Appendix A.1. Figures Codes
# Stacy Burr XII
# pdf
pdf.SBXII <- function(par, x) {
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
f1 <- b*c*d^a/(s^c*gamma(a/b)) * x^(c-1) * (1+(x/s)^c)^(-1)
f2 <- log(1+(x/s)^c)^(a-1) * exp(-(d*log((1+(x/s)^c)))^b)
fd <- f1 * f2
fd
}
# cdf
cdf.SBXII <- function(par, x) {
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
fa <- pgamma((d*log(1+(x/s)^c))^b, a/b)
fa
}
# hrf
hrf.SBXII <- function(par, x) {
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
fd <- b*c*d^a/(s^c*gamma(a/b)) * x^(c-1) * (1+(x/s)^c)^(-d-1) *
log(1+(x/s)^c)^(a-1) * exp(-(d*log((1+(x/s)^c)))^b) * (1+(x/s)^c)^d
fa <- pgamma((d*log(1+(x/s)^c))^b, a/b)
hr <- fd / (1-fa)
hr
}
# Stacy Uniform
# pdf
pdf.SUnif <- function(par, x){
a <- par[1]
b <- par[2]
c<- par[3]
d <- par[4]
G <- punif(x, c, d)
g <- dunif(x, c, d)
fd <- b/(gamma(a/b)*(d-x)) * (-log((d-x)/(d-c)))^(a-1) * exp(-(-log((d-x)/(d-c)))^b)
fd
}
# cdf
cdf.SUnif <- function(par, x){
a <- par[1]
b <- par[2]
c <- par[3]
d <- par[4]
fa <- pgamma((-log((d-x)/(d-c)))^b, a/b)
fa
}
################
# Stacy-Chen
# pdf
pdf.SC <- function(par, x){
a <- par[1]
b <- par[2]
lambda <- par[3]
beta <- par[4]
b*lambda^a*beta/gamma(a/b)*x^(beta-1) * (exp(x^beta)-1)^(a-1) *
exp(x^beta - (-lambda*(1-exp(x^beta)))^b )
}
# cdf
cdf.SC <- function(par, x){
a <- par[1]
b <- par[2]
lambda <- par[3]
beta <- par[4]
pgamma((-lambda*(1-exp(x^beta)))^b, a/b)
}
# hrf
hrf.SC <- function(par, x){
a <- par[1]
b <- par[2]
lambda <- par[3]
beta <- par[4]
fd = b*lambda^a*beta/gamma(a/b)*x^(beta-1) * (exp(x^beta)-1)^(a-1) *
exp(x^beta - (-lambda*(1-exp(x^beta)))^b )
fa = pgamma((-lambda*(1-exp(x^beta)))^b, a/b)
fd / (1-fa)
}
# Stacy Weibull
# fdp
pdf.SW <- function(par, x){
a <- par[1]
b <- par[2]
beta <- par[3]
alpha <- par[4]
fd <- b*alpha*beta^(a*alpha)/gamma(a/b) * x^(a*alpha-1) * exp(-(beta*x)^(b*alpha))
fd
}
# cdf
cdf.SW <- function(par, x){
a <- par[1]
b <- par[2]
beta <- par[3]
alpha <- par[4]
fa <- pgamma((beta*x)^(b*alpha), a/b)
fa
}
# hrf
hrf.SW <- function(par, x){
a <- par[1]
b <- par[2]
beta <- par[3]
alpha <- par[4]
fd <- b*alpha*beta^(a*alpha)/gamma(a/b) * x^(a*alpha-1) * exp(-(beta*x)^(b*alpha))
fa <- pgamma((beta*x)^(alpha*b), a/b)
hr <- fd / (1-fa)
hr
}
## density curves
par(mfrow=c(1,1))
par(mar=c(2.8, 2.8, .5, .5))
par(mgp=c(1.6, 0.45, 0))
# SBXII
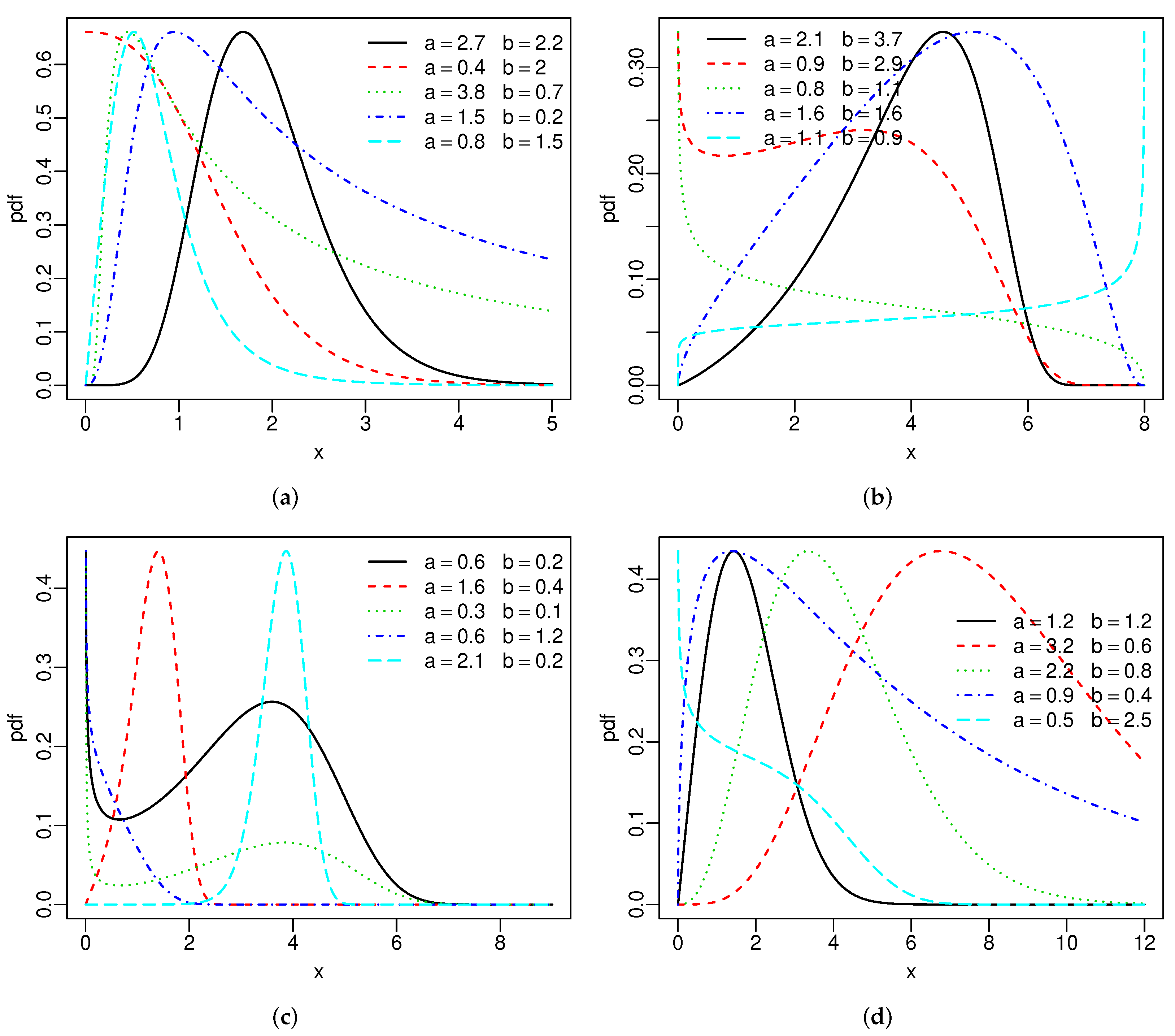
{
curve(pdf.SBXII(c(2.7,2.2,1.2,.7,2.5),x), xlim = c(0,5), col=1, n=2000, ylab = "pdf", lwd=1.5, lty=1)
par(new=T)
curve(pdf.SBXII(c(.4,2,1.2,.7,2.5),x), xlim = c(0,4), col=2, n=2000, axes=F,ann=F, lwd=1.5, lty=2)
par(new=T)
curve(pdf.SBXII(c(3.8,.7,1.2,.7,2.5),x), xlim = c(0,58), col=3, n=2000, axes=F,ann=F, lwd=1.5, lty=3)
par(new=T)
curve(pdf.SBXII(c(1.5,.2,1.2,.7,2.5),x), xlim = c(0,10), col=4, n=2000, axes=F,ann=F, lwd=1.5, lty=4)
par(new=T)
curve(pdf.SBXII(c(.8,1.5,1.2,.7,2.5),x), xlim = c(0,8), col=5, n=2000, axes=F,ann=F, lwd=1.5, lty=5)
legend("topright", legend = c(expression(paste(a==2.7," ", " ", " ", b==2.2)),
expression(paste(a==.4," ", " ", " ", b==2)),
expression(paste(a==3.8," ", " ", " ", b==.7)),
expression(paste(a==1.5," ", " ", " ", b==.2)),
expression(paste(a==.8," ", " ", " ", b==1.5))),
lwd=1.5, bty = "n", col = 1:5, lty=1:5)
}
# SUnif
{
curve(pdf.SUnif(c(2.1,3.7,0,8),x), xlim = c(0,8), col=1, n=2000, ylab = "pdf", lwd=1.5, lty=1)
par(new=T)
curve(pdf.SUnif(c(.9,2.9,0,8),x), xlim = c(0,8), col=2, n=2000, axes=F,ann=F, lwd=1.5, lty=2)
par(new=T)
curve(pdf.SUnif(c(.8,1.1,0,8),x), xlim = c(0,8), col=3, n=2000, axes=F,ann=F, lwd=1.5, lty=3)
par(new=T)
curve(pdf.SUnif(c(1.6,1.6,0,8),x), xlim = c(0,8), col=4, n=2000, axes=F,ann=F, lwd=1.5, lty=4)
par(new=T)
curve(pdf.SUnif(c(1.1,.9,0,8),x), xlim = c(0,8), col=5, n=2000, axes=F,ann=F, lwd=1.5, lty=5)
legend(.2,.64, legend = c(expression(paste(a==2.1," ", " ", " ", b==3.7)),
expression(paste(a==.9," ", " ", " ", b==2.9)),
expression(paste(a==.8," ", " ", " ", b==1.1)),
expression(paste(a==1.6," ", " ", " ", b==1.6)),
expression(paste(a==1.1," ", " ", " ", b==.9))),
lwd=1.5, bty = "n", col=1:5, lty=1:5)
}
# SC
{
curve(pdf.SC(c(.6,.2,1.7,1.3),x), xlim = c(0,9), col=1, n=2000, ylab = "pdf", lwd=1.5, lty=1)
par(new=T)
curve(pdf.SC(c(1.6,.4,1.7,1.3),x), xlim = c(0,15), col=2, n=2000, axes=F,ann=F, lwd=1.5, lty=2)
par(new=T)
curve(pdf.SC(c(.3,.1,1.7,1.3),x), xlim = c(0,15), col=3, n=2000, axes=F,ann=F, lwd=1.5, lty=3)
par(new=T)
curve(pdf.SC(c(.6,1.2,1.7,1.3),x), xlim = c(0,5), col=4, n=2000, axes=F,ann=F, lwd=1.5, lty=4)
par(new=T)
curve(pdf.SC(c(2.1,.2,1.7,1.3),x), xlim = c(0,15), col=5, n=2000, axes=F,ann=F, lwd=1.5, lty=5)
legend("topright", legend = c(expression(paste(a==.6," ", " ", " ", b==.2)),
expression(paste(a==1.6," ", " ", " ", b==.4)),
expression(paste(a==.3," ", " ", " ", b==.1)),
expression(paste(a==.6," ", " ", " ", b==1.2)),
expression(paste(a==2.1," ", " ", " ", b==.2))),
lwd=1.5, bty = "n", col=1:5, lty=1:5)
}
# SW
{
curve(pdf.SW(c(1.2,1.2,.5,1.7),x), xlim = c(0,12), col=1, n=2000, ylab = "pdf", lwd=1.5, lty=1)
par(new=T)
curve(pdf.SW(c(3.2,.6,.5,1.7),x), xlim = c(0,15), col=2, n=2000, axes=F,ann=F, lwd=1.5, lty=2)
par(new=T)
curve(pdf.SW(c(2.2,.8,.5,1.7),x), xlim = c(0,12), col=3, n=2000, axes=F,ann=F, lwd=1.5, lty=3)
par(new=T)
curve(pdf.SW(c(.9,.4,.5,1.7),x), xlim = c(0,12), col=4, n=2000, axes=F,ann=F, lwd=1.5, lty=4)
par(new=T)
curve(pdf.SW(c(.5,2.5,.5,1.7),x), xlim = c(0,5), col=5, n=2000, axes=F,ann=F, lwd=1.5, lty=5)
legend(2.8,1.1, legend = c(expression(paste(a==1.2," ", " ", " ", b==1.2)),
expression(paste(a==3.2," ", " ", " ", b==.6)),
expression(paste(a==2.2," ", " ", " ", b==.8)),
expression(paste(a==.9," ", " ", " ", b==.4)),
expression(paste(a==.5," ", " ", " ", b==2.5))),
lwd=1.5, bty = "n", col=1:5, lty=1:5)
}
# hrfs curves
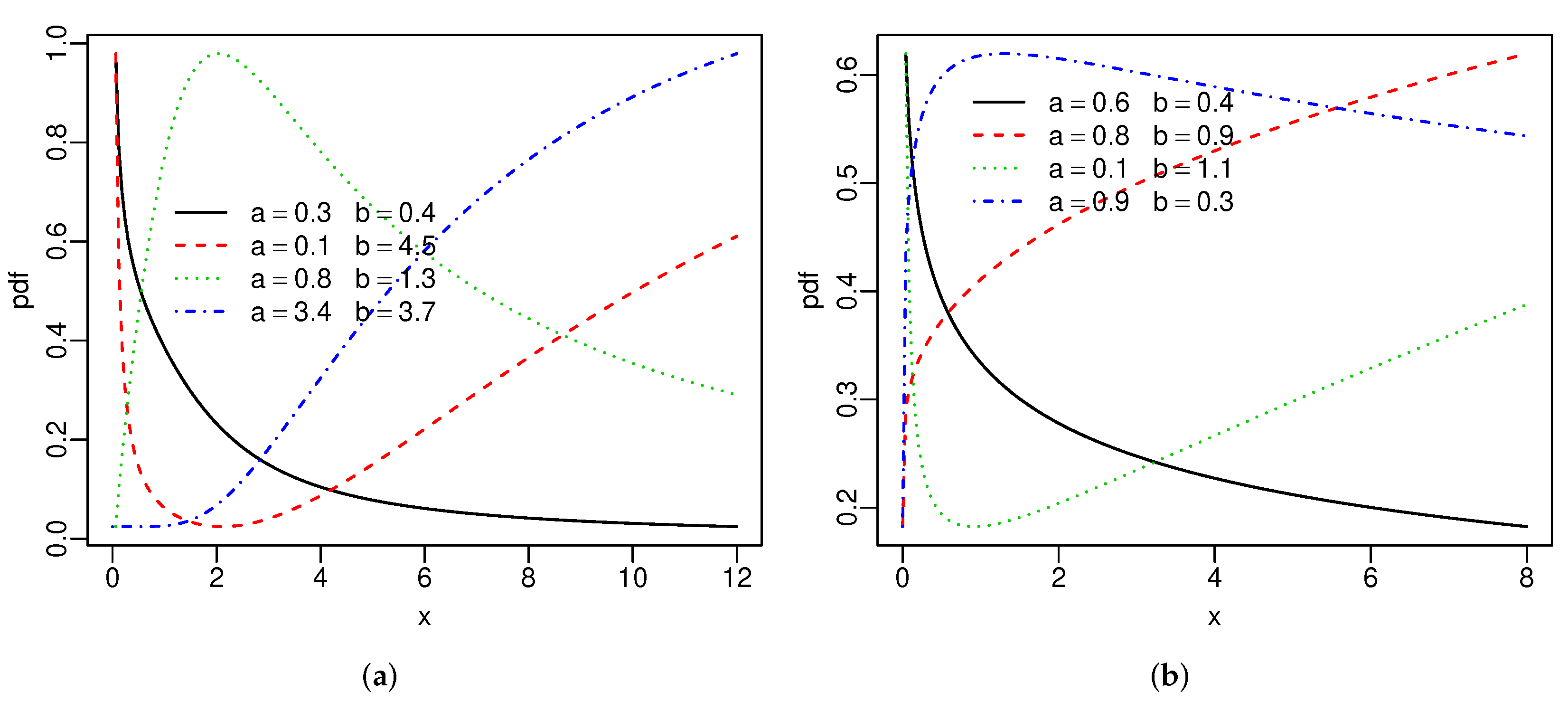
# SBXII
{
curve(hrf.SBXII(c(.3,.4,1.9,.4,2.2),x), xlim = c(0,12), col=1, n=200, ylab = "pdf", lwd=1.5, lty=1)
par(new=T)
curve(hrf.SBXII(c(.1,4.5,1.9,.4,2.2),x), xlim = c(0,12), col=2, n=200, axes=F,ann=F, lwd=1.5, lty=2)
par(new=T)
curve(hrf.SBXII(c(.8,1.3,1.9,.4,2.2),x), xlim = c(0,12), col=3, n=200, axes=F,ann=F, lwd=1.5, lty=3)
par(new=T)
curve(hrf.SBXII(c(3.4,3.7,1.9,.4,2.2),x), xlim = c(0,12), col=4, n=200, axes=F,ann=F, lwd=1.5, lty=4)
legend(.75,.74, legend = c(expression(paste(a==.3," ", " ", " ", b==.4)),
expression(paste(a==.1," ", " ", " ", b==4.5)),
expression(paste(a==.8," ", " ", " ", b==1.3)),
expression(paste(a==3.4," ", " ", " ", b==3.7))),
lwd=1.5, bty = "n", col=1:4, lty=1:4 )
}
# SW
{
curve(hrf.SW(c(.6,.4,.7,1.5),x), xlim = c(0,8), col=1, n=200, ylab = "pdf", lwd=1.5, lty=1)
par(new=T)
curve(hrf.SW(c(.8,.9,.7,1.5),x), xlim = c(0,8), col=2, n=200, axes=F,ann=F, lwd=1.5, lty=2)
par(new=T)
curve(hrf.SW(c(.1,1.1,.7,1.5),x), xlim = c(0,8), col=3, n=200, axes=F,ann=F, lwd=1.5, lty=3)
par(new=T)
curve(hrf.SW(c(.9,.3,.7,1.5),x), xlim = c(0,8), col=4, n=200, axes=F,ann=F, lwd=1.5, lty=4)
#par(new=T)
#curve(hrf.SW(c(.6,.4,.7,1.5),x), xlim = c(0,8), col=7, n=200, axes=F,ann=F)
legend(.6,.061, legend = c(expression(paste(a==.6," ", " ", " ", b==.4)),
expression(paste(a==.8," ", " ", " ", b==.9)),
expression(paste(a==.1," ", " ", " ", b==1.1)),
expression(paste(a==.9," ", " ", " ", b==.3))),
lwd=1.5, bty = "n", col=1:4, lty=1:4)
}
#################################################################################
###################### Assimetria e kurtose da SW #####################
#################################################################################
# momentos centrais
# UWW distribution
momcentral_SW <- function(param, r){
mu1 <- pdf.SW <- function(x, par=param){
a <- par[1]
b <- par[2]
beta <- par[3]
alpha <- par[4]
fd <- b*alpha*beta^(a*alpha)/gamma(a/b) * x^(a*alpha-1) * exp(-(beta*x)^(b*alpha))
fd
}
meanx <- integrate(mu1, 0, Inf, subdivisions = 1e6)$value
mu2.central <- function(x, par=param, mu=meanx){
a <- par[1]
b <- par[2]
beta <- par[3]
alpha <- par[4]
fd1 <- b*alpha*beta^(a*alpha)/gamma(a/b) * x^(a*alpha) * exp(-(beta*x)^(b*alpha))
fd <- fd1 * (x-mu)^2
fd
}
mur.central <- function(x, par=param, mu=meanx,k=r){
a <- par[1]
b <- par[2]
beta <- par[3]
alpha <- par[4]
fd1 <- b*alpha*beta^(a*alpha)/gamma(a/b) * x^(a*alpha-1) * exp(-(beta*x)^(b*alpha))
fd <- fd1 * (x-mu)^k
fd
}
mom2_central <- integrate(mu2.central, 0, Inf, subdivisions = 1e6)$value
momr_central <- integrate(mur.central, 0, Inf, subdivisions = 1e6)$value
# padronizando
mom_central_Padr <- momr_central / mom2_central^(r/2)
mom_central_Padr
#list(mu=meanx)
}
momcentral_SW(c(2,1.4,.2,1.4),0)
momcentral_SW(c(.9,1.4,4.2,.4),0)
a <- seq(from=1,by=.02, to=10)
b <- seq(from=1,by=.02, to=10)
a <- seq(from=1,by=.1, to=10)
b <- seq(from=1,by=.1, to=10)
# assimetria
assimetria_a <- vector()
assimetria_a2 <- vector()
assimetria_a3 <- vector()
assimetria_b <- vector()
assimetria_b2 <- vector()
assimetria_b3 <- vector()
for (i in 1:length(a)) {
print(i)
assimetria_a[i] <- momcentral_SW(c(a[i], 2.5, .4, .8), 3)
assimetria_a2[i] <- momcentral_SW(c(a[i], 4.2, 3.2, 1.5), 3)
assimetria_a3[i] <- momcentral_SW(c(a[i], 1.1, 1.3, .4), 3)
assimetria_b[i] <- momcentral_SW(c(.5, b[i], 1.2, 1.4), 3)
assimetria_b2[i] <- momcentral_SW(c(1.6, b[i], .6, .9), 3)
assimetria_b3[i] <- momcentral_SW(c(3.3, b[i], 2.9, 2.5), 3)
}
# curvas
# para a
{
plot(a,assimetria_a, type = "l", ylab = "skewness", col=2, lwd=1.5, lty=1)
par(new=T)
plot(a,assimetria_a2, type = "l", ylab = "skewness", col=3, axes = F, ann = F, lwd=1.5, lty=2)
par(new=T)
plot(a,assimetria_a3, type = "l", ylab = "skewness", col=4, axes = F, ann = F, lwd=1.5, lty=3)
legend(4.5,.02, legend = c(
expression(paste(b==2.5, " ", " ", " ", beta==.4, " ", " ", " ", alpha==.8)),
expression(paste(b==4.2, " ", " ", " ", beta==3.2, " ", " ", " ", alpha==1.5)),
expression(paste(b==1.1, " ", " ", " ", beta==1.3, " ", " ", " ", alpha==.4))
),
bty = "n", col = 2:4, lwd=1.5, lty=1:3)
}
# para b
{
plot(b,assimetria_b, type = "l", ylab = "skewness", col=2, lwd=1.5, lty=1)
par(new=T)
plot(b,assimetria_b2, type = "l", ylab = "skewness", col=3, axes = F, ann = F, lwd=1.5, lty=2)
par(new=T)
plot(b,assimetria_b3, type = "l", ylab = "skewness", col=4, axes = F, ann = F, lwd=1.5, lty=3)
legend(4.4,-3.5, legend = c(
expression(paste(a==.5, " ", " ", " ", beta==1.2, " ", " ", " ", alpha==1.4)),
expression(paste(a==1.6, " ", " ", " ", beta==.6, " ", " ", " ", alpha==.9)),
expression(paste(a==3.3, " ", " ", " ", beta==.9, " ", " ", " ", alpha==2.5))
),
bty = "n", col = 2:4, lwd=1.5, lty=1:3)
}
# kurtose
kurtose_a <- vector()
kurtose_a2 <- vector()
kurtose_a3 <- vector()
kurtose_b <- vector()
kurtose_b2 <- vector()
kurtose_b3 <- vector()
for (i in 1:length(a)) {
print(i)
kurtose_a[i] <- momcentral_SW(c(a[i], .9, 2.2, 1.2), 4)
kurtose_a2[i] <- momcentral_SW(c(a[i], 2.5, .5, 4.7), 4)
kurtose_a3[i] <- momcentral_SW(c(a[i], 4.4, 1.7, .7), 4)
kurtose_b[i] <- momcentral_SW(c(.5, b[i], 2.2, 2.2), 4)
kurtose_b2[i] <- momcentral_SW(c(1.4, b[i], 1.7, .6), 4)
kurtose_b3[i] <- momcentral_SW(c(3.1, b[i], .5, 1.9), 4)
}
# curvas
# para a
{
plot(a,kurtose_a, type = "l", ylab = "kurtosis", col=2, lwd=1.5, lty=1)
par(new=T)
plot(a,kurtose_a2, type = "l", ylab = "kurtosis", col=3, axes = F, ann = F, lwd=1.5, lty=2)
par(new=T)
plot(a,kurtose_a3, type = "l", ylab = "kurtosis", col=4, axes = F, ann = F, lwd=1.5, lty=3)
legend(4,36, legend = c(
expression(paste(b==.9, " ", " ", " ", beta==2.2, " ", " ", " ", alpha==1.2)),
expression(paste(b==2.5, " ", " ", " ", beta==.5, " ", " ", " ", alpha==4.7)),
expression(paste(b==4.4, " ", " ", " ", beta==1.7, " ", " ", " ", alpha==.7))
),
bty = "n", col = 2:4, lwd=1.5, lty=1:3)
}
# para b
{
plot(b,kurtose_b, type = "l", ylab = "kurtosis", col=2, lwd=1.5, lty=1)
par(new=T)
plot(b,kurtose_b2, type = "l", ylab = "kurtosis", col=3, axes=F, ann=F, lwd=1.5, lty=2)
par(new=T)
plot(b,kurtose_b3, type = "l", ylab = "kurtosis", col=4, axes=F, ann=F, lwd=1.5, lty=3)
legend(4,.3, legend = c(
expression(paste(a==.5," ", " ", " ", beta==2.2," ", " ", " ", alpha==2.2)),
expression(paste(a==1.4," ", " ", " ", beta==1.7, " ", " ", " ", alpha==.6)),
expression(paste(a==3.1," ", " ", " ", beta==.5, " ", " ", " ", alpha==1.9))
),
bty = "n", col = 2:4, lwd=1.5, lty=1:3)
}
Appendix A.3. Applications Codes
Appendix A.3.1. Uncensored Data
par(mfrow=c(1,1))
par(mar=c(2.8, 2.8, .5, .5)) # margens c(baixo,esq,cima,direia)
par(mgp=c(1.6, 0.45, 0))
## Optim function
OPTIM <- function(starts, pdf, cdf, grad=NULL, method="BFGS", dados){
ll <- function(par, x){
-sum(log(pdf(par, x)))
}
if(method=="BFGS" | method=="B") {
opt <- optim(starts,fn=ll, gr=grad, method = "BFGS",x=dados, hessian = T)
} else if(method=="SANN" | method=="S") {
opt <- optim(starts,fn=ll, gr=grad, method = "SANN",x=dados, hessian = T)
} else if(method=="Nelder-Mead" | method=="NM") {
opt <- optim(starts,fn=ll, gr=grad, method = "Nelder-Mead",x=dados, hessian = T)
}
emv <- opt$par
erro <- sqrt(diag(solve(opt$hessian)))
valor <- -opt$value
convergencia <- opt$convergence
#
p <- length(starts)
n <- length(dados)
llhat <- -1 * ll(emv, dados)
CAIC <- -2 * llhat + 2 * p + 2 * (p * (p + 1))/(n-p - 1)
AIC = -2 * llhat + 2 * p
BIC = -2 * llhat + p * log(n)
HQIC = -2 * llhat + 2 * log(log(n)) * p
data_orderdenados = sort(dados)
v = cdf(as.vector(emv), data_orderdenados)
y = qnorm(v)
y[which(y == Inf)] = 10
u = pnorm((y - mean(y))/sqrt(var(y)))
W_temp <- vector()
A_temp <- vector()
for (i in 1:n) {
W_temp[i] = (u[i] - (2 * i - 1)/(2 * n))^2
A_temp[i] = (2 * i - 1) * log(u[i]) + (2 * n + 1 -
2 * i) * log(1 - u[i])
}
A_2 = -n - mean(A_temp)
W_2 = sum(W_temp) + 1/(12 * n)
W = W_2 * (1 + 0.5/n)
A = A_2 * (1 + 0.75/n + 2.25/n^2)
crit <- cbind(W, A, CAIC, AIC, BIC, HQIC)
list(emv=emv,erro=erro,valor=valor, convergencia=convergencia,criterios=crit)
}
require(survival)
# Stacy Burr XII
# pdf
pdf.SBXII <- function(par, x) {
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
f1 <- b*c*d^a/(s^c*gamma(a/b)) * x^(c-1) * (1+(x/s)^c)^(-1)
f2 <- log(1+(x/s)^c)^(a-1) * exp(-(d*log((1+(x/s)^c)))^b)
fd <- f1 * f2
fd
}
# cdf
cdf.SBXII <- function(par, x) {
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
fa <- pgamma((d*log(1+(x/s)^c))^b, a/b)
fa
}
## competitives distributions
# BXII
# pdf
pdf.BXII <- function(par, x) {
s <- par[1]
d <- par[2]
c <- par[3]
c*d/s^c * x^(c-1) * (1+(x/s)^c)^(-d-1)
}
# cdf
cdf.BXII <- function(par, x) {
s <- par[1]
d <- par[2]
c <- par[3]
1 - (1+(x/s)^c)^(-d)
}
# gamma Burr XII
# pdf
pdf.GBXII <- function(par, x) {
a <- par[1]
b <- 1
s <- par[2]
d <- par[3]
c <- par[4]
f1 <- b*c*d^a/(s^c*gamma(a/b)) * x^(c-1) * (1+(x/s)^c)^(-d-1)
f2 <- log(1+(x/s)^c)^(a-1) * exp(-(d*log((1+(x/s)^c)))^b) * (1+(x/s)^c)^d
fd <- f1 * f2
fd
}
# cdf
cdf.GBXII <- function(par, x) {
a <- par[1]
s <- par[2]
d <- par[3]
c <- par[4]
fa <- pgamma(d*log(1+(x/s)^c), a)
fa
}
# Weibull BXII
# pdf
pdf.WBXII <- function(par,x){
a <- par[1]
b <- par[2]
d <- par[3]
c <- par[4]
fd <- a*b*c*d * x^(c-1) * (1+x^c)^(b*d-1) *
(1-(1+x^c)^(-d))^(b-1) * exp(-a*((1+x^c)^d -1 )^b)
fd
}
# cdf
cdf.WBXII <- function(par,x){
a <- par[1]
b <- par[2]
d <- par[3]
c <- par[4]
fa <- 1 - exp(-a*((1+x^c)^d -1 )^b)
fa
}
# beta BXII
# pdf
pdf.BBXII <- function(par,x){
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
fd <- c*d/(s^c*beta(a,b)) * x^(c-1) * (1+(x/s)^c)^(-b*d-1) *
(1 - (1+(x/s)^c)^(-d) )^(a-1)
fd
}
# cdf
cdf.BBXII <- function(par,x){
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
G <- 1 - (1+(x/s)^c)^(-d)
fa <- pbeta(G, a, b)
fa
}
# Kumaraswamy BXII
# pdf
pdf.KBXII <- function(par,x){
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
G <- 1 - (1+(x/s)^c)^(-d)
fd <- a*b*c*d*s^(-c)*x^(c-1)*(1+(x/s)^c)^(-d-1)*G^(a-1)*(1-G^a)^(b-1)
fd
}
# cdf
cdf.KBXII <- function(par,x){
a <- par[1]
b <- par[2]
s <- par[3]
d <- par[4]
c <- par[5]
G <- 1 - (1+(x/s)^c)^(-d)
fa <- 1 - (1-G^a)^b
fa
}
######################################################################################
# dados
# The Generalized Odd Gamma-G Family of
#Distributions: Properties and Applications
# Hosseini et al
hosseini <- c(28.0, 98, 89.0, 68.9, 69.9, 109.0, 52.3, 52.8, 46.7, 82.7,
42.3, 109.1, 96.8, 98.3, 103.6, 110.2, 98.1, 57.0, 43.1,
71.1, 29.7, 96.3, 102.8, 80.3, 122.1, 71.3, 200.8, 80.6,
65.3, 78.0, 65.9, 38.9, 56.5, 104.6, 74.9, 90.4, 54.6, 131.9,
68.3, 52.0, 40.8, 34.3, 44.8, 105.7, 126.4, 83.0, 106.9, 88.2,
33.8, 47.6, 42.7, 41.5, 34.6, 30.9, 100.7, 80.3, 91.0, 156.6,
95.4, 43.5, 61.9, 35.2, 50.9, 31.8, 44.0, 56.8, 75.2, 76.2,
101.1, 47.5, 46.2, 38.2, 49.2, 49.6, 34.5, 37.5, 75.9, 87.2,
52.6, 126.4, 55.6, 73.9, 43.5, 61.8, 88.9, 31.0, 37.6, 52.8,
97.9, 111.1, 114.0, 62.9, 36.8, 56.8, 46.5, 48.3, 32.6, 31.7,
47.8, 75.1, 110.7, 70.0, 52.5, 67, 41.6, 34.8, 61.8, 31.5, 36.6,
76.0, 65.1, 74.7, 77.0, 62.6, 41.1, 58.9, 60.2, 43.0, 32.6, 48,
61.2, 171.1, 113.5, 148.9, 49.9, 59.4, 44.5, 48.1, 61.1, 31.0,
41.9, 75.6, 76.8, 99.8, 80.1, 57.9, 48.4, 41.8, 44.5, 43.8, 33.7,
30.9, 43.3, 117.8, 80.3, 156.6, 109.6, 50.0, 33.7, 54.0, 54.2, 30.3,
52.8, 49.5, 90.2, 109.5, 115.9, 98.5, 54.6, 50.9, 44.7, 41.8, 38.0,
43.2, 70.0, 97.2, 123.6, 181.7, 136.3, 42.3, 40.5, 64.9, 34.1, 55.7,
113.5, 75.7, 99.9, 91.2, 71.6, 103.6, 46.1, 51.2, 43.8, 30.5, 37.5,
96.9, 57.7, 125.9, 49.0, 143.5, 102.8, 46.3, 54.4, 58.3, 34.0, 112.5,
49.3, 67.2, 56.5, 47.6, 60.4, 34.9)
## Cordeiro et al (2019)
# The odd Lomax generator of distributions: Properties,
# estimation and applications
cordeiroetal2019 <- c(1.6,2.0,2.6,3.0,3.5,3.9,4.5,4.6,4.8,5.0,5.1,5.3,5.4,
5.6,5.8,6.0,6.0,6.1,6.3,6.5,6.5,6.7,7.0,7.1,7.3,7.3,
7.3,7.7,7.7,7.8,7.9,8.0,8.1, 8.3, 8.4,8.4, 8.6, 8.7,
8.8, 9.0)
#############################################################################
# aplication 1
SG.dados1 <- hosseini
emv.SBXII <- OPTIM(c(1.1,1,1,1,1), pdf=pdf.SBXII, cdf=cdf.SBXII, dados=SG.dados1)
emv.GBXII <- OPTIM(c(1,1,.1,.1), pdf=pdf.GBXII, cdf=cdf.GBXII, dados=SG.dados1)
emv.BXII <- OPTIM(c(.1,.1,.1), pdf=pdf.BXII, cdf=cdf.BXII, dados=SG.dados1)
emv.BBXII <- OPTIM(c(1,1,1,.1,.1), pdf=pdf.BBXII, cdf=cdf.BBXII, dados=SG.dados1)
emv.KBXII <- OPTIM(c(1,1,1,.1,.1), pdf=pdf.KBXII, cdf=cdf.KBXII, dados=SG.dados1)
emv.WBXII <- OPTIM(c(1,1,.1,.1), pdf=pdf.WBXII, cdf=cdf.WBXII, dados=SG.dados1)
# coefs
SG.coef1 <- rbind(
# BXII
c(NA, NA, emv.BXII$emv), c(NA, NA, emv.BXII$erro),
# GBXII
c(emv.GBXII$emv, NA), c(emv.GBXII$erro, NA),
# SBXII
emv.SBXII$emv, emv.SBXII$erro,
# BBXII
emv.BBXII$emv, emv.BBXII$erro,
# KBXII
emv.KBXII$emv, emv.KBXII$erro,
# WBXII
c(emv.WBXII$emv, NA), c(emv.WBXII$erro, NA)
)
rownames(SG.coef1) <- c("BXII", " ", "GBXII", " ", "SBXII", " ", "BBXII", " ",
"KBXII", " ", "WBXII", " ")
SG.coef1
# criterios
SG.criterios1 <- rbind(
# BXII
emv.BXII$criterios,
# GBXII
emv.GBXII$criterios,
# SBXII
emv.SBXII$criterios,
# BBXII
emv.BBXII$criterios,
# KBXII
emv.KBXII$criterios,
# WBXII
emv.WBXII$criterios
)
SG.criterios1
rownames(SG.criterios1) <- c("BXII", "GBXII", "SBXII", "BBXII", "KBXII", "WBXII")
sort(SG.criterios1[,1])
sort(SG.criterios1[,2])
# LR TEST
2*(emv.SBXII$valor - emv.GBXII$valor)
2*(emv.SBXII$valor - emv.BXII$valor)
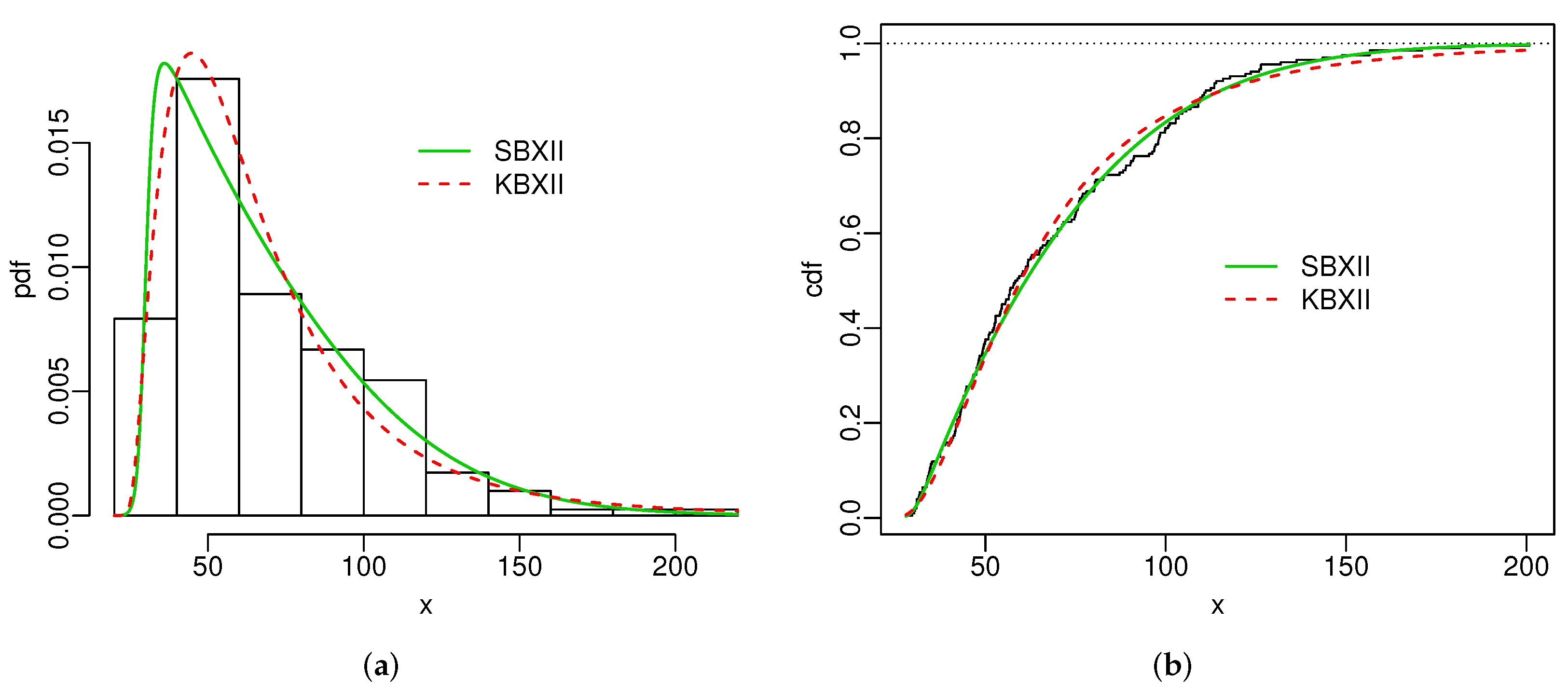
# pdf
{
hist(SG.dados1, freq = F, xlab = "x", ylab = "pdf", main = "", ylim = c(0,.019))
curve(pdf.SBXII(emv.SBXII$emv,x), col=3, add=T, n=500, lty=1, lwd=1.5)
curve(pdf.KBXII(emv.KBXII$emv,x), col=2, add=T, n=500, lty=2, lwd=1.5)
legend(110,.016, lwd = 1.5, col = c(3,2), bty = "n", legend = c("SBXII", "KBXII"), lty=1:2)
}
# cdf
# kaplan-meier
{
SGdados1_KP <- survfit(Surv(SG.dados1) ~ 1)
plot(SGdados1_KP$time, 1-SGdados1_KP$surv, xlab = "x", ylab = "cdf", main = "", type = "s")
abline(h=1, lty=9)
curve(cdf.SBXII(emv.SBXII$emv,x), col=3, add=T, n=500, lty=1, lwd=1.5)
curve(cdf.KBXII(emv.KBXII$emv,x), col=2, add=T, n=500, lty=2, lwd=1.5)
legend(110,.6, lwd = 1.5, col = c(3,2), bty = "n", legend = c("SBXII", "KBXII"), lty=1:2)
}
###########################################################################
# aplication 2
SG.dados2 <- cordeiroetal2019
emv.SBXII2 <- OPTIM(c(1.,.1,1.,1.1,.1), pdf=pdf.SBXII, cdf=cdf.SBXII, dados=SG.dados2)
emv.GBXII2 <- OPTIM(c(1,1.1,1,1), pdf=pdf.GBXII, cdf=cdf.GBXII, dados=SG.dados2)
emv.BXII2 <- OPTIM(c(.1,.1,1), pdf=pdf.BXII, cdf=cdf.BXII, dados=SG.dados2)
emv.BBXII2 <- OPTIM(c(1,1,1,.1,1), pdf=pdf.BBXII, cdf=cdf.BBXII, dados=SG.dados2)
emv.KBXII2 <- OPTIM(c(1,1,1,.1,.1), pdf=pdf.KBXII, cdf=cdf.KBXII, dados=SG.dados2)
emv.WBXII2 <- OPTIM(c(1,1,.1,1), pdf=pdf.WBXII, cdf=cdf.WBXII, dados=SG.dados2)
# coefs
SG.coef2 <- rbind(
# BXII
c(NA, NA, emv.BXII2$emv), c(NA, NA, emv.BXII2$erro),
# GBXII
c(emv.GBXII2$emv, NA), c(emv.GBXII2$erro, NA),
# SBXII
emv.SBXII2$emv, emv.SBXII2$erro,
# BBXII
emv.BBXII2$emv, emv.BBXII2$erro,
# KBXII
emv.KBXII2$emv, emv.KBXII2$erro,
# WBXII
c(emv.WBXII2$emv, NA), c(emv.WBXII2$erro, NA)
)
rownames(SG.coef2) <- c("BXII", " ", "GBXII", " ", "SBXII", " ", "BBXII", " ",
"KBXII", " ", "WBXII", " ")
SG.coef2
# criterios
SG.criterios2 <- rbind(
# BXII
emv.BXII2$criterios,
# GBXII
emv.GBXII2$criterios,
# SBXII
emv.SBXII2$criterios,
# BBXII
emv.BBXII2$criterios,
# KBXII
emv.KBXII2$criterios,
# WBXII
emv.WBXII2$criterios
)
SG.criterios2
rownames(SG.criterios2) <- c("BXII", "GBXII", "SBXII", "BBXII", "KBXII", "WBXII")
SG.criterios2
sort(SG.criterios2[,1])
sort(SG.criterios2[,2])
# LR TEST
2*(emv.SBXII2$valor - emv.GBXII2$valor)
2*(emv.SBXII2$valor - emv.BXII2$valor)
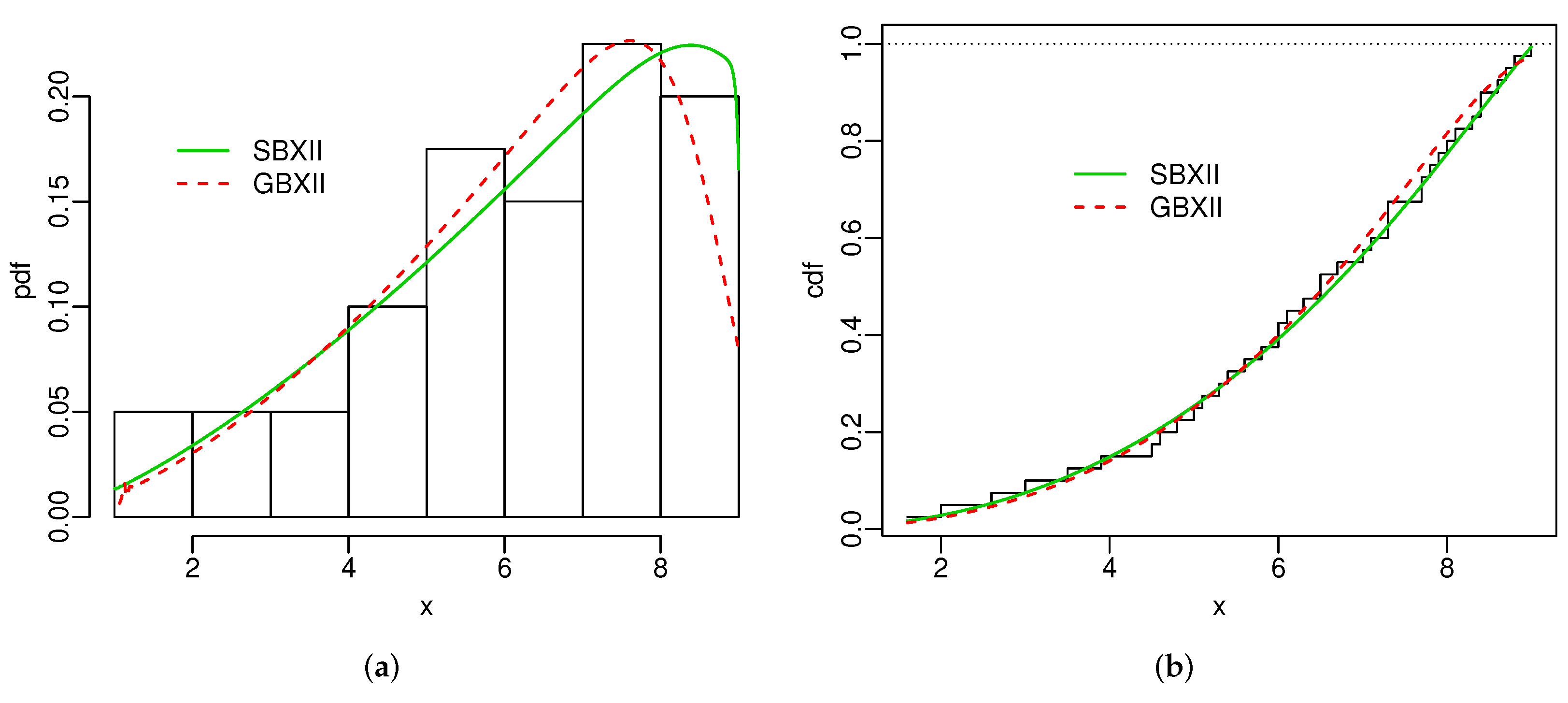
# curves
# pdf
{
hist(SG.dados2, freq = F, xlab = "x", ylab = "pdf", main = "")
curve(pdf.SBXII(emv.SBXII2$emv,x), col=3, add=T, n=500, lwd=1.5, lty=1)
curve(pdf.GBXII(emv.GBXII2$emv,x), col=2, add=T, n=500, lwd=1.5, lty=2)
legend(1.5,.19, lwd = 1.5, col = c(3,2), bty = "n", legend = c("SBXII", "GBXII"), lty=1:2 )
}
# cdf
# kaplan-meier
{
SGdados2_KP <- survfit(Surv(SG.dados2) ~ 1)
plot(SGdados2_KP$time, 1-SGdados2_KP$surv, xlab = "x", ylab = "cdf", main = "", type = "s")
abline(h=1, lty=9)
curve(cdf.SBXII(emv.SBXII2$emv,x), col=3, add=T, n=500, lwd=1.5, lty=1)
curve(cdf.GBXII(emv.GBXII2$emv,x), col=2, add=T, n=500, lwd=1.5, lty=2)
legend(3.3,.8, lwd = 1.5, col = c(3,2), bty = "n", legend = c("SBXII", "GBXII"), lty=1:2 )
}
#########################################################################
############## Figures of the likelihoods in MLEs ################
#########################################################################
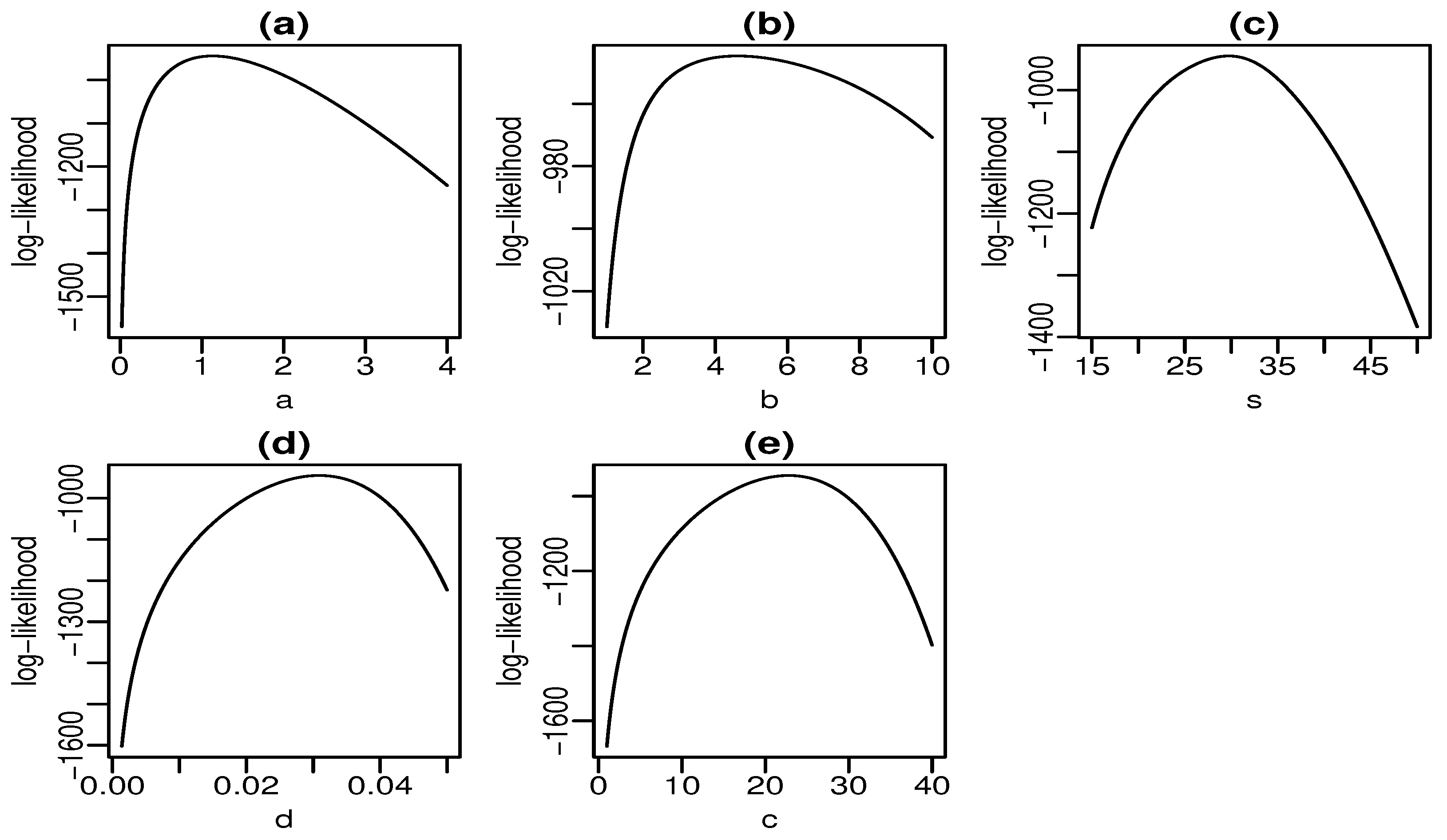
# aplication 1
emvs1 <- emv.SBXII$emv
emv.SBXII$emv
xa <- seq(from=.02, to=4, length.out=1000)
xb <- seq(from=1, to=10, length.out=1000)
xs <- seq(from=15, to=50, length.out=1000)
xd <- seq(from=.0014, to=.05, length.out=1000)
xc <- seq(from=1, to=40, length.out=1000)
emv_a = vector()
emv_b = vector()
emv_s = vector()
emv_d = vector()
emv_c = vector()
for (i in 1:length(xa)) {
emv_a[i] = sum(log(pdf.SBXII(c(xa[i], emvs1[2:5]), SG.dados1)))
}
for (i in 1:length(xb)) {
emv_b[i] = sum(log(pdf.SBXII(c(emvs1[1], xb[i], emvs1[3:5]), SG.dados1)))
}
for (i in 1:length(xs)) {
emv_s[i] = sum(log(pdf.SBXII(c(emvs1[1:2], xs[i], emvs1[4:5]), SG.dados1)))
}
for (i in 1:length(xd)) {
emv_d[i] = sum(log(pdf.SBXII(c(emvs1[1:3], xd[i], emvs1[5]), SG.dados1)))
}
for (i in 1:length(xc)) {
emv_c[i] = sum(log(pdf.SBXII(c(emvs1[1:4], xc[i]), SG.dados1)))
}
par(mfrow=c(2,3))
par(mar=c(2.8, 2.8, 1.5, .5))
par(mgp=c(1.6, .45, 0))
plot(xa, emv_a, type="l", xlab=expression(a), ylab="log-likelihood", main="(a)")
plot(xb, emv_b, type="l", xlab=expression(b), ylab="log-likelihood", main="(b)")
plot(xs, emv_s, type="l", xlab=expression(s), ylab="log-likelihood", main="(c)")
plot(xd, emv_d, type="l", xlab=expression(d), ylab="log-likelihood", main="(d)")
plot(xc, emv_c, type="l", xlab=expression(c), ylab="log-likelihood", main="(e)")
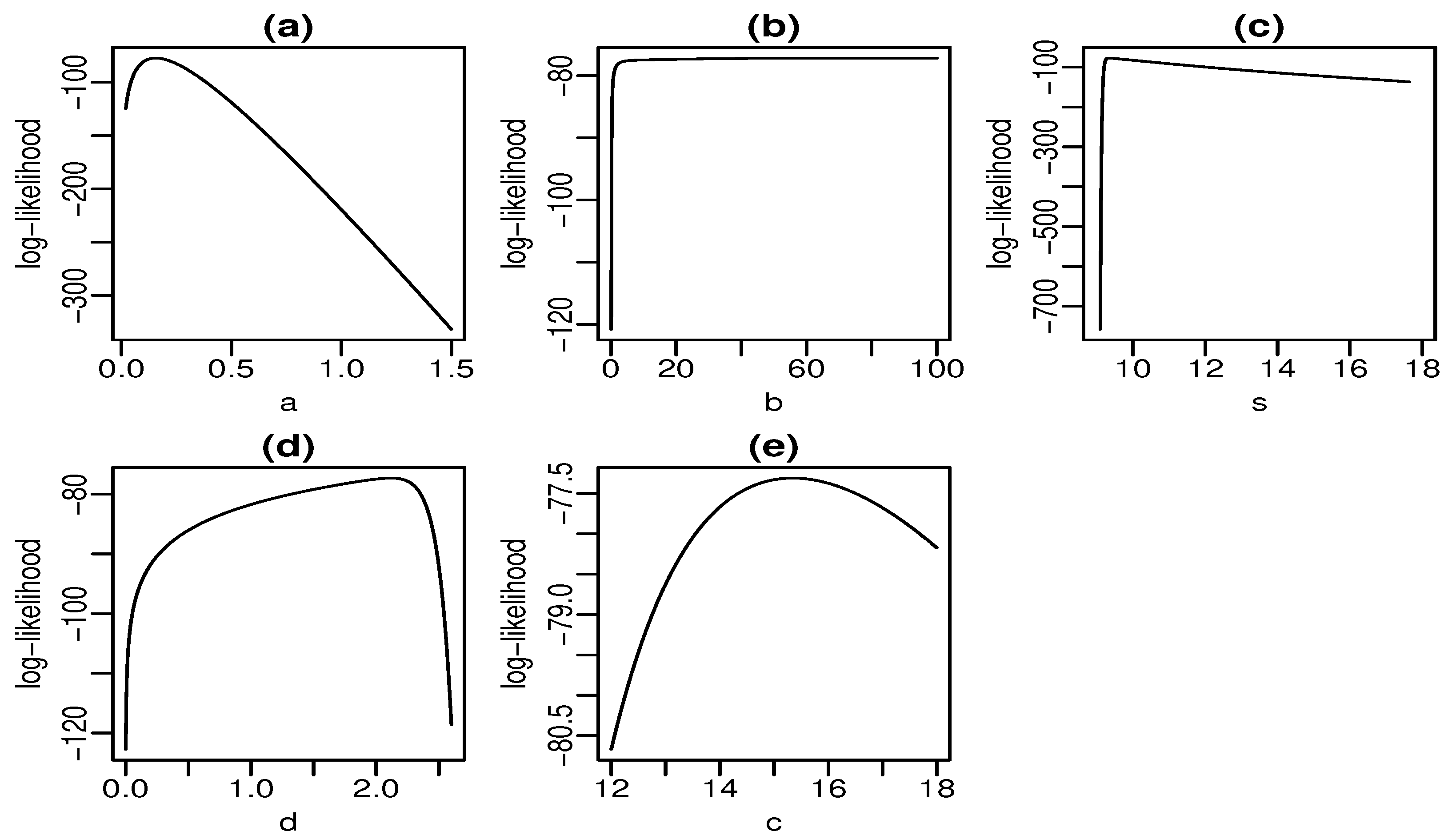
############################################################
# aplication 2
emvs2 <- emv.SBXII2$emv
xa2 <- seq(from=.02, to=1.5, length.out=1000)
xb2 <- seq(from=.09, to=100, length.out=1000)
xs2 <- seq(from=9, to=18, length.out=1000)
xd2 <- seq(from=.0014, to=2.6, length.out=1000)
xc2 <- seq(from=12, to=18, length.out=1000)
emv_a2 = vector()
emv_b2 = vector()
emv_s2 = vector()
emv_d2 = vector()
emv_c2 = vector()
for (i in 1:length(xa2)) {
emv_a2[i] = sum(log(pdf.SBXII(c(xa2[i], emvs2[2:5]), SG.dados2)))
}
for (i in 1:length(xb2)) {
emv_b2[i] = sum(log(pdf.SBXII(c(emvs2[1], xb2[i], emvs2[3:5]), SG.dados2)))
}
for (i in 1:length(xs2)) {
emv_s2[i] = sum(log(pdf.SBXII(c(emvs2[1:2], xs2[i], emvs2[4:5]), SG.dados2)))
}
for (i in 1:length(xd2)) {
emv_d2[i] = sum(log(pdf.SBXII(c(emvs2[1:3], xd2[i], emvs2[5]), SG.dados2)))
}
for (i in 1:length(xc2)) {
emv_c2[i] = sum(log(pdf.SBXII(c(emvs2[1:4], xc2[i]), SG.dados2)))
}
par(mfrow=c(2,3))
par(mar=c(2.8, 2.8, 1.5, .5))
par(mgp=c(1.6, .45, 0))
plot(xa2, emv_a2, type="l", xlab=expression(a), ylab="log-likelihood", main="(a)")
plot(xb2, emv_b2, type="l", xlab=expression(b), ylab="log-likelihood", main="(b)")
plot(xs2, emv_s2, type="l", xlab=expression(s), ylab="log-likelihood", main="(c)")
plot(xd2, emv_d2, type="l", xlab=expression(d), ylab="log-likelihood", main="(d)")
plot(xc2, emv_c2, type="l", xlab=expression(c), ylab="log-likelihood", main="(e)")
Appendix A.3.2. Censored Data
# LSBXII
OPTIM.LSBXII <- function(starts, grad=NULL, cens=NULL, method="BFGS", y, X){
if(is.null(cens)) cens <- rep(1,length(y))
if(length(cens)!=length(y)) stop("variavel censura nao corresponde aos dados")
ll <- function(par, y, X){
a <- par[1]
b <- par[2]
d <- par[3]
sigma <- par[4]
beta <- cbind(par[5:length(par)])
mu <- X %*% beta
z <- (y - mu) / sigma
f1 <- exp(z) * (1+exp(z))^(-1) * log(1+exp(z))^(a-1)
f2 <- exp(-(d*log(1+exp(z)))^b)
# pdf
fd <- b*d^a/(sigma*gamma(a/b)) * f1 * f2
# sdf
sf <- 1 - pgamma((d*log(1+exp(z)))^b, a/b )
lk <- sum(cens*log(fd)) + sum((1-cens)*log(sf))
return(-lk)
}
if(method=="BFGS" | method=="B") {
opt <- optim(starts,fn=ll, gr=grad, method = "BFGS", y=y, X=X, hessian = T)
} else if(method=="SANN" | method=="S") {
opt <- optim(starts,fn=ll, gr=grad, method = "SANN", y=y, X=X, hessian = T)
} else if(method=="Nelder-Mead" | method=="NM") {
opt <- optim(starts,fn=ll, gr=grad, method = "Nelder-Mead", y=y, X=X, hessian = T)
}
emv <- opt$par
erro <- sqrt(diag(solve(opt$hessian)))
valor <- -opt$value
convergencia <- opt$convergence
#
p <- length(starts)
n <- length(y)
#llhat <- -1 * ll(emv, dados)
llhat <- valor
CAIC <- -2 * llhat + 2 * p + 2 * (p * (p + 1))/(n-p - 1)
AIC = -2 * llhat + 2 * p
BIC = -2 * llhat + p * log(n)
HQIC = -2 * llhat + 2 * log(log(n)) * p
crit <- cbind(CAIC, AIC, BIC, HQIC)
list(emv=emv,erro=erro,valor=valor, convergencia=convergencia, criterios=crit)
}
# LBXII
OPTIM.LBXII <- function(starts, grad=NULL, cens=NULL, method="BFGS", y, X){
if(is.null(cens)) cens <- rep(1,length(y))
if(length(cens)!=length(y)) stop("variavel censura nao corresponde aos dados")
ll <- function(par, y, X){
d <- par[1]
sigma <- par[2]
beta <- cbind(par[3:length(par)])
mu <- X %*% beta
z <- (y - mu) / sigma
f1 <- exp(z) * (1+exp(z))^(-1)
f2 <- exp(-d*log(1+exp(z)))
# pdf
fd <- d/(sigma*gamma(1)) * f1 * f2
# sdf
sf <- 1 - pgamma(d*log(1+exp(z)), 1)
#lk <- sum(log(fd)^cens) + sum(log(sf)^(1-cens))
lk <- sum(cens*log(fd)) + sum((1-cens)*log(sf))
return(-lk)
}
if(method=="BFGS" | method=="B") {
opt <- optim(starts,fn=ll, gr=grad, method = "BFGS", y=y, X=X, hessian = T)
} else if(method=="SANN" | method=="S") {
opt <- optim(starts,fn=ll, gr=grad, method = "SANN", y=y, X=X, hessian = T)
} else if(method=="Nelder-Mead" | method=="NM") {
opt <- optim(starts,fn=ll, gr=grad, method = "Nelder-Mead", y=y, X=X, hessian = T)
}
emv <- opt$par
erro <- sqrt(diag(solve(opt$hessian)))
valor <- -opt$value
convergencia <- opt$convergence
#
p <- length(starts)
n <- length(y)
#llhat <- -1 * ll(emv, dados)
llhat <- valor
CAIC <- -2 * llhat + 2 * p + 2 * (p * (p + 1))/(n-p - 1)
AIC = -2 * llhat + 2 * p
BIC = -2 * llhat + p * log(n)
HQIC = -2 * llhat + 2 * log(log(n)) * p
crit <- cbind(CAIC, AIC, BIC, HQIC)
list(emv=emv,erro=erro,valor=valor, convergencia=convergencia, criterios=crit)
}
# logistic
OPTIM.logistic <- function(starts, grad=NULL, cens=NULL, method="BFGS", y, X){
if(is.null(cens)) cens <- rep(1,length(y))
if(length(cens)!=length(y)) stop("variavel censura nao corresponde aos dados")
ll <- function(par, y, X){
d <- 1
sigma <- par[1]
beta <- cbind(par[2:length(par)])
mu <- X %*% beta
z <- (y - mu) / sigma
f1 <- exp(z) * (1+exp(z))^(-1)
f2 <- exp(-d*log(1+exp(z)))
# pdf
fd <- d/(sigma*gamma(1)) * f1 * f2
# sdf
sf <- 1 - pgamma(d*log(1+exp(z)), 1)
#lk <- sum(log(fd)^cens) + sum(log(sf)^(1-cens))
lk <- sum(cens*log(fd)) + sum((1-cens)*log(sf))
return(-lk)
}
if(method=="BFGS" | method=="B") {
opt <- optim(starts,fn=ll, gr=grad, method = "BFGS", y=y, X=X, hessian = T)
} else if(method=="SANN" | method=="S") {
opt <- optim(starts,fn=ll, gr=grad, method = "SANN", y=y, X=X, hessian = T)
} else if(method=="Nelder-Mead" | method=="NM") {
opt <- optim(starts,fn=ll, gr=grad, method = "Nelder-Mead", y=y, X=X, hessian = T)
}
emv <- opt$par
erro <- sqrt(diag(solve(opt$hessian)))
valor <- -opt$value
convergencia <- opt$convergence
#
p <- length(starts)
n <- length(y)
#llhat <- -1 * ll(emv, dados)
llhat <- valor
CAIC <- -2 * llhat + 2 * p + 2 * (p * (p + 1))/(n-p - 1)
AIC = -2 * llhat + 2 * p
BIC = -2 * llhat + p * log(n)
HQIC = -2 * llhat + 2 * log(log(n)) * p
crit <- cbind(CAIC, AIC, BIC, HQIC)
list(emv=emv,erro=erro,valor=valor, convergencia=convergencia, criterios=crit)
}
# dados
# melanona
melanona <- read.table("melanona.txt", header = T)
# melanona
# chutes inciais para betas
XX <- cbind(1, melanona$nodulo)
yy <- melanona$tempo
beta.star <- solve(t(XX)%*%XX)%*% t(XX)%*%yy
beta.star
reg.LSBXII <- OPTIM.LSBXII(c(1,1,1,2,beta.star), cens=melanona$censur, y=yy, X=XX)
reg.LSBXII
# LBXII
reg.LBXII <- OPTIM.LBXII(c(1.4, 2.8, beta.star), cens=melanona$censur, y=yy, X=XX, method = "NM")
reg.LBXII
# logistic
reg.logistic <- OPTIM.logistic(c(2, beta.star), cens=melanona$censur, y=yy, X=XX)
reg.logistic
# LR teste
# LSBXII vs LBXII
w1 <- reg.LSBXII$valor - reg.LBXII$valor
restricao1 <- length(reg.LSBXII$emv) - length(reg.LBXII$emv)
w1.pvalue <- 1 - pchisq(2*w1, restricao1)
# LSBXII vs Logistic
w2 <- reg.LSBXII$valor - reg.logistic$valor
restricao2 <- length(reg.LSBXII$emv) - length(reg.logistic$emv)
w2.pvalue <- 1 - pchisq(2*w2, restricao2)
# Table (summary)
tab <- rbind(
# LSBXII
reg.LSBXII$emv, reg.LSBXII$erro, 2*pnorm(-abs(reg.LSBXII$emv/reg.LSBXII$erro)),
# LBXII
c(NA, NA, reg.LBXII$emv), c(NA, NA, reg.LBXII$erro),
2*pnorm(-abs(c(NA, NA, reg.LBXII$emv)/c(NA, NA, reg.LBXII$erro))),
# logistic
c(NA, NA, NA, reg.logistic$emv), c(NA, NA, NA, reg.logistic$erro),
2*pnorm(-abs(c(NA, NA, NA, reg.logistic$emv)/c(NA, NA, NA, reg.logistic$erro)))
)
rownames(tab) <- c("LSBXII", " ", " ", "LBXII", " ", " ", "Logistic", " ", " ")
tab
# criteria
crit <- rbind(
# LSBXII
reg.LSBXII$criterios,
# LBXII
reg.LBXII$criterios,
# logistic
reg.logistic$criterios
)
rownames(crit) <- c("LSBXII", "LBXII", "Logistic")
crit
# LR
LR.table <- rbind(c(NA, w1, w1.pvalue), c(NA, w2, w2.pvalue))
rownames(LR.table) <- c("LSBXII vs LBXII", "LSBXII vs Logistic")
colnames(LR.table) <- c("hyphotesis", "LR", "p-value")
LR.table