Abstract
Preterm infants are prone to NeuroDevelopmental Impairment (NDI). Some previous works have identified clinical variables that can be potential predictors of NDI. However, machine learning (ML)-based models still present low predictive capabilities when addressing this problem. This work attempts to evaluate the application of ML techniques to predict NDI using clinical data from a cohort of very preterm infants recruited at birth and assessed at 2 years of age. Six different classification models were assessed, using all features, clinician-selected features, and mutual information feature selection. The best results were obtained by ML models trained using mutual information-selected features and employing oversampling, for cognitive and motor impairment prediction, while for language impairment prediction the best setting was clinician-selected features. Although the performance indicators in this local cohort are consistent with similar previous works and still rather poor. This is a clear indication that, in order to obtain better performance rates, further analysis and methods should be considered, and other types of data should be taken into account together with the clinical variables.
1. Introduction
Very preterm infants are at high risk of developing neurodevelopmental impairments (NDI) that include a range of language, cognitive, sensory, and motor impairments [1]. Those born at or before 32 weeks of gestational age are at a higher risk of NDI. Research studies have identified clinical conditions that have been associated with adverse outcomes, such as prenatal, perinatal, and comorbidities factors. Prenatal risk factors include infections [2], hypertension [3], and malnutrition [4]. Perinatal risk factors include gestational age [5], sex [6], and Apgar score [7]. Moreover, comorbidities also may impact the long-term neurodevelopmental outcome, such as necrotizing enterocolitis [8], sepsis [9], bronchopulmonary dysplasia [10] among others. These risk factors can be prospective predictors of NDI, and provide a better understanding of the potential pathways to adverse outcomes in preterm infants.
Previous models using traditional statistical methods have been developed to predict NDI in preterm infants using antenatal and neonatal clinical data [11,12,13]. These studies have demonstrated the significance of predictive models that can assist neonatologists in early diagnosis and decision-making. Despite these efforts, drawbacks have been considered such as the lack of diversity in the variables, treating the variables as independent, and measuring a risk combination of NDI and/or death, among others. Nowadays, owing to the exponential increase in data, advantages can be obtained by applying machine learning (ML) models that can help to support these statistical models. Previous studies have used clinical features as predictors for NDI in preterm infants and extremely low birth weight infants using ML models. Research performed by Ambalavanan et al. [14] used antenatal and perinatal variables to predict neurodevelopmental outcomes at 18 months, using Neural Networks. Some of the limitations of this model were the small sample size and the selection of variables, as they only used variables selected with reported risk.
Furthermore, Ambalavanan et al. [15], used antenatal and postnatal data to predict NDI using classification tree models, among the drawbacks they consider is that the model predicted a combined outcome of NDI or death. Moreover, some of the classification tree nodes were based on very small subsets (e.g., specific treatment), from the original dataset and were less accurate than the ones based on larger subsets. Another study performed by Juul et al. [16], aimed to predict NDI outcomes at 2 years by using Bayesian Additive Regression Trees using three subsets of selected variables. This research provided meaningful conclusions such as using a dichotomous version of the NDI outcome performed better than its original version and that total transfusion volume was the most important predictor for their cohort.
Despite the efforts, to the best of our knowledge there is still no ML-based solution that performs well in this task in small cohorts and highly imbalanced datasets by using clinical data from very preterm infants. This study aims to evaluate the application of ML techniques to predict NDI in very preterm infants using clinical data from a local cohort acquired at the Hospital Puerta del Mar, in Cádiz, Spain, which includes prenatal, perinatal, and comorbidities records. The authors’ main goal is to determine which is the best set of ML techniques, together with the set of clinical variables, that better predicts the NDI outcomes in very preterm infants. The main contributions of this work are next summarized:
- Develop supervised ML models to predict NDI in very preterm infants.
- Analyze the performance of these ML models when using all clinical features available, a subset of them guided by experts in this field, and mutual information-selected features.
- Apply a commonly used data augmentation technique to deal with data scarcity and class imbalance issues.
The rest of the paper is organized as follows. Section 2 describes the study design and data collection. Section 3 explains feature selection methods and ML classifiers implemented in this research. Section 4 describes strategies for the generation of synthetic data, and evaluation metrics that have been considered. Section 5 details and discusses results that were obtained with the experimental design and the chosen methods. Finally, Section 6 gives an overview of the conclusions of this study and the approaches that need to be considered for future work.
2. Dataset
2.1. Study Design and Participants
Data was prospectively collected in a cohort study including very preterm infants from May 2018 to January 2021 at Puerta del Mar University Hospital, Cádiz, Spain. Research and Ethics Committee approval and informed consent of participants were obtained. Inclusion criteria were very preterm infants born at ≤32 weeks and/or very-low-birth-weight infants (≤1500 g).
A total of 52 clinical features were used in this study which include prenatal, perinatal, and comorbidities features. Prenatal variables refer to the mother’s health records at the time of pregnancy such as age, hypertension, hypothyroidism, chorioamnionitis, gestational diabetes, preeclampsia, cesarean delivery, IV fertilization, etc. Perinatal variables refer to clinical records of the preterm infant during delivery, such as gestational age, sex, Apgar score at 1 and 5 min of life, Clinical Risk Index for Babies (CRIB Index), intubation at the delivery room, head circumference at birth, small for gestational age (birth weight bellow 10th centile), etc. Comorbidities include patent ductus arteriosus, bronchopulmonary dysplasia, days of oxygen therapy, and mechanical ventilation, among others.
2.2. Neurodevelopmental Assessments at 2 Years Corrected Age
In this study, we analyzed a dataset of 180 very preterm infants, each assessed for neurodevelopmental outcomes at 2 years of corrected age. The corrected age is the chronological age reduced by the number of weeks born before 40 weeks of gestation. Assessments were conducted using the Bayley Scales of Infant Development, 3rd Edition (Bayley III) [17]. This test evaluates three different areas of neurodevelopment: motor, cognitive, and language. The scores are independent for each area and evaluation is performed by a qualified clinical psychologist. The Bayley score is a quantitative variable, that is categorized in the following threshold: ≥85 normal neurodevelopment, <85 mild impairment, and <70 severe impairment.
To address the imbalance in our dataset, we simplified the problem to binary classification: values ≥ 85 were considered normal neurodevelopment and values < 85 as mild to severe impairment. For cognitive impairment, 155 patients had a normal neurodevelopmental outcome, while 25 had mild to severe impairment. Meanwhile, for motor impairment, 156 patients obtained a normal neurodevelopmental outcome, while 24 patients had a mild to severe impairment. For language impairment, 139 patients had a normal neurodevelopmental outcome, while 41 obtained mild to severe impairment.
2.3. Data Curation and Pre-Processing
From the 52 features considered in this study, 32 features had less than 6% of missing values, while 6 features had around 30% to 40% of missing values. Features with missing values were imputed using two simple methods according to the type of feature. Imputation approaches used in this study, are commonly simple, and fast methods to impute; however, considering the complexity of clinical features, suggestions from the clinicians were considered to perform these imputation methods.
- Numerical features: imputation was performed using the mean value, where missing values were replaced with the mean of all known values in each feature. Moreover, normalization was applied, where each feature was scaled to a range of 0–1. Scaling was adjusted on the training dataset, whereas the test set was normalized based on the training data.
- Categorical features: the most frequent value imputation was performed, where missing values were substituted with the most frequent value in each feature. Subsequently, these features were transformed using dummy encoding, where each feature was converted into k classes, yielding a total of k − 1 [18].
3. Methodology
3.1. Feature Selection
Clinician-based feature selection was performed by clinicians, based on their expertise in following preterm infants across the years, this feature selection ended up reducing the number of 22 features.
Additionally, the mutual information method was employed as an alternative feature selection approach. This method calculates the mutual information between each feature and the target variable [19]. To determine the optimal number of features, an additional variable with random values was generated and analyzed together with the rest of the variables through this feature selection method. Then, only features with a mutual information coefficient greater than the one corresponding to this random variable are retained.
3.2. Classifiers
We evaluated different classification models, which are listed and defined below.
- Logistic regression: a type of regression model used in binary classification problems. It models the dependent variable as a linear combination of the independent variables and employs the logistic function to transform these linear combinations into a probability value between 0 and 1 [20].
- AdaBoost: a meta-estimator that starts by fitting a classifier on the initial dataset and then fits multiple copies of the classifier on the same dataset, but modifies the weights of instances that are mistakenly categorized so that succeeding classifiers focus more on difficult cases [21].
- Decision Trees: a model that creates predictions of the target variable by learning simple choice rules based on the characteristics of the data components. The root node serves as the base of the model and is continued by the leaf and intermediate nodes [20].
- Random Forest: a tree-based ensemble model that combines tree predictors, where each tree in a random forest depends on the values of a random vector that was sampled randomly and along the same distribution for all trees in the forest [22].
- Gaussian Naive Bayes: a model based on the probabilistic method and Gaussian distribution. The classification process considers the likelihood that each feature is Gaussian and that each feature can independently predict the target feature [23].
- K-nearest Neighbor: a model that classifies based on the similarity of the data, where k is the number of closest neighbors. In this case, the technique to find the K-nearest Neighbor is performed using Euclidean distance [20].
4. Experimental Design
4.1. Evaluation Strategy
The models were evaluated using leave-one-out cross-validation to ensure robustness and minimize bias. Additionally, hyperparameter tuning was conducted using grid search combined with 5-fold cross-validation, enabling the identification of optimal parameters in each iteration. The entire process, including data pre-processing and model evaluation, was implemented in Python 3.11.6, and models from the scikit-learn 1.4 package were imported. The subsequent performance metrics were computed as follows, where: TN: True negatives; TP: True positives; FN: False negatives; FP: False positives.
- Accuracy: this metric is calculated by dividing the total number of correct predictions by the total number of observations.
- Recall: this metric is calculated by the fraction of all the positive cases that were correctly classified.
- ROC AUC: this metric indicates how well the model can distinguish between the positive and negative samples. When AUC = 1, the model perfectly distinguished the positive and negative classes.
4.2. Dealing with Data Imbalance Issues
The oversampling method SMOTE-NC (Synthetic Minority Over-sampling Method for Nominal and Continuous) [24] was used because of the class imbalance of the dataset. This method is a variation of SMOTE and it creates additional synthetic data points for the minority class by interpolating between existing samples. SMOTE-NC randomly chooses an example from the minority class (mild/severe class), identifies its k-nearest neighbors, chooses one of them, and linearly interpolates between the chosen example and the neighbor to produce a new example.
5. Results and Discussion
In this study, we worked with a small and class-imbalanced dataset (approximately 80% vs. 20%) and evaluated the performance of different models. These results indicate that performing an oversampling method such as SMOTE-NC yields better results in cognitive and motor impairments for all features Table A1 and Table A2, clinician-selected features Table A3 and Table A4, and mutual information-selected features Table A5 and Table A6. Given our aim to predict the positive class (mild/severe impairment), our primary focus was on models that achieved better results for the recall, as shown in Figure 1. We are particularly interested in a model that can accurately identify the true positives, meaning that when the model indicates a patient has an impairment, this prediction should be correct most of the time.
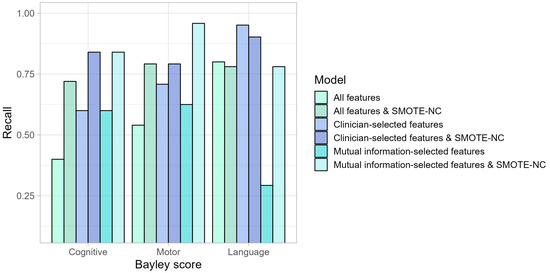
Figure 1.
Model performance based on recall metric. Mutual information-selected features and SMOTE-NC obtained the highest recall for motor and cognitive impairment prediction. Followed by clinical-selected features in language impairment prediction.
Moreover, according to the best model performance (Gaussian Naives) in motor impairment prediction, we present the ROC curve in Figure 2, which achieved an AUC value of 0.69.
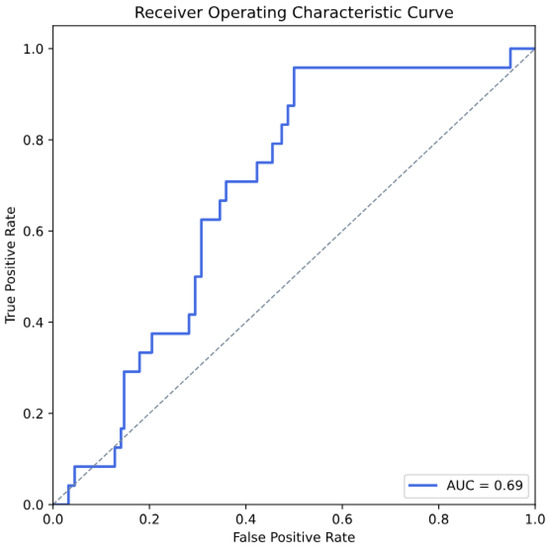
Figure 2.
Receiver Operating Characteristic (ROC) curve demonstrating the performance of the Gaussian Naive Bayes model for predicting motor impairment.
Clinician-selected features performed better based on the recall metric compared with all features for the three types of impairment prediction. However, performing mutual information-selected features and SMOTE-NC outperformed the other settings in cognitive and motor impairment prediction. In contrast, for language impairment prediction, the best setting was clinician-selected features. In this sense, the best model performance according to recall was Gaussian Naive Bayes for all types of neurodevelopmental impairment predictions. Moreover, implementing SMOTE-NC increased its prediction of cognitive and motor impairments across all settings compared to no oversampling settings. Meanwhile, in language impairment prediction, applying SMOTE-NC obtained notable results in the setting of mutual information-selected features. Any feature selection employed in this work was inconsistent for all three cases, indicating reevaluation, because recent studies have proposed more sophisticated and smarter approaches for addressing this task [16].
Furthermore, it is important to mention that even though prediction for the mild/severe class was the goal of this analysis, joining both classes due to the lack of patients might imply a loss of clinical information regarding the state and future of each patient; therefore, future analysis should be considered to work with these classes independently. Moreover, it has been previously stated by Juul et al. [16] that even by applying advanced methods the field is still not able to predict complex long-term outcomes such as the Bayley score. However, in this study, we will be approaching extra features that previously have been considered that are predictors for NDI such as socioeconomic features [25] and image-based features such as brain volumes [26,27]. Moreover, multimodal data integration does seem a greater alternative for predicting NDI outcomes by the use of machine learning methods, research on this scope has been focused on integrating clinical data and image-based data to predict NDI in preterm infants [28,29].
6. Conclusions and Future Work
In this work we assessed a small and class-imbalanced dataset to predict the neurodevelopmental impairments at two years in very preterm infants, using six different classification models, employing all features and feature selection by an expert and by mutual information and performing SMOTE-NC oversampling. Besides, this evaluation was performed using clinical data from a local cohort acquired in a Spanish public hospital. Results are indicative that using mutual information-selected features and SMOTE-NC techniques was the best setting for cognitive and motor impairment prediction, while for language impairment prediction the best setting was clinician-selected features. The feature selection performed by experts in this field should be reconsidered for further studies, implementing a more complex approach as recent studies have suggested.
To this end, this work could be extended by applying further methods, adding more heterogeneous features such as image-based features, and centering the prediction into regression methods so that the Bayley score can be predicted in its original version. Moreover, it is important to note that this is a small dataset, and it is expected to include more patients and validate the model on external cohorts.
Author Contributions
Conceptualization, A.O.-L., D.U. and I.J.T.; methodology, A.O.-L. and A.G.; software, A.O.-L.; validation, A.O.-L. and A.G.; formal analysis, A.O.-L., D.U. and I.J.T.; investigation, A.O.-L. and A.G.; resources, I.B.-F., A.S.-A., D.U. and I.J.T.; data curation, A.O.-L., I.B.-F. and A.S.-A.; writing—original draft preparation, A.O.-L. and A.G.; writing—review and editing, A.O.-L., D.U., I.J.T. and I.B.-F.; visualization, A.O.-L.; supervision, D.U., I.J.T. and I.B.-F.; project administration, I.B.-F., D.U. and I.J.T.; funding acquisition, D.U. and I.J.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the PARENT project that has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Innovative Training Network 2020. Grant Agreement Nº 956394.
Institutional Review Board Statement
Research and Ethics Committee approval was obtained from Hospital Universitario Puerta del Mar, Cádiz, Spain.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The used data in this study are not publicly available. However, access to the data can be granted upon a reasonable request.
Conflicts of Interest
Author Arnaud Gucciardi was employed by the company TOELT LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| NDI | NeuroDevelopmental Impairment |
| ML | Machine Learning |
Appendix A

Table A1.
Performances of models using all features and no oversampling.
Table A1.
Performances of models using all features and no oversampling.
| Model | Accuracy | Recall | ROC AUC |
|---|---|---|---|
| Cognitive | |||
| AdaBoost | 0.83 | 0.20 | 0.61 |
| Decision Tree | 0.81 | 0.24 | 0.65 |
| Gaussian NB | 0.66 | 0.40 | 0.42 |
| KNN | 0.89 | 0.28 | 0.65 |
| Logistic regression | 0.81 | 0.28 | 0.72 |
| RandomForest | 0.86 | 0.16 | 0.74 |
| Motor | |||
| AdaBoost | 0.84 | 0.08 | 0.50 |
| Decision Tree | 0.84 | 0.17 | 0.60 |
| Gaussian NB | 0.54 | 0.54 | 0.57 |
| KNN | 0.84 | 0.21 | 0.58 |
| Logistic regression | 0.80 | 0.25 | 0.65 |
| RandomForest | 0.86 | 0.08 | 0.69 |
| Language | |||
| AdaBoost | 0.76 | 0.32 | 0.66 |
| Decision Tree | 0.70 | 0.27 | 0.55 |
| Gaussian NB | 0.35 | 0.80 | 0.58 |
| KNN | 0.74 | 0.27 | 0.66 |
| Logistic regression | 0.71 | 0.37 | 0.72 |
| RandomForest | 0.77 | 0.20 | 0.67 |

Table A2.
Performances of models using all features and SMOTE-NC.
Table A2.
Performances of models using all features and SMOTE-NC.
| Model | Accuracy | Recall | ROC AUC |
|---|---|---|---|
| Cognitive | |||
| AdaBoost | 0.73 | 0.40 | 0.62 |
| Decision Tree | 0.74 | 0.52 | 0.69 |
| Gaussian NB | 0.41 | 0.72 | 0.63 |
| KNN | 0.72 | 0.64 | 0.69 |
| Logistic regression | 0.79 | 0.36 | 0.72 |
| RandomForest | 0.82 | 0.20 | 0.71 |
| Motor | |||
| AdaBoost | 0.80 | 0.29 | 0.63 |
| Decision Tree | 0.66 | 0.83 | 0.76 |
| Gaussian NB | 0.66 | 0.58 | 0.57 |
| KNN | 0.70 | 0.63 | 0.67 |
| Logistic regression | 0.73 | 0.29 | 0.72 |
| RandomForest | 0.83 | 0.17 | 0.76 |
| Language | |||
| AdaBoost | 0.64 | 0.71 | 0.66 |
| Decision Tree | 0.60 | 0.46 | 0.57 |
| Gaussian NB | 0.37 | 0.78 | 0.74 |
| KNN | 0.63 | 0.66 | 0.57 |
| Logistic regression | 0.72 | 0.49 | 0.74 |
| RandomForest | 0.73 | 0.32 | 0.64 |

Table A3.
Performances of models using clinician-selected features and no oversampling.
Table A3.
Performances of models using clinician-selected features and no oversampling.
| Model | Accuracy | Recall | ROC AUC |
|---|---|---|---|
| Cognitive | |||
| AdaBoost | 0.81 | 0.24 | 0.51 |
| Decision Tree | 0.85 | 0.20 | 0.70 |
| Gaussian NB | 0.41 | 0.60 | 0.52 |
| KNN | 0.88 | 0.48 | 0.71 |
| Logistic regression | 0.83 | 0.32 | 0.73 |
| RandomForest | 0.87 | 0.28 | 0.82 |
| Motor | |||
| AdaBoost | 0.85 | 0.33 | 0.62 |
| Decision Tree | 0.85 | 0.13 | 0.63 |
| Gaussian NB | 0.44 | 0.71 | 0.63 |
| KNN | 0.84 | 0.33 | 0.63 |
| Logistic regression | 0.83 | 0.33 | 0.73 |
| RandomForest | 0.86 | 0.13 | 0.70 |
| Language | |||
| AdaBoost | 0.73 | 0.32 | 0.54 |
| Decision Tree | 0.74 | 0.17 | 0.62 |
| Gaussian NB | 0.28 | 0.95 | 0.57 |
| KNN | 0.75 | 0.41 | 0.63 |
| Logistic regression | 0.73 | 0.34 | 0.68 |
| RandomForest | 0.74 | 0.22 | 0.67 |

Table A4.
Performances of classification models using clinician-selected features and SMOTE-NC.
Table A4.
Performances of classification models using clinician-selected features and SMOTE-NC.
| Model | Accuracy | Recall | ROC AUC |
|---|---|---|---|
| Cognitive | |||
| AdaBoost | 0.53 | 0.48 | 0.53 |
| Decision Tree | 0.71 | 0.44 | 0.63 |
| Gaussian NB | 0.42 | 0.84 | 0.41 |
| KNN | 0.84 | 0.76 | 0.80 |
| Logistic regression | 0.76 | 0.60 | 0.75 |
| RandomForest | 0.89 | 0.44 | 0.84 |
| Motor | |||
| AdaBoost | 0.82 | 0.58 | 0.76 |
| Decision Tree | 0.73 | 0.58 | 0.72 |
| Gaussian NB | 0.57 | 0.67 | 0.53 |
| KNN | 0.76 | 0.79 | 0.81 |
| Logistic regression | 0.78 | 0.58 | 0.72 |
| RandomForest | 0.88 | 0.38 | 0.74 |
| Language | |||
| AdaBoost | 0.62 | 0.76 | 0.64 |
| Decision Tree | 0.64 | 0.68 | 0.64 |
| Gaussian NB | 0.29 | 0.90 | 0.43 |
| KNN | 0.72 | 0.63 | 0.72 |
| Logistic regression | 0.67 | 0.51 | 0.68 |
| RandomForest | 0.76 | 0.41 | 0.69 |

Table A5.
Performances of classification models using mutual information-selected features.
Table A5.
Performances of classification models using mutual information-selected features.
| Model | Accuracy | Recall | ROC AUC |
|---|---|---|---|
| Cognitive | |||
| AdaBoost | 0.84 | 0.24 | 0.55 |
| Decision Tree | 0.83 | 0.16 | 0.67 |
| Gaussian NB | 0.68 | 0.60 | 0.60 |
| KNN | 0.85 | 0.44 | 0.68 |
| Logistic regression | 0.81 | 0.36 | 0.61 |
| RandomForest | 0.87 | 0.16 | 0.76 |
| Motor | |||
| AdaBoost | 0.82 | 0.21 | 0.72 |
| Decision Tree | 0.83 | 0.13 | 0.62 |
| Gaussian NB | 0.64 | 0.63 | 0.60 |
| KNN | 0.81 | 0.21 | 0.57 |
| Logistic regression | 0.79 | 0.25 | 0.68 |
| RandomForest | 0.82 | 0.04 | 0.72 |
| Language | |||
| AdaBoost | 0.73 | 0.17 | 0.52 |
| Decision Tree | 0.72 | 0.07 | 0.52 |
| Gaussian NB | 0.71 | 0.29 | 0.62 |
| KNN | 0.71 | 0.15 | 0.52 |
| Logistic regression | 0.73 | 0.17 | 0.64 |
| RandomForest | 0.72 | 0.07 | 0.55 |

Table A6.
Performances of classification models using mutual information-selected features and SMOTE-NC.
Table A6.
Performances of classification models using mutual information-selected features and SMOTE-NC.
| Model | Accuracy | Recall | ROC AUC |
|---|---|---|---|
| Cognitive | |||
| AdaBoost | 0.58 | 0.68 | 0.68 |
| Decision Tree | 0.52 | 0.60 | 0.59 |
| Gaussian NB | 0.49 | 0.84 | 0.74 |
| KNN | 0.63 | 0.68 | 0.71 |
| Logistic regression | 0.66 | 0.56 | 0.63 |
| RandomForest | 0.82 | 0.48 | 0.75 |
| Motor | |||
| AdaBoost | 0.51 | 0.67 | 0.58 |
| Decision Tree | 0.56 | 0.63 | 0.61 |
| Gaussian NB | 0.54 | 0.96 | 0.69 |
| KNN | 0.68 | 0.67 | 0.70 |
| Logistic regression | 0.69 | 0.54 | 0.66 |
| RandomForest | 0.79 | 0.33 | 0.71 |
| Language | |||
| AdaBoost | 0.57 | 0.78 | 0.64 |
| Decision Tree | 0.57 | 0.63 | 0.61 |
| Gaussian NB | 0.54 | 0.76 | 0.64 |
| KNN | 0.64 | 0.44 | 0.53 |
| Logistic regression | 0.62 | 0.76 | 0.63 |
| RandomForest | 0.67 | 0.51 | 0.65 |
References
- Adams-Chapman, I.; Heyne, R.J.; DeMauro, S.B.; Duncan, A.F.; Hintz, S.R.; Pappas, A.; Vohr, B.R.; McDonald, S.A.; Das, A.; Newman, J.E.; et al. Neurodevelopmental Impairment Among Extremely Preterm Infants in the Neonatal Research Network. Pediatrics 2018, 141, e20173091. [Google Scholar] [CrossRef] [PubMed]
- Leviton, A.; Allred, E.N.; Kuban, K.C.K.; O’Shea, T.M.; Paneth, N.; Onderdonk, A.B.; Fichorova, R.N.; Dammann, O. The Development of Extremely Preterm Infants Born to Women Who Had Genitourinary Infections During Pregnancy. Am. J. Epidemiol. 2016, 183, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Ushida, T.; Nakatochi, M.; Kobayashi, Y.; Moriyama, Y.; Imai, K.; Nakano-Kobayashi, T.; Hayakawa, M.; Kajiyama, H.; Kikkawa, F.; et al. Mortality and neurological outcomes in extremely and very preterm infants born to mothers with hypertensive disorders of pregnancy. Sci. Rep. 2021, 11, 1729. [Google Scholar] [CrossRef]
- Cortés-Albornoz, M.C.; García-Guáqueta, D.P.; Velez-van Meerbeke, A.; Talero-Gutiérrez, C. Maternal Nutrition and Neurodevelopment: A Scoping Review. Nutrients 2021, 13, 3530. [Google Scholar] [CrossRef] [PubMed]
- Salas, A.A.; Carlo, W.A.; Ambalavanan, N.; Nolen, T.L.; Stoll, B.J.; Das, A.; Higgins, R.D. Gestational age and birthweight for risk assessment of neurodevelopmental impairment or death in extremely preterm infants. Arch. Dis. Child.-Fetal Neonatal Ed. 2016, 101, F494–F501. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Shi, L.; Rajadurai, V.S.; Zheng, Q.; Yang, P.H.; Khoo, P.C.; Quek, B.H.; Daniel, L.M. Factors affecting neurodevelopmental outcome at 2 years in very preterm infants below 1250 g: A prospective study. J. Perinatol. 2018, 38, 1093–1100. [Google Scholar] [CrossRef]
- Khorram, B.; Kilmartin, K.C.; Dahan, M.; Zhong, Y.J.; Abdelmageed, W.; Wintermark, P.; Shah, P.S. Outcomes of Neonates with a 10-min Apgar Score of Zero: A Systematic Review and Meta-Analysis. Neonatology 2022, 119, 669–685. [Google Scholar] [CrossRef]
- Matei, A.; Montalva, L.; Goodbaum, A.; Lauriti, G.; Zani, A. Neurodevelopmental impairment in necrotising enterocolitis survivors: Systematic review and meta-analysis. Arch. Dis. Child.-Fetal Neonatal Ed. 2020, 105, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.M.; Austin, N.C.; Inder, T.E.; Bora, S.; Woodward, L.J. Neonatal Infection and Later Neurodevelopmental Risk in the Very Preterm Infant. J. Pediatr. 2016, 170, 97–104. [Google Scholar] [CrossRef]
- Vliegenthart, R.J.S.; Kaam, A.H.V.; Aarnoudse-Moens, C.S.H.; Wassenaer, A.G.V.; Onland, W. Duration of mechanical ventilation and neurodevelopment in preterm infants. Arch. Dis. Child.-Fetal Neonatal Ed. 2019, 104, F631–F635. [Google Scholar] [CrossRef]
- Ambalavanan, N.; Carlo, W.A.; Tyson, J.E.; Langer, J.C.; Walsh, M.C.; Parikh, N.A.; Das, A.; Van Meurs, K.P.; Shankaran, S.; Stoll, B.J.; et al. Outcome Trajectories in Extremely Preterm Infants. Pediatrics 2012, 130, e115–e125. [Google Scholar] [CrossRef] [PubMed]
- Kiechl-Kohlendorfer, U.; Ralser, E.; Peglow, U.P.; Reiter, G.; Trawöger, R. Adverse neurodevelopmental outcome in preterm infants: Risk factor profiles for different gestational ages. Acta Paediatr. 2009, 98, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Suenaga, H.; Uchiyama, A.; Kono, Y.; Kusuda, S.; Neonatal Research Network, Japan. Trends in the neurodevelopmental outcomes among preterm infants from 2003–2012: A retrospective cohort study in Japan. J. Perinatol. Off. J. Calif. Perinat. Assoc. 2018, 38, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Ambalavanan, N.; Nelson, K.G.; Alexander, G.; Johnson, S.E.; Biasini, F.; Carlo, W.A. Prediction of Neurologic Morbidity in Extremely Low Birth Weight Infants. J. Perinatol. 2000, 20, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Ambalavanan, N.; Baibergenova, A.; Carlo, W.A.; Saigal, S.; Schmidt, B.; Thorpe, K.E. Early prediction of poor outcome in extremely low birth weight infants by classification tree analysis. J. Pediatr. 2006, 148, 438–444.e1. [Google Scholar] [CrossRef] [PubMed]
- Juul, S.E.; Wood, T.R.; German, K.; Law, J.B.; Kolnik, S.E.; Puia-Dumitrescu, M.; Mietzsch, U.; Gogcu, S.; Comstock, B.A.; Li, S.; et al. Predicting 2-year neurodevelopmental outcomes in extremely preterm infants using graphical network and machine learning approaches. eClinicalMedicine 2023, 56, 101782. [Google Scholar] [CrossRef]
- Bayley, N. Bayley Scales of Infant and Toddler Development 2006. Available online: https://journals.sagepub.com/doi/10.1177/0734282906297199 (accessed on 5 July 2024).
- Butcher, B.; Smith, B.J. Feature Engineering and Selection: A Practical Approach for Predictive Models. Am. Stat. 2020, 74, 308–309. [Google Scholar] [CrossRef]
- Kraskov, A.; Stögbauer, H.; Grassberger, P. Estimating mutual information. Phys. Rev. E 2004, 69, 066138. [Google Scholar] [CrossRef] [PubMed]
- Géron, A. Hands-on Machine Learning with Scikit-Learn, Keras, and TensorFlow; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2022. [Google Scholar]
- Freund, Y.; Schapire, R.; Abe, N. A short introduction to boosting. J.-Jpn. Soc. Artif. Intell. 1999, 14, 1612. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chan, T.F.; Golub, G.H.; LeVeque, R.J. Updating Formulae and a Pairwise Algorithm for Computing Sample Variances. In COMPSTAT 1982 5th Symposium Held at Toulouse 1982: Part I: Proceedings in Computational Statistics; Caussinus, H., Ettinger, P., Tomassone, R., Eds.; Physica-Verlag HD: Heidelberg, Germany, 1982; pp. 30–41. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Benavente-Fernández, I.; Synnes, A.; Grunau, R.E.; Chau, V.; Ramraj, C.; Glass, T.; Cayam-Rand, D.; Siddiqi, A.; Miller, S.P. Association of Socioeconomic Status and Brain Injury With Neurodevelopmental Outcomes of Very Preterm Children. JAMA Netw. Open 2019, 2, e192914. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Loukas, S.; Lazeyras, F.; Hüppi, P.; Meskaldji, D.; Borradori Tolsa, C. Longitudinal study of neonatal brain tissue volumes in preterm infants and their ability to predict neurodevelopmental outcome. NeuroImage 2019, 185, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Keunen, K.; Išgum, I.; van Kooij, B.J.; Anbeek, P.; van Haastert, I.C.; Koopman-Esseboom, C.; Fieret-van Stam, P.C.; Nievelstein, R.A.; Viergever, M.A.; de Vries, L.S.; et al. Brain Volumes at Term-Equivalent Age in Preterm Infants: Imaging Biomarkers for Neurodevelopmental Outcome through Early School Age. J. Pediatr. 2016, 172, 88–95. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Li, H.; Chen, M.; Wang, J.; Altaye, M.; Dillman, J.R.; Parikh, N.A. Deep Multimodal Learning From MRI and Clinical Data for Early Prediction of Neurodevelopmental Deficits in Very Preterm Infants. Front. Neurosci. 2021, 15, 753033. [Google Scholar] [CrossRef]
- Wagner, M.W.; So, D.; Guo, T.; Erdman, L.; Sheng, M.; Ufkes, S.; Grunau, R.E.; Synnes, A.; Branson, H.M.; Chau, V.; et al. MRI based radiomics enhances prediction of neurodevelopmental outcome in very preterm neonates. Sci. Rep. 2022, 12, 11872. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).