degrees of freedom. Note that
is a squared Mahalanobis distance.
It is common to implement a Wald-type test using
as
if
is true, where the
symmetric positive definite matrix
. Hence
is the wrong dispersion matrix, and
does not have a
distribution when
is true. Ref. [
36] showed how to bootstrap Wald tests with the wrong dispersion matrix. When
, the bootstrap tests often became conservative as
g increased to
n. For some of these tests, the
m out of
n bootstrap, which draws a sample of size
m without replacement from the
n, works better than the nonparametric bootstrap. Sampling without replacement is also known as subsampling and the delete
d jackknife. For some methods, better high-dimensional tests are reviewed by [
37].
Using a high-dimensional dispersion estimator with considerable outlier resistance is another useful technique. Let W be a data matrix, where the rows correspond to the cases. For example, or . One of the simplest outlier detection methods uses the Euclidean distances of the from the coordinatewise median Concentration type steps compute the weighted median : the coordinatewise median computed from the “half set” of cases with where . We often use (no concentration type steps) or . Let . Let if where and is the default choice. Let , otherwise. Using ensures that at least half of the cases get weight 1. This weighting corresponds to the weighting that would be used in a one sided metrically trimmed mean (Huber-type skipped mean) of the distances. Here, the sample median absolute deviation is , where is the sample median of .
Let the
covmb2 set B of at least
cases correspond to the cases with weight
. Then the [
38] (p. 120)
covmb2 estimator
is the sample mean and sample covariance matrix applied to the cases in set
B. If
, then
This estimator was built for speed, applications, and outlier resistance. In low dimensions, the population dispersion matrix is the population covariance matrix of a spherically truncated distribution. In high dimensions, spherical truncation is still used, but the sample weighted median varies about the population weighted median by Equation (3).
A useful application is to apply high- (and low)-dimensional methods to the cases that get weight 1. If the ith case where , then this application can be used if all of the variables are continuous. For a variant, let the continuous predictors from be denoted by for . Apply the covmb2 estimator to the , and then run the method on the m cases corresponding to the covmb2 set B indices , where . If the estimator has large sample theory “conditional” on the predictors x, then typically the same theory applies for the “robust estimator” since the response variables were not used to select the cases in B. These two applications can be used for regression, classification, neural networks, et cetera.
Another method to get an outlier-resistant estimator
is to use the following identity. If
X and
Y are random variables, then
Then replace Var(
by
, where
is a robust estimator of scale or standard deviation and
or
. We used
where
Hence
In low dimensions, the [
38] Olive (2017) RMVN or RFCH estimator of dispersion can be used.