Emerging Technologies for the Assessment of Natural Killer Cell Activity
Abstract
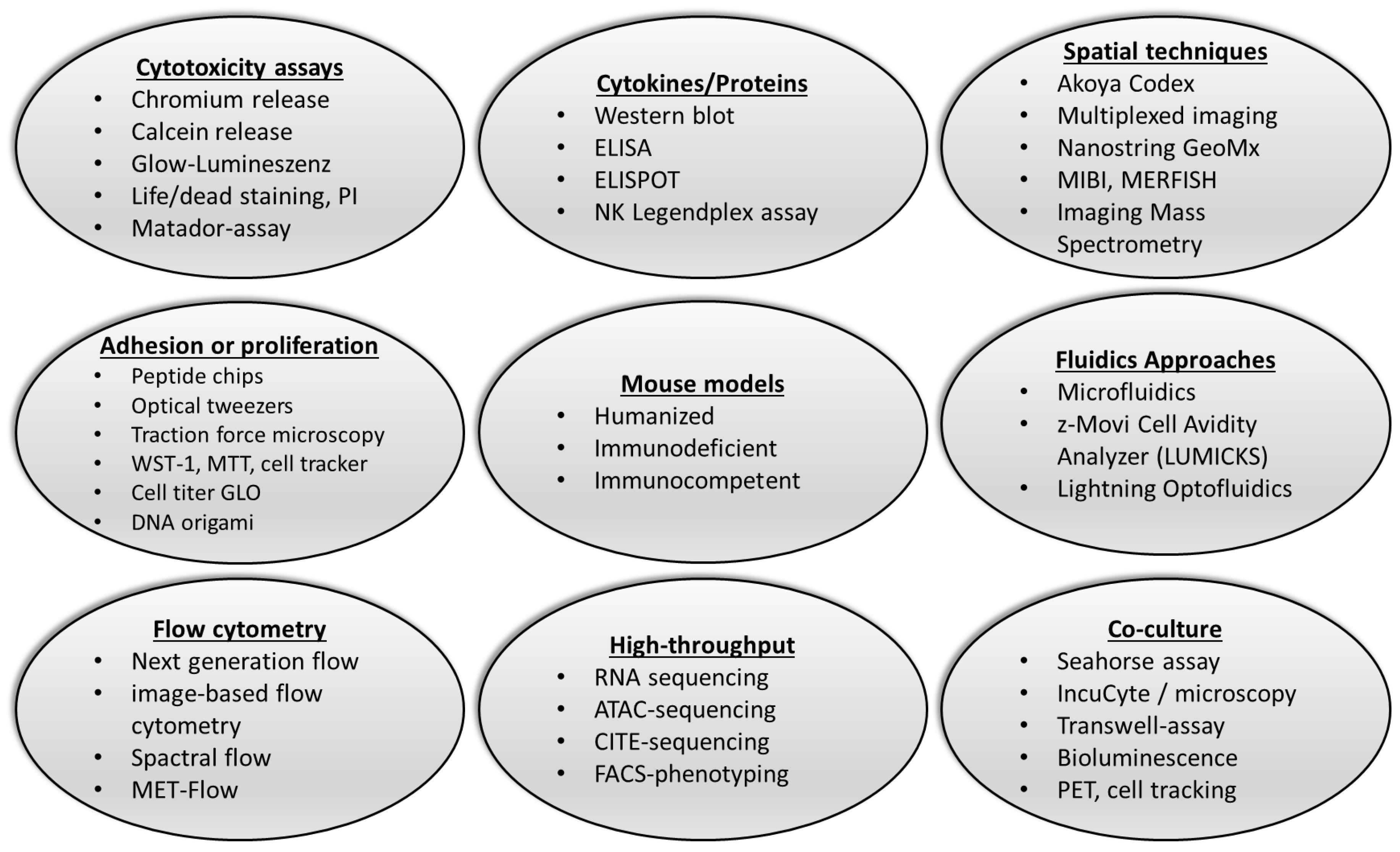
:1. Introduction
2. Traditional Methods Like Chromium Release Assays Assess NK Cell Function
3. Novel Approaches to the Detection of NK Cell Function
3.1. Flow Cytometry as Tool to Characterize NK Cell Function and Metabolic State
3.2. Impedance-Based Real-Time Cell Analysis Systems for Label-Free Monitoring of Interactions Between NK Cells and Target Cells
3.3. Cutting-Edge Microfluidic Approaches to the Analysis of Individual NK Cell–Target Cell Interactions
3.4. Bioluminescence Cell Tracking and PET Techniques Enable Real-Time Monitoring of NK Cell Activity
3.5. Advanced Multiplexed Imaging Systems for NK Cell Functionality and Metabolism
3.6. High-Throughput Screening Platforms for NK Cell Functional Assessment
3.7. Murine Models for Studying NK Cell Biology and Function
4. Discussion
5. Conclusions
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, C.; Siebert, J.R.; Burns, R.; Gerbec, Z.J.; Bonacci, B.; Rymaszewski, A.; Rau, M.; Riese, M.J.; Rao, S.; Carlson, K.-S.; et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 2019, 10, 3931. [Google Scholar] [CrossRef] [PubMed]
- Netskar, H.; Pfefferle, A.; Goodridge, J.P.; Sohlberg, E.; Dufva, O.; Teichmann, S.A.; Brownlie, D.; Michaëlsson, J.; Marquardt, N.; Clancy, T.; et al. Pan-cancer profiling of tumor-infiltrating natural killer cells through transcriptional reference mapping. Nat. Immunol. 2024, 25, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Terunuma, H.; Nieda, M. Immunosurveillance of Cancer and Viral Infections with Regard to Alterations of Human NK Cells Originating from Lifestyle and Aging. Biomedicines 2021, 9, 557. [Google Scholar] [CrossRef] [PubMed]
- Carayol, G.; Robin, C.; Bourhis, J.H.; Bennaceur-Griscelli, A.; Chouaib, S.; Coulombel, L.; Caignard, A. NK cells differentiated from bone marrow, cord blood and peripheral blood stem cells exhibit similar phenotype and functions. Eur. J. Immunol. 1998, 28, 1991–2002. [Google Scholar] [CrossRef]
- Roda-Navarro, P. Assembly and function of the natural killer cell immune synapse. Front. Biosci. (Landmark Ed.) 2009, 14, 621–633. [Google Scholar] [CrossRef]
- Chockley, P.J.; Ibanez-Vega, J.; Krenciute, G.; Talbot, L.J.; Gottschalk, S. Synapse-tuned CARs enhance immune cell anti-tumor activity. Nat. Biotechnol. 2023, 41, 1434–1445. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Fenis, A.; Demaria, O.; Gauthier, L.; Vivier, E.; Narni-Mancinelli, E. New immune cell engagers for cancer immunotherapy. Nat. Rev. Immunol. 2024, 24, 471–486. [Google Scholar] [CrossRef]
- Zheng, X.; Qian, Y.; Fu, B.; Jiao, D.; Jiang, Y.; Chen, P.; Shen, Y.; Zhang, H.; Sun, R.; Tian, Z.; et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat. Immunol. 2019, 20, 1656–1667. [Google Scholar] [CrossRef]
- Ran, G.h.; Lin, Y.q.; Tian, L.; Zhang, T.; Yan, D.m.; Yu, J.h.; Deng, Y.c. Natural killer cell homing and trafficking in tissues and tumors: From biology to application. Signal Transduct. Target. Ther. 2022, 7, 205. [Google Scholar] [CrossRef]
- Vivier, E.; Rebuffet, L.; Narni-Mancinelli, E.; Cornen, S.; Igarashi, R.Y.; Fantin, V.R. Natural killer cell therapies. Nature 2024, 626, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Gao, X.; Zhang, L.; Yang, E.; Li, Y.; Yu, L. The Advances and Challenges of NK Cell-Based Cancer Immunotherapy. Curr. Oncol. 2021, 28, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Carotta, S. Targeting NK Cells for Anticancer Immunotherapy: Clinical and Preclinical Approaches. Front. Immunol. 2016, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front. Immunol. 2015, 6, 368. [Google Scholar] [CrossRef]
- Gauthier, L.; Morel, A.; Anceriz, N.; Rossi, B.; Blanchard-Alvarez, A.; Grondin, G.; Trichard, S.; Cesari, C.; Sapet, M.; Bosco, F.; et al. Multifunctional Natural Killer Cell Engagers Targeting NKp46 Trigger Protective Tumor Immunity. Cell 2019, 177, 1701–1713.e1716. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; He, Z.; Li, L.; Liu, S.; Jiang, M.; Zhao, B.; Deng, M.; Wang, W.; Mi, X.; et al. Breakthrough of solid tumor treatment: CAR-NK immunotherapy. Cell Death Discov. 2024, 10, 40. [Google Scholar] [CrossRef]
- Peng, L.; Sferruzza, G.; Yang, L.; Zhou, L.; Chen, S. CAR-T and CAR-NK as cellular cancer immunotherapy for solid tumors. Cell. Mol. Immunol. 2024, 21, 1089–1108. [Google Scholar] [CrossRef]
- Marin, D.; Li, Y.; Basar, R.; Rafei, H.; Daher, M.; Dou, J.; Mohanty, V.; Dede, M.; Nieto, Y.; Uprety, N.; et al. Safety, efficacy and determinants of response of allogeneic CD19-specific CAR-NK cells in CD19+ B cell tumors: A phase 1/2 trial. Nat. Med. 2024, 30, 772–784. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Kerbauy, L.N.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef]
- Pomeroy, E.J.; Hunzeker, J.T.; Kluesner, M.G.; Lahr, W.S.; Smeester, B.A.; Crosby, M.R.; Lonetree, C.L.; Yamamoto, K.; Bendzick, L.; Miller, J.S.; et al. A Genetically Engineered Primary Human Natural Killer Cell Platform for Cancer Immunotherapy. Mol. Ther. 2020, 28, 52–63. [Google Scholar] [CrossRef]
- Poznanski, S.M.; Singh, K.; Ritchie, T.M.; Aguiar, J.A.; Fan, I.Y.; Portillo, A.L.; Rojas, E.A.; Vahedi, F.; El-Sayes, A.; Xing, S.; et al. Metabolic flexibility determines human NK cell functional fate in the tumor microenvironment. Cell Metab. 2021, 33, 1205–1220.e1205. [Google Scholar] [CrossRef] [PubMed]
- Gurshaney, S.; Morales-Alvarez, A.; Ezhakunnel, K.; Manalo, A.; Huynh, T.-H.; Abe, J.-I.; Le, N.-T.; Weiskopf, D.; Sette, A.; Lupu, D.S.; et al. Metabolic dysregulation impairs lymphocyte function during severe SARS-CoV-2 infection. Commun. Biol. 2023, 6, 374. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.L.; Finlay, D.K. Immunometabolism and natural killer cell responses. Nat. Rev. Immunol. 2019, 19, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Jhaveri, N.; Ben Cheikh, B.; Nikulina, N.; Ma, N.; Klymyshyn, D.; DeRosa, J.; Mihani, R.; Pratapa, A.; Kassim, Y.; Bommakanti, S.; et al. Mapping the Spatial Proteome of Head and Neck Tumors: Key Immune Mediators and Metabolic Determinants in the Tumor Microenvironment. GEN Biotechnol. 2023, 2, 418–434. [Google Scholar] [CrossRef]
- Alter, G.; Malenfant, J.M.; Altfeld, M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 2004, 294, 15–22. [Google Scholar] [CrossRef]
- Crinier, A.; Milpied, P.; Escalière, B.; Piperoglou, C.; Galluso, J.; Balsamo, A.; Spinelli, L.; Cervera-Marzal, I.; Ebbo, M.; Girard-Madoux, M.; et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49, 971–986.e975. [Google Scholar] [CrossRef]
- Hsieh, W.-C.; Budiarto, B.R.; Wang, Y.-F.; Lin, C.-Y.; Gwo, M.-C.; So, D.K.; Tzeng, Y.-S.; Chen, S.-Y. Spatial multi-omics analyses of the tumor immune microenvironment. J. Biomed. Sci. 2022, 29, 96. [Google Scholar] [CrossRef]
- Schürch, C.M.; Bhate, S.S.; Barlow, G.L.; Phillips, D.J.; Noti, L.; Zlobec, I.; Chu, P.; Black, S.; Demeter, J.; McIlwain, D.R.; et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 2020, 182, 1341–1359.e1319. [Google Scholar] [CrossRef]
- Poznanski, S.M.; Ashkar, A.A. What Defines NK Cell Functional Fate: Phenotype or Metabolism? Front. Immunol. 2019, 10, 1414. [Google Scholar] [CrossRef]
- Buller, C.W.; Mathew, S.O. NK Cell Isolation and Cytotoxicity by Radioactive Chromium Release Assay and DELFIA-EuTDA Cytotoxicity Assay. Methods Mol. Biol. 2022, 2463, 221–233. [Google Scholar] [CrossRef]
- Sjaastad, F.V. Assay for Cell Death: Chromium Release Assay of Cytotoxic Ability. JoVE Sci. Educ. Database Immunol. 2023, e10505. Available online: https://www.jove.com/v/10505/chromium-release-assay-for-testing-cytotoxicity (accessed on 31 July 2024).
- Tognarelli, S.; Jacobs, B.; Staiger, N.; Ullrich, E. Flow Cytometry-based Assay for the Monitoring of NK Cell Functions. J. Vis. Exp. 2016, 116, e54615. [Google Scholar] [CrossRef]
- Lee, S.H.; Shin, D.J.; Kim, Y.; Kim, C.J.; Lee, J.J.; Yoon, M.S.; Uong, T.N.T.; Yu, D.; Jung, J.Y.; Cho, D.; et al. Comparison of Phenotypic and Functional Characteristics Between Canine Non-B, Non-T Natural Killer Lymphocytes and CD3+CD5dimCD21− Cytotoxic Large Granular Lymphocytes. Front. Immunol. 2018, 9, 841. [Google Scholar] [CrossRef] [PubMed]
- Biswas, B.K.; Guru, S.A.; Sumi, M.P.; Jamatia, E.; Gupta, R.K.; Lali, P.; Konar, B.C.; Saxena, A.; Mir, R. Natural Killer Cells Expanded and Preactivated Exhibit Enhanced Antitumor Activity against Different Tumor Cells in Vitro. Asian Pac. J. Cancer Prev. 2020, 21, 1595–1605. [Google Scholar] [CrossRef]
- Coronnello, C.; Busà, R.; Cicero, L.; Comelli, A.; Badami, E. A Radioactive-Free Method for the Thorough Analysis of the Kinetics of Cell Cytotoxicity. J. Imaging 2021, 7, 222. [Google Scholar] [CrossRef]
- Kim, J.M.; Yi, E.; Cho, H.; Choi, W.S.; Ko, D.H.; Yoon, D.H.; Hwang, S.H.; Kim, H.S. Assessment of NK Cell Activity Based on NK Cell-Specific Receptor Synergy in Peripheral Blood Mononuclear Cells and Whole Blood. Int. J. Mol. Sci. 2020, 21, 8112. [Google Scholar] [CrossRef]
- Cossarizza, A.; Chang, H.D.; Radbruch, A.; Abrignani, S.; Addo, R.; Akdis, M.; Andrä, I.; Andreata, F.; Annunziato, F.; Arranz, E.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur. J. Immunol. 2021, 51, 2708–3145. [Google Scholar] [CrossRef]
- Ahl, P.J.; Hopkins, R.A.; Xiang, W.W.; Au, B.; Kaliaperumal, N.; Fairhurst, A.-M.; Connolly, J.E. Met-Flow, a strategy for single-cell metabolic analysis highlights dynamic changes in immune subpopulations. Commun. Biol. 2020, 3, 305. [Google Scholar] [CrossRef]
- Sivori, S.; Della Chiesa, M.; Carlomagno, S.; Quatrini, L.; Munari, E.; Vacca, P.; Tumino, N.; Mariotti, F.R.; Mingari, M.C.; Pende, D.; et al. Inhibitory Receptors and Checkpoints in Human NK Cells, Implications for the Immunotherapy of Cancer. Front. Immunol. 2020, 11, 2156. [Google Scholar] [CrossRef]
- Holicek, P.; Truxova, I.; Kasikova, L.; Vosahlikova, S.; Salek, C.; Rakova, J.; Holubova, M.; Lysak, D.; Cremer, I.; Spisek, R.; et al. Assessment of NK cell-mediated cytotoxicity by flow cytometry after rapid, high-yield isolation from peripheral blood. Methods Enzymol. 2020, 631, 277–287. [Google Scholar] [CrossRef]
- Truong, D.; Lamhamedi-Cherradi, S.; Porter, R.; Krishnan, S.; Swaminathan, J.; Gibson, A.; Lazar, A.; Livingston, J.; Gopalakrishnan, V.; Gordon, N.; et al. Dissociation protocols used for sarcoma tissues bias the transcriptome observed in single-cell and single-nucleus RNA sequencing. BMC Cancer 2023, 23, 488. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Lu, C.; Zhang, B.; Li, H.; Zhao, X.; Chen, H.; Liu, Y.; Cui, X. Advances in metabolic reprogramming of NK cells in the tumor microenvironment on the impact of NK therapy. J. Transl. Med. 2024, 22, 229. [Google Scholar] [CrossRef]
- Keating, S.E.; Zaiatz-Bittencourt, V.; Loftus, R.M.; Keane, C.; Brennan, K.; Finlay, D.K.; Gardiner, C.M. Metabolic Reprogramming Supports IFN-γ Production by CD56bright NK Cells. J. Immunol. 2016, 196, 2552–2560. [Google Scholar] [CrossRef] [PubMed]
- Sohn, H.; Cooper, M.A. Metabolic regulation of NK cell function: Implications for immunotherapy. Immunometabolism 2023, 5, e00020. [Google Scholar] [CrossRef]
- Poznanski, S.M.; Barra, N.G.; Ashkar, A.A.; Schertzer, J.D. Immunometabolism of T cells and NK cells: Metabolic control of effector and regulatory function. Inflamm. Res. 2018, 67, 813–828. [Google Scholar] [CrossRef]
- Schraivogel, D.; Kuhn, T.M.; Rauscher, B.; Rodríguez-Martínez, M.; Paulsen, M.; Owsley, K.; Middlebrook, A.; Tischer, C.; Ramasz, B.; Ordoñez-Rueda, D.; et al. High-speed fluorescence image-enabled cell sorting. Science 2022, 375, 315–320. [Google Scholar] [CrossRef]
- Llufrio, E.M.; Wang, L.; Naser, F.J.; Patti, G.J. Sorting cells alters their redox state and cellular metabolome. Redox Biol. 2018, 16, 381–387. [Google Scholar] [CrossRef]
- Fasbender, F.; Watzl, C. Impedance-based analysis of Natural Killer cell stimulation. Sci. Rep. 2018, 8, 4938. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, X.; Xu, X.; Abassi, Y.A. Dynamic and label-free monitoring of natural killer cell cytotoxic activity using electronic cell sensor arrays. J. Immunol. Methods 2006, 309, 25–33. [Google Scholar] [CrossRef]
- Subedi, N.; Van Eyndhoven, L.C.; Hokke, A.M.; Houben, L.; Van Turnhout, M.C.; Bouten, C.V.C.; Eyer, K.; Tel, J. An automated real-time microfluidic platform to probe single NK cell heterogeneity and cytotoxicity on-chip. Sci. Rep. 2021, 11, 17084. [Google Scholar] [CrossRef]
- Hipolito, J.; Peretz-Soroka, H.; Zhang, M.; Yang, K.; Karimi-Abdolrezaee, S.; Lin, F.; Kung, S.K.P. A New Microfluidic Platform for Studying Natural Killer Cell and Dendritic Cell Interactions. Micromachines 2019, 10, 851. [Google Scholar] [CrossRef] [PubMed]
- Arshavsky-Graham, S.; Segal, E. Lab-on-a-Chip Devices for Point-of-Care Medical Diagnostics. In Microfluidics in Biotechnology; Bahnemann, J., Grünberger, A., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 247–265. [Google Scholar] [CrossRef]
- Utharala, R.; Grab, A.; Vafaizadeh, V.; Peschke, N.; Ballinger, M.; Turei, D.; Tuechler, N.; Ma, W.; Ivanova, O.; Ortiz, A.G.; et al. A microfluidic Braille valve platform for on-demand production, combinatorial screening and sorting of chemically distinct droplets. Nat. Protoc. 2022, 17, 2920–2965. [Google Scholar] [CrossRef]
- Antona, S.; Platzman, I.; Spatz, J.P. Droplet-Based Cytotoxicity Assay: Implementation of Time-Efficient Screening of Antitumor Activity of Natural Killer Cells. ACS Omega 2020, 5, 24674–24683. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Truttschel, R.; Gong, M.; Humayun, M.; Virumbrales-Muñoz, M.; Vitek, R.; Felder, M.; Gillies, S.; Sondel, P.; Wisinski, K.; et al. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. OncoImmunology 2018, 8, 1553477. [Google Scholar] [CrossRef]
- Bogojevic, D.; Chamberlain, M.D.; Barbulovic-Nad, I.; Wheeler, A.R. A digital microfluidic method for multiplexed cell-based apoptosis assays. Lab A Chip 2012, 12, 627–634. [Google Scholar] [CrossRef]
- Kolmar, L.; Autour, A.; Ma, X.; Vergier, B.; Eduati, F.; Merten, C.A. Technological and computational advances driving high-throughput oncology. Trends Cell Biol. 2022, 32, 947–961. [Google Scholar] [CrossRef]
- Carannante, V.; Wiklund, M.; Onfelt, B. In vitro models to study natural killer cell dynamics in the tumor microenvironment. Front. Immunol. 2023, 14, 1135148. [Google Scholar] [CrossRef]
- Mayoh, C.; Mao, J.; Xie, J.; Tax, G.; Chow, S.-O.; Cadiz, R.; Pazaky, K.; Barahona, P.; Ajuyah, P.; Trebilcock, P.; et al. High-Throughput Drug Screening of Primary Tumor Cells Identifies Therapeutic Strategies for Treating Children with High-Risk Cancer. Cancer Res. 2023, 83, 2716–2732. [Google Scholar] [CrossRef]
- Reed, R.G.; Al-Attar, A.; Presnell, S.R.; Lutz, C.T.; Segerstrom, S.C. A longitudinal study of the stability, variability, and interdependencies among late-differentiated T and NK cell subsets in older adults. Exp. Gerontol. 2019, 121, 46–54. [Google Scholar] [CrossRef]
- Medina, M.A.; Fuentes-Villalobos, F.; Quevedo, C.; Aguilera, F.; Riquelme, R.; Rioseco, M.L.; Barria, S.; Pinos, Y.; Calvo, M.; Burbulis, I.; et al. Longitudinal Transcriptional Changes Reveal Genes from the Natural Killer Cell-Mediated Cytotoxicity Pathway as Critical Players Underlying COVID-19 Progression; Cold Spring Harbor Laboratory: New York, NY, USA, 2024. [Google Scholar] [CrossRef]
- Chen, M.; Zhou, K.; Dai, S.-Y.; Tadepalli, S.; Balakrishnan, P.B.; Xie, J.; Rami, F.E.I.; Dai, T.; Cui, L.; Idoyaga, J.; et al. In vivo bioluminescence imaging of granzyme B activity in tumor response to cancer immunotherapy. Cell Chem. Biol. 2022, 29, 1556–1567.e1556. [Google Scholar] [CrossRef]
- Gangadaran, P.; Ahn, B.C. Molecular Imaging: A Useful Tool for the Development of Natural Killer Cell-Based Immunotherapies. Front. Immunol. 2017, 8, 1090. [Google Scholar] [CrossRef] [PubMed]
- Shapovalova, M.; Pyper, S.R.; Moriarity, B.S.; LeBeau, A.M. The Molecular Imaging of Natural Killer Cells. Mol. Imaging 2018, 17, 1536012118794816. [Google Scholar] [CrossRef] [PubMed]
- Ichise, H.; Tsukamoto, S.; Hirashima, T.; Konishi, Y.; Oki, C.; Tsukiji, S.; Iwano, S.; Miyawaki, A.; Sumiyama, K.; Terai, K.; et al. Functional visualization of NK cell-mediated killing of metastatic single tumor cells. Elife 2022, 11, e76269. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, T.M.; Aalipour, A.; Schürch, C.M.; Gambhir, S.S. PET Imaging of the Natural Killer Cell Activation Receptor NKp30. J. Nucl. Med. 2020, 61, 1348–1354. [Google Scholar] [CrossRef]
- Pham, T.T.; Chenoweth, A.; Patel, N.; Banu, A.; Osborn, G.; Blower, P.J.; Karagiannis, S.N.; Ma, M.T. In Vivo PET Imaging of (89)Zr-Labeled Natural Killer Cells and the Modulating Effects of a Therapeutic Antibody. J. Nucl. Med. 2024, 65, 1035–1042. [Google Scholar] [CrossRef]
- Fares, J.; Davis, Z.B.; Rechberger, J.S.; Toll, S.A.; Schwartz, J.D.; Daniels, D.J.; Miller, J.S.; Khatua, S. Advances in NK cell therapy for brain tumors. NPJ Precis. Oncol. 2023, 7, 17. [Google Scholar] [CrossRef]
- Elhanani, O.; Ben-Uri, R.; Keren, L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 2023, 41, 404–420. [Google Scholar] [CrossRef]
- Bost, P.; Schulz, D.; Engler, S.; Wasserfall, C.; Bodenmiller, B. Optimizing multiplexed imaging experimental design through tissue spatial segregation estimation. Nat. Methods 2023, 20, 418–423. [Google Scholar] [CrossRef]
- Einhaus, J.; Rochwarger, A.; Mattern, S.; Gaudillière, B.; Schürch, C.M. High-multiplex tissue imaging in routine pathology—Are we there yet? Virchows Arch. 2023, 482, 801–812. [Google Scholar] [CrossRef]
- Human Tumor Atlas Network. Catching up with multiplexed tissue imaging. Nat. Methods 2022, 19, 259. [Google Scholar] [CrossRef]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Piyadasa, H.; Angelo, M.; Bendall, S.C. Spatial proteomics of tumor microenvironments reveal why location matters. Nat. Immunol. 2023, 24, 565–566. [Google Scholar] [CrossRef] [PubMed]
- De Souza, N.; Zhao, S.; Bodenmiller, B. Multiplex protein imaging in tumour biology. Nat. Rev. Cancer 2024, 24, 171–191. [Google Scholar] [CrossRef] [PubMed]
- Rebuffet, L.; Melsen, J.E.; Escalière, B.; Basurto-Lozada, D.; Bhandoola, A.; Björkström, N.K.; Bryceson, Y.T.; Castriconi, R.; Cichocki, F.; Colonna, M.; et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat. Immunol. 2024, 25, 1474–1488. [Google Scholar] [CrossRef]
- Vayrynen, J.P.; Haruki, K.; Lau, M.C.; Vayrynen, S.A.; Ugai, T.; Akimoto, N.; Zhong, R.; Zhao, M.; Dias Costa, A.; Borowsky, J.; et al. Spatial Organization and Prognostic Significance of NK and NKT-like Cells via Multimarker Analysis of the Colorectal Cancer Microenvironment. Cancer Immunol. Res. 2022, 10, 215–227. [Google Scholar] [CrossRef]
- Janesick, A.; Shelansky, R.; Gottscho, A.D.; Wagner, F.; Williams, S.R.; Rouault, M.; Beliakoff, G.; Morrison, C.A.; Oliveira, M.F.; Sicherman, J.T.; et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 2023, 14, 8353. [Google Scholar] [CrossRef]
- Flores, B.C.T.; Chawla, S.; Ma, N.; Sanada, C.; Kujur, P.K.; Yeung, R.; Bellon, M.B.; Hukari, K.; Fowler, B.; Lynch, M.; et al. Microfluidic live tracking and transcriptomics of cancer-immune cell doublets link intercellular proximity and gene regulation. Commun. Biol. 2022, 5, 1231. [Google Scholar] [CrossRef]
- Patwa, A.; Yamashita, R.; Long, J.; Risom, T.; Angelo, M.; Keren, L.; Rubin, D.L. Multiplexed imaging analysis of the tumor-immune microenvironment reveals predictors of outcome in triple-negative breast cancer. Commun. Biol. 2021, 4, 852. [Google Scholar] [CrossRef]
- Millian, D.E.; Saldarriaga, O.A.; Wanninger, T.; Burks, J.K.; Rafati, Y.N.; Gosnell, J.; Stevenson, H.L. Cutting-Edge Platforms for Analysis of Immune Cells in the Hepatic Microenvironment-Focus on Tumor-Associated Macrophages in Hepatocellular Carcinoma. Cancers 2022, 14, 1861. [Google Scholar] [CrossRef]
- Monkman, J.; Taheri, T.; Ebrahimi Warkiani, M.; O’Leary, C.; Ladwa, R.; Richard, D.; O’Byrne, K.; Kulasinghe, A. High-Plex and High-Throughput Digital Spatial Profiling of Non-Small-Cell Lung Cancer (NSCLC). Cancers 2020, 12, 3551. [Google Scholar] [CrossRef]
- Kobelt, D.; Walther, W.; Stein, U.S. Real-Time Cell Migration Monitoring to Analyze Drug Synergism in the Scratch Assay Using the IncuCyte System. Methods Mol. Biol. 2021, 2294, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, D.; Hippen, K.L.; Lemire, A.; Hying, S.; Luo, X.; Lenvik, T.; Curtsinger, J.; Davis, Z.; Zhang, B.; Cooley, S.; et al. Adaptive NK Cells Resist Regulatory T-cell Suppression Driven by IL37. Cancer Immunol. Res. 2018, 6, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Poggi, A. Generation of Tumor Spheroids to Evaluate T Cell and NK Cell Cytotoxicity. Curr. Protoc. 2022, 2, e366. [Google Scholar] [CrossRef] [PubMed]
- Herms, A.; Fernandez-Antoran, D.; Alcolea, M.P.; Kalogeropoulou, A.; Banerjee, U.; Piedrafita, G.; Abby, E.; Valverde-Lopez, J.A.; Ferreira, I.S.; Caseda, I.; et al. Self-sustaining long-term 3D epithelioid cultures reveal drivers of clonal expansion in esophageal epithelium. Nat. Genet. 2024, 56, 2158–2173. [Google Scholar] [CrossRef]
- Li, Y.; Hermanson, D.L.; Moriarity, B.S.; Kaufman, D.S. Human iPSC-Derived Natural Killer Cells Engineered with Chimeric Antigen Receptors Enhance Anti-tumor Activity. Cell Stem Cell 2018, 23, 181–192.e185. [Google Scholar] [CrossRef]
- Damodharan, S.N.; Walker, K.L.; Forsberg, M.H.; McDowell, K.A.; Bouchlaka, M.N.; Drier, D.A.; Sondel, P.M.; DeSantes, K.B.; Capitini, C.M. Analysis of ex vivo expanded and activated clinical-grade human NK cells after cryopreservation. Cytotherapy 2020, 22, 450–457. [Google Scholar] [CrossRef]
- Stefańczyk, S.A.; Hagelstein, I.; Lutz, M.S.; Müller, S.; Holzmayer, S.J.; Jarjour, G.; Zekri, L.; Heitmann, J.S.; Salih, H.R.; Märklin, M. Induction of NK cell reactivity against acute myeloid leukemia by Fc-optimized CD276 (B7-H3) antibody. Blood Cancer J. 2024, 14, 67. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef]
- Freund-Brown, J.; Chirino, L.; Kambayashi, T. Strategies to enhance NK cell function for the treatment of tumors and infections. Crit. Rev. Immunol. 2018, 38, 105–130. [Google Scholar] [CrossRef]
- Grandi, F.C.; Modi, H.; Kampman, L.; Corces, M.R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 2022, 17, 1518–1552. [Google Scholar] [CrossRef]
- Hu, Y.; Wan, S.; Luo, Y.; Li, Y.; Wu, T.; Deng, W.; Jiang, C.; Jiang, S.; Zhang, Y.; Liu, N.; et al. Benchmarking algorithms for single-cell multi-omics prediction and integration. Nat. Methods 2024. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.I.; Li, J.H.; Riggan, L.; Chen, B.; Tafti, R.Y.; Chin, S.; Ma, F.; Pellegrini, M.; Hrncir, H.; Arnold, A.P.; et al. The X-linked epigenetic regulator UTX controls NK cell-intrinsic sex differences. Nat. Immunol. 2023, 24, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Wahlen, S.; Matthijssens, F.; Van Loocke, W.; Taveirne, S.; Kiekens, L.; Persyn, E.; Van Ammel, E.; De Vos, Z.; De Munter, S.; Matthys, P.; et al. The transcription factor RUNX2 drives the generation of human NK cells and promotes tissue residency. eLife 2022, 11, e80320. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Kang, W.; Jiang, S.; Li, K.; Ray, S.; Luther, E.; Ivanov, A.R.; Fu, Y.; Konry, T. Machine learning-aided quantification of antibody-based cancer immunotherapy by natural killer cells in microfluidic droplets. Lab. Chip 2020, 20, 2317–2327. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lastra, S.; Di Santo, J.P. Modeling Natural Killer Cell Targeted Immunotherapies. Front. Immunol. 2017, 8, 370. [Google Scholar] [CrossRef]
- Kim, J.T.; Bresson-Tan, G.; Zack, J.A. Current Advances in Humanized Mouse Models for Studying NK Cells and HIV Infection. Microorganisms 2023, 11, 1984. [Google Scholar] [CrossRef]
- Brown, Z.J.; Heinrich, B.; Greten, T.F. Mouse models of hepatocellular carcinoma: An overview and highlights for immunotherapy research. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 536–554. [Google Scholar] [CrossRef]
- Niu, Z.; Zhou, X.; Li, Y.; Jin, J.; Shen, Y. 17 Humanized NKP46 mouse models for testing novel NK cell-based immunotherapies. J. ImmunoTherapy Cancer 2023, 11 (Suppl. S1), A19. [Google Scholar] [CrossRef]
- Chu, Y.; Hochberg, J.; Yahr, A.; Ayello, J.; van de Ven, C.; Barth, M.; Czuczman, M.; Cairo, M.S. Targeting CD20+ Aggressive B-cell Non-Hodgkin Lymphoma by Anti-CD20 CAR mRNA-Modified Expanded Natural Killer Cells In Vitro and in NSG Mice. Cancer Immunol. Res. 2015, 3, 333–344. [Google Scholar] [CrossRef]
- Katano, I.; Nishime, C.; Ito, R.; Kamisako, T.; Mizusawa, T.; Ka, Y.; Ogura, T.; Suemizu, H.; Kawakami, Y.; Ito, M.; et al. Long-term maintenance of peripheral blood derived human NK cells in a novel human IL-15- transgenic NOG mouse. Sci. Rep. 2017, 7, 17230. [Google Scholar] [CrossRef]
- Nagatani, M.; Kodera, T.; Suzuki, D.; Igura, S.; Fukunaga, Y.; Kanemitsu, H.; Nakamura, D.; Mochizuki, M.; Kemi, M.; Tamura, K.; et al. Comparison of biological features between severely immuno-deficient NOD/Shi-scid Il2rg(null) and NOD/LtSz-scid Il2rg(null) mice. Exp. Anim. 2019, 68, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Rascle, P.; Woolley, G.; Jost, S.; Manickam, C.; Reeves, R.K. NK cell education: Physiological and pathological influences. Front. Immunol. 2023, 14, 1087155. [Google Scholar] [CrossRef] [PubMed]
- Ireson, C.R.; Alavijeh, M.S.; Palmer, A.M.; Fowler, E.R.; Jones, H.J. The role of mouse tumour models in the discovery and development of anticancer drugs. Br. J. Cancer 2019, 121, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Mace, E.M.; Orange, J.S. Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol. Rev. 2019, 287, 202–225. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Zhang, H.; Meng, Y.; Jiao, S.; An, L.; Zhou, Z. Modeling human gastric cancers in immunocompetent mice. Cancer Biol. Med. 2024, 21, 553–570. [Google Scholar] [CrossRef]
- Zeng, Z.; Wong, C.J.; Yang, L.; Ouardaoui, N.; Li, D.; Zhang, W.; Gu, S.; Zhang, Y.; Liu, Y.; Wang, X.; et al. TISMO: Syngeneic mouse tumor database to model tumor immunity and immunotherapy response. Nucleic Acids Res. 2022, 50, D1391–D1397. [Google Scholar] [CrossRef]
- Lampreht Tratar, U.; Horvat, S.; Cemazar, M. Transgenic Mouse Models in Cancer Research. Front. Oncol. 2018, 8, 268. [Google Scholar] [CrossRef]
- Clinthorne, J.F.; Beli, E.; Duriancik, D.M.; Gardner, E.M. NK cell maturation and function in C57BL/6 mice are altered by caloric restriction. J. Immunol. 2013, 190, 712–722. [Google Scholar] [CrossRef]
- Glasner, A.; Isaacson, B.; Viukov, S.; Neuman, T.; Friedman, N.; Mandelboim, M.; Sexl, V.; Hanna, J.H.; Mandelboim, O. Increased NK cell immunity in a transgenic mouse model of NKp46 overexpression. Sci. Rep. 2017, 7, 13090. [Google Scholar] [CrossRef]
- Williams, B.A.; Wang, X.H.; Leyton, J.V.; Maghera, S.; Deif, B.; Reilly, R.M.; Minden, M.D.; Keating, A. CD16+NK-92 and anti-CD123 monoclonal antibody prolongs survival in primary human acute myeloid leukemia xenografted mice. Haematologica 2018, 103, 1720–1729. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Guan, D.; Wang, S.; Chai, L.Y.A.; Xu, S.; Lam, K.P. Glycolysis and Oxidative Phosphorylation Play Critical Roles in Natural Killer Cell Receptor-Mediated Natural Killer Cell Functions. Front. Immunol. 2020, 11, 202. [Google Scholar] [CrossRef] [PubMed]
- van der Windt, G.J.W.; Chang, C.H.; Pearce, E.L. Measuring Bioenergetics in T Cells Using a Seahorse Extracellular Flux Analyzer. Curr. Protoc. Immunol. 2016, 113, 3.16b.11–13.16b.14. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, C.; Le, A. The limitless applications of single-cell metabolomics. Curr. Opin. Biotechnol. 2021, 71, 115–122. [Google Scholar] [CrossRef]
- Campbell, K.S.; Hasegawa, J. Natural killer cell biology: An update and future directions. J. Allergy Clin. Immunol. 2013, 132, 536–544. [Google Scholar] [CrossRef]
- Weber, L.K.; Isse, A.; Rentschler, S.; Kneusel, R.E.; Palermo, A.; Hubbuch, J.; Nesterov-Mueller, A.; Breitling, F.; Loeffler, F.F. Antibody fingerprints in lyme disease deciphered with high density peptide arrays. Eng. Life Sci. 2017, 17, 1078–1087. [Google Scholar] [CrossRef]
- Palermo, A.; Weber, L.K.; Rentschler, S.; Isse, A.; Sedlmayr, M.; Herbster, K.; List, V.; Hubbuch, J.; Löffler, F.F.; Nesterov-Müller, A.; et al. Identification of a Tetanus Toxin Specific Epitope in Single Amino Acid Resolution. Biotechnol. J. 2017, 12, 1700197. [Google Scholar] [CrossRef]
- Jenne, F.; Biniaminov, S.; Biniaminov, N.; Marquardt, P.; Von Bojnicic-Kninski, C.; Popov, R.; Seckinger, A.; Hose, D.; Nesterov-Mueller, A. Resemblance-Ranking Peptide Library to Screen for Binders to Antibodies on a Peptidomic Scale. Int. J. Mol. Sci. 2022, 23, 3515. [Google Scholar] [CrossRef]
- Jenne, F.; Berezkin, I.; Tempel, F.; Schmidt, D.; Popov, R.; Nesterov-Mueller, A. Screening for Primordial RNA-Peptide Interactions Using High-Density Peptide Arrays. Life 2023, 13, 796. [Google Scholar] [CrossRef]
- Antonescu, O.N.; Rasmussen, A.; Damm, N.A.M.; Heidemann, D.F.; Popov, R.; Nesterov-Mueller, A.; Johansson, K.E.; Winther, J.R. Substitutional landscape of a split fluorescent protein fragment using high-density peptide microarrays. PLoS ONE 2021, 16, e0241461. [Google Scholar] [CrossRef]
- Sonnentag, S.J.; Jenne, F.; Orian-Rousseau, V.; Nesterov-Mueller, A. High-throughput screening for cell binding and repulsion peptides on multifunctionalized surfaces. Commun. Biol. 2024, 7, 870. [Google Scholar] [CrossRef] [PubMed]
- Legutki, J.B.; Zhao, Z.-G.; Greving, M.; Woodbury, N.; Johnston, S.A.; Stafford, P. Scalable high-density peptide arrays for comprehensive health monitoring. Nat. Commun. 2014, 5, 4785. [Google Scholar] [CrossRef] [PubMed]
- Chapel, A.; Garcia-Beltran, W.F.; Hölzemer, A.; Ziegler, M.; Lunemann, S.; Martrus, G.; Altfeld, M. Peptide-specific engagement of the activating NK cell receptor KIR2DS1. Sci. Rep. 2017, 7, 2414. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Guedes, S.; Costa, J.P.D.; Kašička, V. Microfluidics for Peptidomics, Proteomics, and Cell Analysis. Nanomaterials 2021, 11, 1118. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, X.; Peng, Y.; Joyee, A.G.; Bai, H.; Wang, S.; Yang, J.; Zhao, W.; Yang, X. Differential modulating effect of natural killer (NK) T cells on interferon-γ production and cytotoxic function of NK cells and its relationship with NK subsets in Chlamydia muridarum infection. Immunology 2011, 134, 172–184. [Google Scholar] [CrossRef]
- Hood, S.P.; Foulds, G.A.; Imrie, H.; Reeder, S.; McArdle, S.E.B.; Khan, M.; Pockley, A.G. Phenotype and Function of Activated Natural Killer Cells from Patients with Prostate Cancer: Patient-Dependent Responses to Priming and IL-2 Activation. Front. Immunol. 2019, 9, 3169. [Google Scholar] [CrossRef]
- Maerkle, F.; Loeffler, F.F.; Schillo, S.; Foertsch, T.; Muenster, B.; Striffler, J.; Schirwitz, C.; Bischoff, F.R.; Breitling, F.; Nesterov-Mueller, A. High-density peptide arrays with combinatorial laser fusing. Adv. Mater. 2014, 26, 3730–3734. [Google Scholar] [CrossRef]
- Maia, A.; Tarannum, M.; Lerias, J.R.; Piccinelli, S.; Borrego, L.M.; Maeurer, M.; Romee, R.; Castillo-Martin, M. Building a Better Defense: Expanding and Improving Natural Killer Cells for Adoptive Cell Therapy. Cells 2024, 13, 451. [Google Scholar] [CrossRef]
- Lim, O.; Jung, M.Y.; Hwang, Y.K.; Shin, E.C. Present and Future of Allogeneic Natural Killer Cell Therapy. Front. Immunol. 2015, 6, 286. [Google Scholar] [CrossRef]
- Page, A.; Chuvin, N.; Valladeau-Guilemond, J.; Depil, S. Development of NK cell-based cancer immunotherapies through receptor engineering. Cell. Mol. Immunol. 2024, 21, 315–331. [Google Scholar] [CrossRef]
- Hong, M.; Chen, Y.Y. Killer fatigue: Transition to NK-cell-like phenotype is a signature of CAR-T cell exhaustion. Cell 2021, 184, 6017–6019. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, J. Emerging roles of CAR-NK cell therapies in tumor immunotherapy: Current status and future directions. Cell Death Discov. 2024, 10, 318. [Google Scholar] [CrossRef]

| Technique | Required Cell Count | Cell Isolation | Staining Technique | Kinetics | Long-Term Culture | Interaction with Microenvironment | Ref. |
|---|---|---|---|---|---|---|---|
| Chromium release assay | +++ | Yes | Radioactive labeling | Limited | No | No | [30,31] |
| FACS | ++ | Yes | Fluorescent antibodies | Good | No | Limited | [32,40] |
| Impedance monitoring | + | Yes | None | Excellent | Yes | Limited | [48] |
| Microfluidics | + | Yes | Varies | Excellent | Limited | Yes | [50,51,52,53,54] |
| Bioluminescence imaging | ++ | Yes | Bioluminescent lable | Good | Limited | Yes | [62] |
| PET | +++ | No | Radiotracker | Good | No | Yes | [66,67] |
| Multiplexed imaging | ++ | No | Antibodies | Limited | No | Yes | [70] |
| Lunaphore | ++ | No | Antibodies | Limited | No | Yes | [68,69] |
| Codex akpya | ++ | No | Antibodies + Dyes | Limited | No | Yes | [70] |
| Geo Mx Profiler | + | No | Antibodies | Limited | No | Yes | [72,73] |
| Cite-seq | + | Yes | Antibody-oligo conjugates | Limited | No | Limited | [90] |
| RNA-seq | + | Yes | None | Limited | No | Limited | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grab, A.L.; Nesterov-Müller, A. Emerging Technologies for the Assessment of Natural Killer Cell Activity. J 2024, 7, 457-471. https://doi.org/10.3390/j7040027
Grab AL, Nesterov-Müller A. Emerging Technologies for the Assessment of Natural Killer Cell Activity. J. 2024; 7(4):457-471. https://doi.org/10.3390/j7040027
Chicago/Turabian StyleGrab, Anna Luise, and Alexander Nesterov-Müller. 2024. "Emerging Technologies for the Assessment of Natural Killer Cell Activity" J 7, no. 4: 457-471. https://doi.org/10.3390/j7040027
APA StyleGrab, A. L., & Nesterov-Müller, A. (2024). Emerging Technologies for the Assessment of Natural Killer Cell Activity. J, 7(4), 457-471. https://doi.org/10.3390/j7040027






