Abstract
The discovery of natural products has been pivotal in drug development, providing a vast reservoir of bioactive compounds from various biological sources. This narrative review addresses a critical research gap: the largely underexplored role of gut microbiota in the mediation and biotransformation of medicinal herb-derived natural products for therapeutic use. By examining the interplay between gut microbiota and natural products, this review highlights the potential of microbiota-mediated biotransformation to unveil novel therapeutic agents. It delves into the mechanisms by which gut microbes modify and enhance the efficacy of natural products, with a focus on herbal medicines from Ayurveda and traditional Chinese medicine, known for their applications in treating metabolic and inflammatory diseases. The review also discusses recent advances in microbiota-derived natural product research, including innovative methodologies such as culturomics, metagenomics, and metabolomics. By exploring the intricate interactions between gut microorganisms and their substrates, this review uncovers new strategies for leveraging gut microbiota-mediated processes in the development of groundbreaking therapeutics.
1. Introduction
Natural products represent a vast reservoir of bioactive compounds that have played a pivotal role in the development of numerous drugs and pharmaceuticals. Natural products, characterized by their origin from living organisms, continue to interest researchers due to their pharmacological diversity, and they play a crucial role in drug development, agriculture, and various industrial applications. The rich biodiversity of our planet offers an immense reservoir of potentially valuable compounds with diverse biological activities. Traditional approaches to natural products discovery have focused on isolation from plant, marine, and microbial sources, yielding a plethora of structurally diverse molecules with diverse pharmacological properties [1]. The process of natural product discovery involves the identification, isolation, characterization, and optimization of these compounds for various applications.
The discovery of natural products and their derivatives has contributed significantly to drug development, providing treatments for a wide range of diseases, including cancer, infectious diseases, and neurological disorders [2]. With the increasing prevalence of drug-resistant pathogens and emerging diseases, there is a growing need for innovative approaches to natural products discovery. Recent advances in microbiome research have shed light on the remarkable metabolic potential of microbial communities residing in the gastrointestinal tract, offering new opportunities for natural products discovery and drug development [3]. This narrative review explores the current relevance of natural products in drug discovery and the relationship between gut microbiota and natural products metabolism and highlights the multifaceted activities of microbial communities in the context of the biotransformation of bioactive compounds derived from medicinal herbs from Ayurveda and traditional Chinese medicine. The field of natural products discovery has witnessed significant advancements fueled by technological innovations, interdisciplinary collaborations, and a deeper understanding of biological systems. This review discusses the future directions and emerging trends in natural products discovery, focusing on cutting-edge methodologies, challenges, and the potential impact on drug development and various industries.
The central research gap this review addresses is the underexplored potential of gut microbiota as a source and mediator of natural products with therapeutic applications, specifically in the context of drug discovery. While significant advances have been made in understanding natural product biosynthesis from traditional sources like plants and marine organisms, the role of gut microbiota in biosynthesizing and biotransforming natural products has not been fully investigated or utilized in drug development. Current research has largely focused on the general metabolic activities of gut microbiota, but there remains a substantial gap in comprehensively understanding how these microbial communities can be routinely harnessed to discover novel bioactive compounds from medicinal herbs, which could lead to innovative therapeutic strategies. This gap is particularly evident in the biotransformation processes where gut microbes convert dietary and herbal components into bioactive metabolites, a process that could reveal new drug candidates with enhanced efficacy and bioavailability. This review provides a detailed examination of the microbiome-mediated modification of medicinal herbs, highlighting the mechanisms by which gut microbes influence the pharmacokinetics and pharmacodynamics of these compounds. This work offers a novel perspective on the integration of microbiome research with natural product discovery, proposing new methodologies and future directions to explore this underexplored territory.
In this narrative review, we provide an in-depth exploration of microbiota-mediated natural products discovery, emphasizing the pivotal role of gut microbiota in the biotransformation of medicinal herbs and its impact on drug development. This review is organized into several key sections. We begin by discussing the traditional and emerging applications of natural products in drug discovery. This is followed by an examination of natural product sources, including terrestrial plants, marine organisms, and microbial sources, with a particular emphasis on the gut microbiome’s role in natural product biotransformation. We then delve into specific examples of the microbiome-mediated modification of medicinal herbs used in Ayurveda and traditional Chinese medicine, highlighting classes such as polyphenols, terpenes, alkaloids, and polysaccharides. Subsequent sections detail the methods and recent advances in natural products discovery, particularly those derived from gut microbiota, using modern techniques like culturomics, metagenomics, and metabolomics. We conclude with a discussion of the future directions in the field, considering the challenges and opportunities that lie ahead, particularly in the context of technological advancements and ethical considerations.
2. Applications and Sources of Natural Products
Natural products remain an essential component of drug discovery efforts, with their diverse chemical structures and biological activities holding promise for the treatment of various diseases. In recent years, advances in technology and methodologies have revitalized interest in natural products research, leading to exciting discoveries and clinical prospects. Natural products have been leveraged for a wide array of applications. For example, they have contributed to the development of numerous drugs, including antibiotics, anticancer agents, and immunosuppressants. In addition, natural products play a crucial role in the development of biopesticides and plant growth regulators and are utilized in the cosmetic and nutraceutical industries due to their perceived health benefits.
Natural products are derived from living organisms. Terrestrial plants remain a primary source of natural products, with secondary metabolites like alkaloids, flavonoids, and terpenoids exhibiting a wide range of bioactivities. In addition, bacteria, fungi, and actinomycetes are prolific producers of bioactive compounds. Importantly, microbial natural products have contributed significantly to antibiotics, immunosuppressants, and anticancer agents [1]. The oceans harbor a vast array of organisms that produce unique compounds. Marine natural products have also shown promise in drug discovery, with applications in antiviral, antibacterial, and anticancer drug development [4]. Importantly, the gut microbiome and its activities, while becoming more appreciated in this context, remains an essentially untapped reservoir of potential natural products.
Recent advances in microbiome research have shed light on the intricate interactions between microorganisms and their hosts, revealing the diverse metabolic pathways involved in natural product biosynthesis. Natural products synthesized and biotransformed by gut microbes are of key importance across many pathways and functions (Figure 1). Gut microbiota-derived natural products can be classified into three groups based on their biogenesis: those biosynthesized de novo by gut microbiota, those derived from the modification of host metabolites and nutrients, or those generated by the biotransformation of host dietary components such as foods and herbal medicines. The relevance of this dietary exposure of medicinal herbs and the mechanisms of action are starting to be revealed. Thus, the current review highlights recent advances in gut microbiota-derived natural products, including strategies for discovering such bioactive compounds, and the gut microbiota-mediated biotransformation of natural products derived from medicinal herbs.
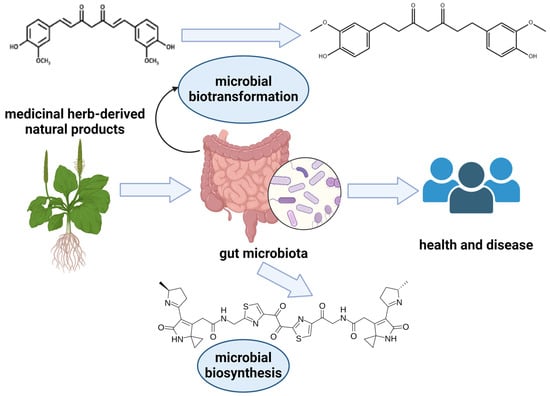
Figure 1.
Medicinal herb-derived natural products and the gut microbiota. The gut microbiota mediates the biosynthesis and biotransformation of natural products derived from medicinal herbs to impact both health and disease. Figure created with Biorender.com; URL accessed on 19 July 2024.
3. Microbiome-Mediated Modification of Medicinal Herbs
Natural products derived from medicinal herbs interact intricately with the gut microbiota, as revealed by advancements in sequencing technologies targeting gut microbiota. These natural products can modify both the composition and structure of the gut microbiota, leading to beneficial health outcomes through either enhancing or inhibiting specific microbial populations. Additionally, certain gut microbes have the ability to metabolize ingested natural products, thereby increasing their bioavailability or transforming them into biologically active compounds within the body.
Numerous studies have established strong connections between the gut microbiota and the therapeutic effects of natural products from medicinal herbs [5]. However, significant questions remain unanswered, particularly concerning the specific gut microbes responsible for mediating the therapeutic effects, especially for natural products that are poorly absorbed orally. Understanding these mechanisms has been catalyzed by advanced culturing and metabolomics approaches, which have provided insights into how the gut microbiota influence various physiological pathways such as the gut–brain axis.
Recent studies have highlighted the role of the gut microbiome in modulating the bioavailability and pharmacological activity of medicinal herbs. Gut microbes can metabolize herbal compounds, leading to the formation of bioactive metabolites with enhanced pharmacological properties [6]. For example, certain bacterial strains can hydrolyze the glycosidic bonds in herbal compounds, releasing aglycones that are more readily absorbed by the body. Additionally, gut microbes can biotransform herbal compounds into secondary metabolites, which may exhibit different pharmacological activities compared with the parent compounds.
Several studies have demonstrated the microbiome-mediated modification of medicinal herbs and their constituents. For instance, the gut microbiota can metabolize the flavonoids, alkaloids, polysaccharides, and terpenoids found in many medicinal herbs (Figure 2) [7]. In one study, gut bacteria could convert flavonoid glycosides into aglycones, increasing their bioavailability and antioxidant activity [8]. Similarly, certain bacterial species have been shown to metabolize alkaloids, such as berberine and sanguinarine, into biologically active metabolites with enhanced pharmacological effects [9].
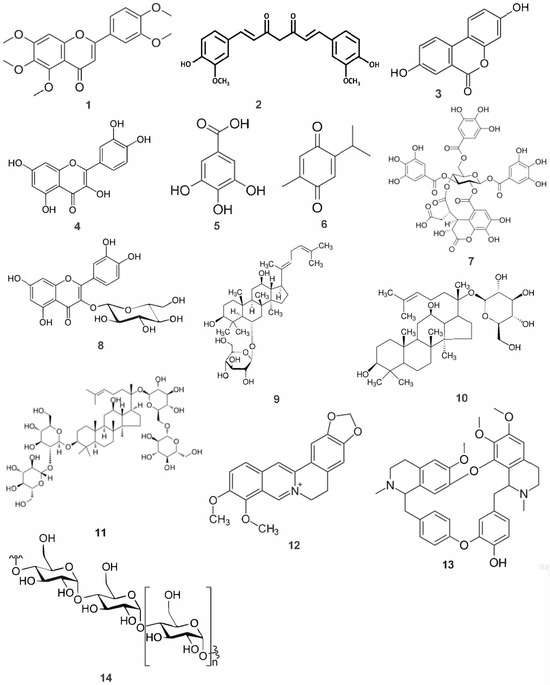
Figure 2.
Structure of medicinal herb-derived metabolites with gut microbiota-mediated action mechanisms. The compounds displayed are 1. sinensetin, 2. curcumin, 3. urolithin A, 4. quercetin, 5. gallic acid, 6. thymoquinone, 7. chebulinic acid, 8. isoquercetin, 9. ginsenoside Rh4, 10. ginsenoside compound K, 11. ginsenoside Rb1, 12. berberine, 13. berbamine, and 14. basic polysaccharide structure.
3.1. Polyphenols
The absorption of polyphenols in the gastrointestinal tract is often limited, which poses difficulties in elucidating their mechanisms of action. Recent studies have begun to identify the processes behind the interactions between polyphenols derived from medicinal herbs and gut microorganisms [10,11]. Animal models have demonstrated that polyphenols derived from herbal medicines can effectively treat metabolic and inflammatory illnesses. For example, mice who were made obese by their diet and given more polymethoxyflavones (PMFs) found in citrus fruits, such as sinensetin, nobiletin, heptamethoxyflavone, and tangeretin, had notable decreases in both body weight and adipose tissue (Figure 2) [10]. PMFs also enhanced the prevalence of Bacteroides in the gut microbiota and impacted the metabolism of BCAAs. In addition, the enrichment of B. ovatus by PMFs contributed to a reduction in metabolic disorders and levels of BCAAs in both the circulation and feces. This study highlights the essential function of the gut microbiota in regulating the equilibrium of BCAAs. A further investigation discovered that phlorizin had anti-obesity properties in diet-induced obese mice via altering the gut flora and elevating levels of short-chain fatty acids (SCFAs) [12]. Theabrownins derived from Pu-erh tea have been discovered to decrease cholesterol levels and modify the gut microbiota by diminishing bile-salt hydrolase activity and enhancing primary bile acids [13]. These modifications are thought to play a role in the metabolic advantages of theabrownins, with the FXR-FGF15 signaling pathway recognized as a crucial factor in lowering cholesterol.
Polyphenols derived from medicinal herbs have exhibited anti-inflammatory properties and have been found to be advantageous in mitigating symptoms associated with inflammatory bowel disease (IBD), which features chronic inflammation of the gut mucosa that is associated with gut dysbiosis [14]. For example, urolithin A, a molecule derived from ellagitannin and generated by bacteria in the gut, decreases inflammation and improves the function of the intestinal barrier by specifically affecting the AhR-Nrf2 signaling pathways (Figure 2) [15]. Ellagitannins are present in many Ayurvedic herbal medicines such as amalaki (Emblica officinalis or Phyllanthus emblica), haritaki (Terminalia chebula), and arjuna (Terminalia arjuna) [16,17]. For example, arjuna extract reduced the expression of pro-inflammatory cytokines, decreased oxidative stress, increased plasma zinc levels, and improved the gut microbiota community structure in a murine model of colitis [18]. Rhein, an anthraquinone and primary constituent of rhubarb, effectively ameliorated chronic colitis in a study using dextran sodium sulfate (Figure 2) [19]. Through fecal microbiota transplantation, the involvement of gut microbiota in the anti-inflammatory effects of rhein has been observed. This process leads to an elevation in Lactobacillus levels, which subsequently reduces the levels of uric acid that harm the functions of the intestinal barrier.
Several Ayurvedic herbs, amalaki (Emblica officinalis), neem (Azadirachta indica), turmeric (Curcuma longa), and tulsi (Ocimum sanctum), contain the flavanol quercetin, which is considered the most potent flavonoid [16,20,21,22]. For example, quercetin-rich plants such as amalaki can promote platelet recovery in serious conditions such as chemotherapy-induced thrombocytopenia (Figure 2) [23]. In an animal model, tulsi exhibited a protective role and reduced biochemical and histopathological changes via antioxidant activities [24]. Quercetin derived from amalaki has shown promise as a drug with antidiabetic and antihyperglycemic effects, influencing glucose, cholesterol, and triglyceride levels, as demonstrated by both in silico and in vivo studies [25]. Phytochemicals in the Ayurvedic formulation triphala (a combination of amalaki (Phyllanthus emblica), bibhitaki (Terminalia bellirica), and haritaki (Terminalia chebula) in equal parts), such as quercetin and gallic acid, foster the growth of beneficial gut bacteria such as Bifidobacteria and Lactobacillus species, while suppressing harmful bacteria like E. coli (Figure 2). Lactic acid bacteria can enzymatically degrade plant tannins, including the gallic acid found in triphala [26]. Triphala polyphenols, such as chebulinic acid, are converted by the gut microbiota into metabolites like urolithins, which can prevent oxidative damage (Figure 2) [27]. Some of the gut bacterial species known to be involved in the biotransformation of quercetin into active metabolites include Bacteroides fragilis, Eubacterium ramulus, Clostridium perfringens, Bacteroides, Bifidobacterium, Lactobacillus, and Streptococcus [28,29]. This suggests that the bioactivity of triphala is mediated by the gut microbiome, enhancing the production and diversity of anti-inflammatory compounds.
Chronic kidney disease (CKD) is another common inflammatory disorder. Isoquercetin, present in many herbal medicines, decreased the formation of indole in the intestine and lowered the levels of indoxyl sulfate in the plasma of both normal and CKD mice (Figure 2) [30]. The quercetin compound decreased the activity of complex I in the electron transport chain of the gut bacteria, resulting in a decrease in the H proton potential and the inhibition of tryptophan transport into the bacteria. This indicates a potentially effective approach to slow down the course of CKD by manipulating the gut microbiota to reduce the levels of uremic toxins originating from the gut. Another example, curcumin, a diarylheptanoid derived from turmeric, has been shown to positively modulate inflammation, lipid peroxidation, and gut microbiota community structure in CKD (Figure 2) [31]. Despite mounting evidence, the influence of curcumin on the intestinal microbiota remains unclear. However, current data suggest that curcumin is biotransformed in the gut to various bioactive metabolites through pathways including demethylation, hydroxylation, and demethoxylation [32]. Many gut microbes, including E. coli, E. fergusonii, Blautia sp., Bifidobacteria longum, Bifidobacteria pseudocatenulaum, Lactobacillus casei, Lactobacillus acidophilus, and Enterococcus faecalis, are biologically relevant in the biotransformation of curcumin [33]. The curcumin metabolites, along with changes in the microbiota composition and diversity induced by curcumin, suggest potential indirect health benefits. Previous research on plant polyphenols has frequently failed to identify the crucial gut bacteria responsible for polyphenol metabolism and the synthesis of active compounds such as bile acids and SCFAs. Future studies should focus on addressing this gap in knowledge.
3.2. Terpenes
Terpenes are bioactive compounds present in many medicinal plants. Recent advances in the study of terpenes with gut microbiota-linked therapeutic benefits have identified critical mechanisms and roles of gut microbiota in regulating host pathways. The essential oil extracted from black cumin seed (Nigella sativa) (common name: kalonji or black cumin seed), commonly used as an edible oil in India and the Middle East, is known for numerous pharmacological benefits, such as antioxidant, anti-inflammatory, antimicrobial, immune-enhancing, and anticancer properties [34]. These beneficial effects are primarily attributed to thymoquinone, a bioactive monoterpene in the oil, which has demonstrated free-radical scavenging activity and inhibition of various inflammatory mediators (Figure 2). In an animal model of cadmium exposure, treatment with black cumin seed oil inhibited the overgrowth of pathogenic Staphylococci, E. coli, Pseudomonas, and Enterococci, which are commonly increased with cadmium toxicity [34]. It was concluded that protective mechanisms were mediated by a combination of major bioactive components including thymoquinone and the diverse fatty acids identified in the seed oil.
Ginsenosides are a well-studied group of glycosylated triterpenes, also known as saponins, that are primary bioactive compounds found in the root Panax ginseng [35]. Ginsenoside Rh4 has demonstrated potential in regulating intestinal inflammation and correcting gut microbiota dysbiosis in colorectal cancer (CRC) (Figure 2) [36]. One strategy to elucidate the mechanisms by which Rh4 modulates the gut microbiota to exert its inhibitory effects on CRC utilized a mouse model along with transcriptomics, genomics, and metabolomics. Additional experiments included antibiotic treatments and fecal microbiota transplantation (FMT) to determine the role of gut microbiota in Rh4’s effects. With this approach, the anti-CRC effects of Rh4 were found to be dependent on the gut microbiota. The compound corrected the dysbiosis of gut microbiota associated with CRC and regulated bile acid metabolism mediated by Akkermansia muciniphila. The study concluded that Rh4 effectively inhibits CRC through a gut microbiota-dependent mechanism. By modulating gut microbiota-mediated bile acid metabolism and promoting the production of ursodeoxycholic acid, Rh4 activates the farnesoid X receptor (FXR) and regulates the TLR4-NF-κB signaling pathway, thereby inhibiting CRC. These findings suggest that Rh4 holds promise as a modulator of gut microbiota for the prevention and treatment of CRC.
Gut microbiota dysbiosis is believed to be linked to inflammatory bowel disease (IBD). Ginsenoside compound K (CK), the primary metabolite of Panax ginseng ginsenosides, has shown efficacy as an anti-inflammatory agent for IBD (Figure 2). Recent findings indicate that CK can suppress the release of proinflammatory cytokines by modulating gut microbiota. Supplementation with CK has been shown to restore gut microbiota balance, particularly by enhancing populations of Lactobacillus and Akkermansia. Moreover, CK significantly increased the levels of tryptophan metabolites derived from Lactobacillus, which can activate the aryl hydrocarbon receptor [37]. Thus, the beneficial effects of CK in alleviating IBD are primarily due to the promotion of Lactobacillus growth and the production of tryptophan metabolites, highlighting CK’s potential as a therapeutic for IBD.
Several studies on ginsenosides have revealed gut microbiota-linked mechanisms and roles of the gut microbiota in regulating the glucose and lipid metabolism of the host. A recent animal study reported that ginsenoside Rb1, compound K, and 20(s)-protopanaxadiol (PPD) attenuate high-fat-diet-induced hyperlipidemia via modulation of the gut microbiota and bile acid metabolism (Figure 2) [38]. Notably, only ginsenoside Rb1 enhanced lipid metabolism by modulating the PPARγ/ACC/FAS signaling pathway and promoting fatty acid β-oxidation. Additionally, all three ginsenosides significantly improved bile acid enterohepatic circulation via the FXR/CYP7A1 pathway. This led to reduced hepatic and serum total bile acids and altered bile acid pool composition, characterized by a decrease in primary/unconjugated bile acids and an increase in conjugated bile acids, which correlated with changes in the gut microbiota. In another study, a ginsenoside extract significantly reduced adiposity and enhanced energy metabolism in obese mice [38]. This effect was associated with an enrichment of Enterococcus faecalis in the gut following oral administration of the extract. The strain of E. faecalis played a role in controlling obesity by producing myristoleic acid, which increased brown adipose tissue activity. However, the specific pathway targeted by the myristoleic acid remained unclear. In studies on medicinal Ganoderma mushrooms, a Ganoderma meroterpene derivative was found to have gut microbiota-associated benefits against metabolic disorders. This was linked to an enrichment of Parabacteroides distasonis. Oral supplementation with live P. distasonis provided metabolic benefits, including reductions in weight gain, hyperglycemia, and hepatic steatosis in mice. Targeted metabolomic analysis indicated that colonization with P. distasonis enhanced the production of lithocholic acid, ursodeoxycholic acid, and succinate in the gut. This subsequently influenced the secondary bile acid-mediated gut–liver axis via the FXR signaling pathway and the succinate-mediated gut–brain axis through the activation of intestinal gluconeogenesis.
3.3. Alkaloids
Several medicinal herb-derived alkaloids, including berberine, piperine, and ajmaline, have exhibited diverse bioactivities with gut microbiota-associated mechanisms. Berberine is an isoquinoline alkaloid derived from various medicinal plants such as Berberis aristata, Berberis aquifolium, Hydrastis canadensis, and Coptis chinensis (Figure 2). In an animal model of glucolipid metabolism disorders, berberine administration increased intestinal barrier function and lowered serum glycolipid levels. Berberine was metabolized by gut microbiota into 12 metabolites, with its primary metabolic pathways including oxidation, demethylation, and hydrogenation. Additionally, berberine significantly improved gut microbiota diversity and promoted the growth of beneficial microbes. Tryptophan metabolites such as indole, indole-3-acetamide, indole-3-acetaldehyde, indole-3-pyruvic acid, and indole-3-acetic acid were altered by berberine treatment. Taken together, the data suggest that berberine alleviates intestinal barrier dysfunction in glucolipid metabolic disorders by modulating gut microbiota and related tryptophan metabolites, suggesting a potential pharmacological mechanism for future therapeutics.
Berberis amurensis contains several alkaloids, including berberine, berbamine, magnoflorine, jatrorrhizine, and palmatine. In an animal model, normal and antibiotic-treated pseudo-germ-free animals were treated with B. amurensis extracts [39]. A total of 46 metabolites were detected in the normal animals with key metabolic pathways including demethylation, dehydrogenation, methylation, hydroxylation, sulfation, and glucuronidation. In contrast, only 29 metabolites were found in the pseudo-germ-free rats, with dehydrogenated and methylated metabolites among those missing. These findings suggest that gut bacteria play a significant role in the metabolism of B. amurensis, which may enhance our understanding of B. amurensis metabolism and its clinical use. In another animal study, the primary components and metabolites were identified in rat intestinal contents and serum samples following the oral administration of a Berberis kansuensis extract to diabetic and pseudo-germ-free diabetic rats. The analysis revealed 16 primary components and 40 metabolites. The main metabolic pathways of the chemical compounds from the B. kansuensis extract included demethylation, desaturation, deglycosylation, reduction, hydroxylation, and various conjugation reactions such as sulfation, glucuronidation, glycosidation, and methylation. Comparing the metabolites between diabetic and pseudo-germ-free diabetic rats revealed that the metabolic transformation of certain components, such as bufotenin, ferulic acid 4-O-β-D-glucopyranoside, magnoflorine, and 8-oxyberberine, was influenced by the gut microbiota. These results highlight the significant role of the gut microbiota in the metabolic biotransformation of the chemical constituents in Berberis kansuensis [40].
In clinical studies, the hypolipidemic effect of berberine was associated with short-chain fatty acid-producing gut microbes such as Alistipes and Blautia [41]. Causality experiments in mice demonstrated that the gut microbiota are both essential and sufficient for mediating the lipid-lowering effects of berberine. Specifically, the absence of Blautia significantly reduced berberine’s ability to lower cholesterol. A clinical trial in type 2 diabetes patients using metagenomics and metabolism methods reported that berberine altered microbial bile acid metabolism [42]. The hypoglycemic effect of berberine was mediated by the inhibition of deoxycholic acid biotransformation by Ruminococcus bromii. Therefore, the data provide a human microbial related mechanism underlying the antidiabetic effect of berberine. Future studies should investigate other gut microbes that may mediate the hypolipidemic and hypoglycemic effects of berberine.
3.4. Polysaccharides
The gut microbiota are crucial for the bioavailability and bioactivity of polysaccharides derived from medicinal herbs. Utilizing a range of carbohydrate-active enzymes, the gut microbiome is essential in the degradation of the complex polysaccharides found in herbal medicines (Figure 2). This degradation produces physiologically active metabolites, such as short-chain fatty acids, and alters the gut microbiota composition and structure to influence host health and disease. These polysaccharides, which serve as beneficial prebiotics, are poorly absorbed in the gut as a result of their high molecular weights and complex polymer structures. However, they significantly influence the gut microbiota and have demonstrated positive effects on conditions such as metabolic syndrome. Our in vitro and clinical work has demonstrated the robust prebiotic potential of medicinal herbs [43,44]. The restructuring of microbial communities with glycan substrates alters community metabolism, potentially impacting host physiology [45]. We analyzed sugar fermentation pathways and predicted products as well as the prebiotic-induced changes in vitamin and amino acid biosynthesis and degradation. The results of the lab model revealed that the sugar composition of the medicinal herbs is not the main driver of the gut microbial changes. The functions of a bacterial gene family called glycosyl hydrolases are more strongly associated with herbal medicine response. These findings underscore the value of combining genome-wide metabolic reconstruction with 16S rRNA sequence-based community profiles to gain insights into community metabolism. This approach also offers a rational method for prioritizing in vivo analyses of prebiotics and medicinal herbs to test hypotheses related to their therapeutic potential in specific diseases. In a pilot clinical trial, the prebiotic effects of turmeric were demonstrated through gut microflora profiles of healthy individuals in a dietary intervention using curcumin tablets, turmeric, or a placebo [46]. The data suggested that the response to turmeric may be due to the metabolism of polysaccharide compounds in the root by a variety of glycosyl hydrolases produced by certain gut species, including Bacteroides, Bifidobacterium, Alistipes, and Parabacteroides, which were found in higher levels in responsive subjects.
Several animal studies have explored the effects of polysaccharides in the context of diseases affecting metabolism. In a high-fat-diet-induced type 2 diabetes model, the effects of Nigella sativa seed polysaccharides on gut microbiota and blood biomarkers were explored [47]. The results demonstrated that high-dose polysaccharides significantly reduced fasting blood glucose, total cholesterol, low-density lipoprotein cholesterol, malondialdehyde, TNF-α, IL-6, and IL-1β. Conversely, it significantly increased insulin, high-density lipoprotein cholesterol, total antioxidant capacity, superoxide dismutase, catalase, and the levels of p-AKT and GLUT4 in the mice. Additionally, high-dose polysaccharides notably increased the abundance of Muribaculaceae and Bacteroides, which had been significantly reduced following streptozotocin treatment. These findings suggest that polysaccharides may ameliorate the diabetic state in mice by modulating the PI3K/AKT signaling pathway and altering the gut microbiota composition. In an immunosuppressive mouse model using cyclophosphamide, the impact of N. sativa seed polysaccharide treatment led to significant increases in the levels of lactate dehydrogenase, acid phosphatase, total antioxidant capacity, superoxide dismutase, IL-2, IL-4, and IL-6, while reducing TNF-α and malondialdehyde levels [48]. Proteomic analysis suggested that the polysaccharide enhances immune responses by modulating key proteins involved in the PI3K and PTEN pathways as well as affecting metabolism-related pathways such as autoimmune diseases and the PI3K-AKT signaling pathway. Gut microbiome analysis suggested that N. sativa polysaccharide improves immune regulation by enhancing the gut microbiota diversity; regulating short-chain fatty acids; and modifying metabolic pathways related to lipid metabolism, polysaccharide synthesis, and signal transduction.
In diet-induced obese mice, polysaccharides from Lyophyllum decastes reduced obesity by altering the gut microbiota community structure and increasing energy expenditure by increasing the abundance of Bacteroides intestinalis and Lactobacillus johnsonii [49]. Supplementation experiments with B. intestinalis and L. johnsonii prevented obesity and hyperlipidemia in obese mice, thus highlighting their causative role in the efficacy of the polysaccharides. In addition, these species increased primary bile acid metabolism and increased secondary bile acid levels in the gut. Thus, one proposed mechanism was the increased energy expenditure in brown adipose tissue driven by the upregulation of the TGR5 pathway activated by secondary bile acids. Further research is needed to identify the specific carbohydrate-active enzymes in the gut microbiota and to more fully understand the mechanisms underlying the effectiveness of polysaccharide-rich medicinal herbs.
The microbiome-mediated modification of medicinal herbs can significantly influence their pharmacokinetic and pharmacodynamic properties. By altering the metabolism and bioavailability of herbal compounds, gut microbes can affect the onset, duration, and intensity of pharmacological effects. Moreover, microbiome-mediated modification may contribute to interindividual variability in response to herbal treatments. Factors such as age, diet, and gut microbiome composition can influence the extent of microbial metabolism and, consequently, the therapeutic efficacy of medicinal herbs.
The gut microbiome plays a critical role in modulating the bioavailability and pharmacological activity of medicinal herbs. By metabolizing herbal compounds, gut microbes can generate bioactive metabolites with enhanced therapeutic potential. By targeting specific gut microbial pathways, it may be possible to enhance the efficacy and safety of herbal treatments. In addition, personalized approaches that take into account individual differences in gut microbiome composition could optimize herbal therapy outcomes [50].
Further research is needed to elucidate the complex interactions between the gut microbiome and medicinal herbs fully. However, these findings hold promise for the development of novel herbal treatments with improved efficacy and safety profiles. The interaction between natural products from medicinal herbs and the gut microbiota is a complex and dynamic process that holds promise for developing targeted therapies. Further research is needed to elucidate the specific roles of gut microbes and the molecular mechanisms underlying their interactions with natural products, particularly in the context of polyphenols, polysaccharides, terpenes, and alkaloids found in herbal medicines.
4. Gut Microbiota Biotransformation of Essential Host Nutrients
The gut microbiota metabolize dietary nutrients into bioactive natural products, and thus provide insights into the effects of food and dietary supplementation on human health. The complex interplay between dietary substrates, gut microbiota, and host metabolism highlights the importance of understanding these interactions for health and disease management. For example, branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, are essential nutrients that play a critical role in protein synthesis and energy metabolism. However, high host serum levels of the BCAAs, which are both nutrients and signaling molecules, are associated with type 2 diabetes risk and insulin resistance [51]. These amino acids are predominantly obtained from protein-rich foods in the diet; however, medicinal herbs such as ashwagandha, shatavari, and amalaki also contain these amino acids. Specific gut bacteria such as Prevotella copri and Bacteroides vulgatus are known to biosynthesize BCAAs within the gut [51]. Conversely, other microbes like Clostridium sporogenes can biotransform BCAAs into immune-modulatory branched short-chain fatty acids such as isobutyrate, 2-methylbutyrate, and isovalerate. These interactions underscore the role of the gut microbiome in modulating BCAA levels and their subsequent effects on host health and disease [52].
Additionally, gut microbes play a significant role in the metabolism of other amino acids such as the aromatic group, which includes tryptophan, tyrosine, and phenylalanine. For example, the gut species Lactobacillus plantarum and Bacteroides ovatus can generate 4-ethylphenyl sulfate (4EPS) from tyrosine, which can impact brain health and behavior [53]. Tryptophan, another essential aromatic amino acid, is metabolized by gut bacteria into various indole derivatives, such as indole-3-lactic acid (ILA), indole-3-propionic acid (IPA), indole-3-acetic acid (IAA), and indole-3-aldehyde (IAld). These metabolites have been associated with inflammatory bowel disease (IBD), tumors, and neuropsychiatric conditions. ILA in particular has been shown to modulate the immune system by increasing the production of the immunoregulatory factor galectin-1 in T helper cells, which reduces T-cell activation. Studies with the ILA-producing Bifidobacterium infantis EVC001 demonstrated similar immune system effects in breastfed infants [54].
Tryptophan metabolites are also involved in tumor development. For instance, the ILA-producing Lactobacillus gallinarum and ILA itself can induce apoptosis and decrease the incidence of colorectal tumors in certain mice models. IPA and IPA-producing gut bacteria have been linked to axonal regeneration, particularly through intermittent fasting therapy, as oral administration of IPA can promote axon regeneration and functional recovery via an immune-regulatory mechanism. An example of host nutrient biotransformation in the context of essential amino acids is that Streptococcus mutans and Eggerthella lenta metabolize histidine into imidazole propionate, a compound found in higher levels in individuals with type 2 diabetes (T2D) that is linked to insulin resistance. This compound impairs oral glucose tolerance through the activation of the p38γ MAPK/p62/mTORC1 signaling pathway. Identifying the specific gut bacteria involved in the metabolism of nutrients in healthy and diseased states will inform the design of more personalized therapeutics.
5. Methods and Advances in Natural Products Discovery
Many innovative methods have been developed and applied in the context of natural products discovery [2] (Table 1). For example, bioprospecting is one commonly used method that systematically explores biodiversity-rich regions for new organisms and their bioactive compounds. A robust laboratory approach called bioassay-guided fractionation isolates and tests fractions from crude extracts based on bioactivity, leading to greater identification of active compounds. Automation and miniaturization of bioassays for the rapid screening of large compound libraries enables high-throughput screening. Other advances include engineering microorganisms to produce specific natural products or analogs with synthetic biology that overcomes the challenges associated with supply and sustainability.

Table 1.
Strategies for natural products discovery. This table provides a comparison of different methods used in natural products discovery, including traditional methods, modern analytical techniques, and computational approaches.
The isolation of natural products from complex mixtures is a crucial step in the discovery process. Traditional isolation techniques, such as solvent extraction, column chromatography, and crystallization, are still widely used. However, recent advances in separation technologies such as high-performance liquid chromatography (HPLC), supercritical fluid extraction (SFE), and solid-phase extraction (SPE) have improved the efficiency and purity of isolated compounds [55].
Spectroscopic techniques, including nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS), and infrared (IR) spectroscopy, play a vital role in the structural elucidation of natural products. NMR spectroscopy in particular is a powerful tool for determining the chemical structure and stereochemistry of complex molecules. MS provides valuable information on the molecular mass to charge ratio, elemental composition, and fragmentation pattern of natural products, while IR spectroscopy can be used to identify functional groups.
Computational methods are increasingly being used to predict the structures and bioactivities of natural products. Molecular docking, virtual screening, and quantitative structure–activity relationship (QSAR) modeling are valuable tools for identifying potential drug candidates from large libraries of natural products [56,57]. In silico screening of natural product databases can significantly reduce the time and cost involved in drug discovery.
Advances in biotechnology have revolutionized natural products discovery by enabling the cultivation and genetic manipulation of microorganisms and plants (Table 2). Metagenomics, genome mining, and synthetic biology approaches have facilitated the discovery of novel biosynthetic gene clusters and the production of bioactive compounds in heterologous hosts. Genomic mining leverages genomic information to predict and identify novel biosynthetic gene clusters for targeted isolation [58]. For example, non-ribosomal proteins (NRPs) often produce or can be engineered to produce compounds that are extremely difficult for chemists to synthesize or engineer. Microbial NRPs have been engineered to produce artemisinic acid, a precursor to artemisinin, in microbial hosts such as yeast. This approach allows for the scalable production of artemisinic acid and has the potential to lower the cost of artemisinin production, making this important antimalarial drug more accessible [59,60]. Metagenomics explores the collective genetic material of microbial communities to access the biosynthetic potential of unculturable organisms [61]. The integration of advanced analytical techniques to analyze the metabolites and genomic data of organisms facilitates targeted isolation. In addition, bioinformatics tools and databases such as antiSMASH, MIBiG, and NORINE are valuable resources for the identification and characterization of natural product biosynthetic pathways [62,63,64].

Table 2.
Recent advances in natural products discovery. This table summarizes the key recent advances in natural products discovery, including new technologies, methodologies, and approaches, that have revolutionized the field.
The bioprospecting of microbial communities offers a promising strategy for the identification and isolation of bioactive compounds with therapeutic potential [65]. By harnessing high-throughput screening platforms and metagenomic libraries, researchers can systematically interrogate the metabolic activities of microbiota and identify lead compounds with desired pharmacological properties [66]. Moreover, innovative screening assays such as phenotypic profiling and functional genomics enable the rapid prioritization of candidate molecules for further characterization and development [67].
Recent advances in natural products discovery have led to the identification of novel bioactive compounds with therapeutic potential. For example, the discovery of teixobactin, a promising antibiotic with activity against drug-resistant bacteria, highlights the importance of innovative screening techniques and cultivation methods [68]. Similarly, the identification of artemisinin from Artemisia annua and its derivatives as potent antimalarial agents underscores the value of traditional herbal medicines in drug discovery [69].
Mass spectrometry-based metabolomics has revolutionized the ability to study the chemical composition of complex biological samples and to conduct natural products discovery. Introduced in 2012, molecular networking has become a cornerstone bioinformatics tool for visualizing and annotating non-targeted mass spectrometry data as it offers a powerful approach to analyze mass spectrometry data beyond spectral matching against reference libraries. The global natural products social (GNPS) molecular networking analysis environment provides a highly efficient platform to perform molecular networking (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp; URL accessed on 19 July 2024) [63]. To build upon the success of classical molecular networking, which is based on the MS-cluster algorithm, a complementary tool named feature-based molecular networking (FBMN) was introduced [70]. FBMN leverages mass spectrometry processing software to incorporate MS1 information such as isotope patterns, retention time, and ion-mobility separation when performed. This method enables the detection of isomers, facilitates spectral annotation, and incorporates relative quantitative information for robust downstream metabolomics statistical analysis. FBMN represents a significant advancement in mass spectrometry-based metabolomics, enabling the characterization of isomers, the incorporation of relative quantification, and the integration of ion mobility data. The FBMN workflow, available on the GNPS web platform, offers automated spectral library search, spectral entry curation, and integration with other annotation tools. This innovative approach promotes data analysis reproducibility, advancing the understanding of the chemical composition of complex biological samples and facilitating rapid discovery of novel natural products.
6. Advances in Methods for Gut Microbiota-Derived Natural Products Discovery
Microbial metabolites and biotransformation products are reservoirs for therapeutic compounds and may also provide information about the involvement of microbes in both health and disease. Currently, three primary strategies are employed to identify and characterize bioactive natural products derived from gut microbiota: culturomics, metabolomics, and metagenomics.
6.1. Culturomics-Based Methodologies
Culturomics, a method that combines diverse culture conditions and advanced isolation techniques, has led to the discovery of several natural products from the human microbiome. This approach allows researchers to cultivate a wide array of microbial species that are often missed by other methods such as metagenomics. Culturomics has significantly increased the number of microbial species recovered from microbiome samples, leading to the identification of many other bioactive compounds with potential therapeutic applications. By providing pure cultures of previously uncultured microbes, this method facilitates the detailed study of microbial physiology and the discovery of novel natural products that could be harnessed for medical and biotechnological applications. By using a variety of culture conditions and advanced isolation techniques such as serial-dilution plating and single-cell microfluidics, approximately 1500–2000 microbial species have been successfully isolated from human fecal samples [71]. This approach allows for the creation of a human gut microbe bank, where strains are selected based on their bioactivity, genomic data, and chemical structure information. These strains are then fermented with conditioned media and extracted for further metabolite profiling and targeted isolation.
One notable discovery using culturomics is the identification of the antimicrobial compound lugdunin from Staphylococcus lugdunensis. This bacterium, isolated from the human nasal cavity, produces lugdunin, which has shown effectiveness against various pathogenic bacteria, including Staphylococcus aureus [72]. Another example includes the identification of isoalloLCA, an anti-infective bile acid derivative produced by certain gut microbes [73]. This compound was discovered through metabolomic-based methods in conjunction with culturomics, demonstrating its potential in protecting against infections.
Despite its potential, culturomics faces challenges such as the low abundance of target compounds in growth media, detection difficulties, and labor-intensive purification processes [74]. Enhancing the understanding of gut microbes’ physiology and biochemical characteristics can optimize culture conditions. Techniques like liquid chromatography–high-resolution mass spectrometry (LC-HRMS), global natural products social (GNPS) molecular networking, in situ MS imaging (MSI), and high-field NMR spectrometry can accelerate the identification of bioactive NPs from gut microbiota [2].
6.2. Metagenomics-Based Methodologies
Metagenomics, the study of genetic material recovered directly from all the organisms (typically microbes) in a community or bulk sample, has significantly advanced the discovery of natural products. This approach also facilitates identification and characterization of bioactive compounds produced by microorganisms that cannot be cultured in the lab. Advanced sequencing and gut metagenome assemblies have uncovered numerous biosynthetic genes within the gut microbiome. Multi-omics analyses combined with machine learning (ML) approaches have been used to discover many microbial natural products.
Computational and synthetic biology tools are effective strategies for mining natural products from biosynthetic gene clusters in the gut microbiome [75]. This method has led to the discovery of compounds such as commendamide, which is a quorum-sensing molecule and GPCR agonist discovered from the human gut microbiome [76]. Commendamide influences host signaling pathways and has potential implications in microbiome–host interactions.
Additionally, deep-learning approaches utilizing natural language processing (NLP) neural network models have been developed to mine antimicrobial peptides from the human gut microbiome [2]. The continued development of suitable hosts for heterologous expression, new gene editing tools, single-cell sequencing, and gene modulation techniques are essential to expedite the identification of natural products derived from gut microbiota. By using metagenomics methods, the vast genetic diversity of microbial communities can be observed to facilitate identification of natural products with potential applications in medicine, agriculture, and biotechnology.
6.3. Metabolomics-Based Methodologies
Gut microbiota-derived natural products enter the bloodstream and affect physiological processes in distant organs. Metabolomic analysis can distinguish health- and disease-related biomarkers by comparing serum and fecal metabolomes in patient or animal cohorts. Untargeted metabolomic analysis initially identifies potentially distinct metabolites, which are then verified through targeted metabolomic detection [77]. Novel metabolites require chemical synthesis or targeted isolation for structural determination [78].
Metabolomics-based methods have led to the discovery of several significant natural products. One notable example is the identification of lugdunin, an antibiotic discovered from the human nasal microbiota. Metabolomics facilitated the analysis of microbial metabolites, leading to this important discovery with potential applications in combating antibiotic-resistant bacteria. The discovery of the anti-infection effects of isoalloLCA in centenarians validated metabolomics-based methods for identifying gut microbiota-derived natural products [73]. This discovery highlights the potential of metabolomics in finding bioactive compounds that could contribute to healthy aging and disease prevention. The integration of artificial intelligence, culturomics, and comprehensive metabolomic and metagenomic analyses is expected to advance the discovery of new bioactive natural products from the gut microbiome and enhance understanding of their biotransformation and biosynthetic pathways.
7. Methods for Identifying Gut Microbiome-Mediated Metabolism of Natural Products
To understand the mechanisms by which gut microbiota-derived natural products exert their effects, a multifaceted systems approach is necessary. Such strategies include RNA sequencing, protein binding studies, signaling transduction analysis, and detection of catalytic functions, similar to the approach used for small-molecule drugs, in the context of in vivo and in vitro assays. There are several advancements in strategies to elucidate these microbiota-driven mechanisms for orally administered natural products with low bioavailability. A general strategy may begin with case control studies in animal or human cohorts that can elucidate the therapeutic benefits of the orally administered natural products. To establish causality, fecal microbiota transplantation (FMT) or animal co-housing experiments alongside therapeutic assays with germ-free or antibiotic-treated animals can be designed. Multi-omics methods can be used to identify the key species, genes, proteins, or metabolites from gut microbiota that mediate phenotypic efficacy. Additionally, signaling pathways involved in gut microbiota–host interactions can be confirmed using strategies such as inoculating the host with the key microbe and its corresponding mutant.
To uncover the mechanisms by which gut microbiota mediate the effects of natural products, several multi-omics approaches can be utilized. For example, global microbiome analysis with metagenomics can be employed to identify candidate biosynthetic genes, bacteria, or microbial communities associated with specific phenotypes. Techniques such as linear discriminant analysis effect size (LEfSe) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis are commonly used to determine which gut bacteria and metabolic pathways are enriched by bioactive natural products [79,80]. These methods have been effective in studies involving polysaccharides and terpene derivatives [81,82].
Additional approaches to identifying gut microbiome-mediated metabolism of natural products include metabolomics and metaproteomics. Both targeted and global untargeted metaproteomics and metabolomics can identify microbial signatures and intervention-specific molecules [77,78]. Significant mediators derived from gut microbiota, including short-chain fatty acids, vitamins (such as folic acid and vitamin B12), secondary bile acids, amino acid derivatives, and relevant microbial enzymes, have been identified through these techniques [42,83]. However, the current limitations in our understanding of gut microbiome metabolism pose challenges to accurately identifying key metabolites and proteins. Artificial intelligence and computational prediction algorithms, when combined with metabolomics, can enhance the identification of gut-microbiome-derived mediators and involved proteins [84,85]. In addition, systems biology approaches that combine metagenomics, metaproteomics, and metabolomics can provide additional power for the discovery of bioactive microbial gene products. For example, one such study systematically identified over 340,000 microbial protein families potentially associated with bioactivity in gut inflammation during inflammatory bowel disease [86].
Culture-based methods and culturomics approaches can isolate specific gut microbes, contributing to observed differences in the metaproteome and metabolome [87]. By culturing enriched gut microbes treated with natural products and comparing their effects with those of natural products, specific microbial contributions can be identified. One notable public resource is the human gut microbial biobank (hGMB), which provides resources for studying interactions between the microbiome, natural products, and the host [71]. In addition, intervention with key metabolites or proteins in animal models can facilitate the determination of the effects of natural product leads.
By leveraging some of these previously mentioned strategies, recent research has revealed that the potential regulatory mechanisms of citrus polysaccharide on cholesterol metabolism are regulated by cholesterol-related gut microbiota and specific metabolites [88]. To understand how citrus polysaccharide influences cholesterol metabolism through the gut microbiota, an integrative analysis using Illumina MiSeq sequencing and non-targeted metabolomics in an in vitro gut microbiota fermentation model was utilized. The addition of POS significantly altered the metabolite profile of the gut microbiota fermentation metabolites, with changes observed in 221 different metabolites and an increase in short-chain fatty acid production. Specifically, metabolites related to cholesterol metabolism such as adenosine monophosphate, cyclic adenosine monophosphate, guanosine, and butyrate were more abundant in the citrus polysaccharide-treated group compared with the control group. Another recent study identified protective effects and underlying mechanisms of Xiaoyu Xiezhuo polyherbal decoction in addressing acute kidney injury caused by ischemia–reperfusion, focusing on the gut–kidney axis [89]. In total, 1778 targets related to acute kidney injury were identified, with 140 of these potentially regulated by the traditional Chinese medicine decoction. The core targets were predominantly associated with inflammation-related pathways. These findings demonstrate the advantages of an integrated strategy for understanding the interactions among natural products, gut microbiota, and the host.
8. Future Directions and Advances in Natural Products Discovery
Continued advancements in technology, such as artificial intelligence and machine learning, hold the potential to revolutionize natural product discovery [90]. Integrating interdisciplinary approaches can further accelerate the identification and development of novel natural products. Natural products continue to be invaluable sources of bioactive compounds with diverse applications. The ongoing exploration of novel molecules from nature is poised to revolutionize the drug discovery, agriculture, dietary supplement, and other industrial sectors.
Metagenomics and microbiome exploration represent powerful tools for the future of natural products discovery. Metagenomics examines the genetic material recovered directly from environmental samples, enabling the exploration of uncultured microorganisms and their biosynthetic potential. Furthermore, drug discovery pipelines should include activities and metabolites of human microbiota and microbiome-derived compounds. For example, a powerful method for natural products discovery in medicinal plants developed in my lab incorporates the microbial metabolism of herb-derived products and microbially generated metabolites. Unraveling the intricate relationship between host-associated microbial communities and the production of bioactive compounds with therapeutic potential will provide new opportunities for therapeutics development.
Microbial communities play a pivotal role in the synthesis and modification of natural products through enzymatic transformations and biotransformations [91]. By harnessing the catalytic activities of microbial enzymes, it is possible to engineer microbial consortia to produce structurally diverse compounds and analogs with enhanced pharmacological profiles [92]. Furthermore, microbial fermentation and biotransformation processes offer scalable and sustainable approaches for the production of bioactive molecules with therapeutic applications [93]. Synthetic biology and metabolic engineering offer tools for precision engineering of biosynthetic pathways in microorganisms for the production of target natural products. In addition, combinatorial biosynthesis, or creating libraries of bioactive compounds through the manipulation of biosynthetic pathways, will allow for the generation of novel analogs.
The integration of -omics technologies leverages multiple global approaches for a systems-based strategy. Rapid advancements in metabolomic techniques for the comprehensive profiling of natural product extracts and the identification of bioactive compounds continues to accelerate discovery. Leveraging genomic and transcriptomic data helps to predict and prioritize the biosynthetic potential of organisms. Combining -omics with high-throughput screening and automation further promotes these investigations. For example, continued miniaturization of bioassays and the development of high-throughput screening platforms allows rapid and efficient compound screening. Integration of robotic systems and automation streamline the isolation and characterization of bioactive compounds.
Future directions in natural products discovery include computational approaches and artificial intelligence. Predictive modeling using computational tools to predict the chemical space of natural products will accelerate the identification of potential drug candidates, as will harnessing machine learning algorithms to analyze large datasets and predict bioactivity based on structural and chemical features. Leveraging artificial intelligence and other models will help navigate the vast combinatorial and complex chemical spaces relevant in natural products discovery.
9. Challenges and Opportunities
While microbiota-mediated natural products discovery holds immense promise for drug development, several challenges must be addressed to realize its full potential. These include the identification of novel microbial strains, optimization of fermentation conditions, and elucidation of biosynthetic pathways [94]. Additionally, ethical considerations surrounding microbial collection and utilization require careful consideration to ensure sustainable and responsible practices [95]. Nonetheless, the advent of cutting-edge technologies including metagenomics, synthetic biology, and machine learning offers unprecedented opportunities for accelerating natural products discovery and innovation [96].
Other challenges to consider in natural product discovery include rediscovery and ethical issues. Some natural products may be rediscovered due to the re-isolation of known compounds from different sources. In addition, supplies are often limited, as isolation of sufficient quantities of rare compounds from natural sources can be challenging. Increased innovation in bioactivity assays will be key to increasing discovery rates. Developing relevant and efficient bioassays to screen for specific biological activities is of high priority in the field. Addressing ethical concerns related to biodiversity conservation and the sustainable sourcing of natural products is an important consideration, especially in the context of the use of synthetic biology. Fair and equitable frameworks for the sharing of benefits derived from the use of genetic resources are vital for access and benefit sharing ethical considerations. Despite the significant progress in natural products discovery, several other challenges remain, including the identification of novel scaffolds, optimization of production methods, and sustainable sourcing of natural resources. Future research should focus on the development of high-throughput screening methods, the exploration of unexplored ecological niches, and the application of innovative biotechnological tools for drug discovery.
Amidst the challenges, great opportunities emerge, such as the positive impact on drug development. Exploring natural products as a source of new antibiotics to combat the rising threat of antibiotic-resistant bacteria is one important opportunity. In addition, the identification of natural compounds with potent anticancer properties and novel mechanisms of action represents another key area of development. Future collaborations and global initiatives are opportunities to accelerate this work, such as strengthening collaborations between researchers, institutions, and industries to foster global advancements in natural products discovery. Moreover, initiatives that promote open-access platforms for sharing data, compounds, and knowledge will further accelerate this research.
While this narrative review provides a comprehensive overview of microbiota-mediated natural products discovery, several limitations must be acknowledged. Firstly, the scope of the review, while extensive, may not cover all emerging methodologies or recent breakthroughs in the rapidly evolving field of microbiome research. The focus on microbiota-mediated biotransformation of medicinal herbs, although central to the review, may exclude other significant sources of natural products and their interactions with the microbiome. Additionally, the review is constrained by the availability and quality of current data, which can vary widely across studies. Some findings may be preliminary or based on limited sample sizes, potentially impacting the generalizability of the conclusions. Furthermore, the review primarily discusses established methods and may not fully explore the potential of newer, innovative techniques in natural products discovery. Finally, the ethical and practical challenges associated with microbiota-based research, including issues of reproducibility and the complexities of translating preclinical findings to clinical applications, are only briefly addressed. Future reviews should aim to integrate these aspects more thoroughly and consider the implications of ongoing technological advancements.
10. Conclusions
Microbiota-mediated natural products discovery represents a frontier in drug development, offering a treasure trove of bioactive compounds with therapeutic potential. By unraveling the metabolic activities and biosynthetic capabilities of microbial communities, researchers can harness the relationship between microorganisms and their environment to unlock novel drug candidates and address unmet medical needs. As we continue this exploration and discovery, collaboration between scientists, biotech, and pharmaceutical companies will be essential for translating microbiota-mediated natural products into next-generation therapeutics.
The future of natural products discovery holds immense promise, driven by the convergence of technological breakthroughs, innovative methodologies, and a collective commitment to harnessing nature’s chemical diversity. Continued exploration and collaboration will undoubtedly pave the way for the development of novel therapeutics, sustainable solutions, and transformative technologies. Methods and advances in natural products discovery have transformed the field of drug discovery, providing a vast array of bioactive compounds with therapeutic potential. From traditional isolation techniques to cutting-edge biotechnological tools, researchers have a wide range of methods at their disposal for the identification and characterization of natural products. By harnessing the power of nature and innovation, the discovery of novel natural products holds great promise for the development of new treatments for a variety of diseases.
In conclusion, the scientific community is on the brink of a new era in natural products discovery, where interdisciplinary approaches and cutting-edge technologies converge to unlock the full potential of nature’s chemical repertoire and clinical potential. The microbiome’s multifaceted impact on natural products discovery underscores the need for a holistic understanding of host–microbe interactions. Leveraging this knowledge holds immense promise for the identification of novel bioactive compounds with applications in pharmaceuticals, agriculture, and beyond. Natural product discovery remains a dynamic and multidisciplinary field, providing a foundation for the development of new medicines and sustainable technologies. Despite challenges, ongoing innovations and collaborative efforts promise a future filled with exciting discoveries from the wealth of nature’s chemical diversity.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences, T.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Dupont, H.L.; Jiang, Z.D.; Dupont, A.W.; Utay, N.S. The Intestinal Microbiome in Human Health and Disease. Trans. Am. Clin. Climatol. Assoc. 2020, 131, 178–197. [Google Scholar] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.M.; Tan, R.X. Interaction between gut microbiota and ethnomedicine constituents. Nat. Prod. Rep. 2019, 36, 788–809. [Google Scholar] [CrossRef] [PubMed]
- Selma, M.V.; Espin, J.C.; Tomas-Barberan, F.A. Interaction between phenolics and gut microbiota: Role in human health. J. Agric. Food Chem. 2009, 57, 6485–6501. [Google Scholar] [CrossRef]
- Zhou, J.; Ma, Y.H.; Zhou, Z.; Chen, Y.; Wang, Y.; Gao, X. Intestinal Absorption and Metabolism of Epimedium Flavonoids in Osteoporosis Rats. Drug Metab. Dispos. 2015, 43, 1590–1600. [Google Scholar] [CrossRef]
- Zhang, S.H.; Wang, Y.Z.; Meng, F.Y.; Li, Y.L.; Li, C.X.; Duan, F.P.; Wang, Q.; Zhang, X.T.; Zhang, C.N. Studies of the microbial metabolism of flavonoids extracted from the leaves of Diospyros kaki by intestinal bacteria. Arch. Pharmacal Res. 2015, 38, 614–619. [Google Scholar] [CrossRef]
- Zielinska, S.; Dziagwa-Becker, M.; Junka, A.; Piatczak, E.; Jezierska-Domaradzka, A.; Brozyna, M.; Paleczny, J.; Sobiecka, A.; Slupski, W.; Mess, E.; et al. Screening Papaveraceae as Novel Antibiofilm Natural-Based Agents. Molecules 2021, 26, 4778. [Google Scholar] [CrossRef]
- Jin, Y.J.; Jang, M.G.; Kim, J.W.; Baek, S.; Ko, H.C.; Hur, S.P.; Kim, S.J. Anti-Obesity Effects of Polymethoxyflavone-Rich Fraction from Jinkyool (Citrus sunki Hort. ex Tanaka) Leaf on Obese Mice Induced by High-Fat Diet. Nutrients 2022, 14, 865. [Google Scholar] [CrossRef]
- Zeng, S.L.; Li, S.Z.; Xiao, P.T.; Cai, Y.Y.; Chu, C.; Chen, B.Z.; Li, P.; Li, J.; Liu, E.H. Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci. Adv. 2020, 6, eaax6208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Chen, J.; Yi, K.; Peng, L.; Xie, J.; Gou, X.; Peng, T.; Tang, L. Phlorizin ameliorates obesity-associated endotoxemia and insulin resistance in high-fat diet-fed mice by targeting the gut microbiota and intestinal barrier integrity. Gut Microbes 2020, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zheng, X.; Ma, X.; Jiang, R.; Zhou, W.; Zhou, S.; Zhang, Y.; Lei, S.; Wang, S.; Kuang, J.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 2019, 10, 4971. [Google Scholar] [CrossRef]
- Chiu, H.F.; Venkatakrishnan, K.; Golovinskaia, O.; Wang, C.K. Gastroprotective Effects of Polyphenols against Various Gastro-Intestinal Disorders: A Mini-Review with Special Focus on Clinical Evidence. Molecules 2021, 26, 2090. [Google Scholar] [CrossRef]
- Singh, R.; Chandrashekharappa, S.; Bodduluri, S.R.; Baby, B.V.; Hegde, B.; Kotla, N.G.; Hiwale, A.A.; Saiyed, T.; Patel, P.; Vijay-Kumar, M.; et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat. Commun. 2019, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Godiyal, S.; Laddha, K. Validated high-performance thin-layer chromatographic method for quantification of gallic acid and ellagic acid in fruits of Terminalia chebula, Phyllanthus emblica, and Quercus infectoria. J. Sep. Sci. 2023, 46, e2200991. [Google Scholar] [CrossRef]
- Thoithoisana Devi, S.; Devika Chanu, K.; Singh, N.B.; Chaudhary, S.K.; Keithellakpam, O.S.; Singh, K.B.; Mukherjee, P.K.; Sharma, N. Chemical Profiling and Therapeutic Evaluation of Standardized Hydroalcoholic Extracts of Terminalia chebula Fruits Collected from Different Locations in Manipur against Colorectal Cancer. Molecules 2023, 28, 2901. [Google Scholar] [CrossRef]
- Cota, D.; Mishra, S.; Shengule, S. Beneficial role of Terminalia arjuna hydro-alcoholic extract in colitis and its possible mechanism. J. Ethnopharmacol. 2019, 230, 117–125. [Google Scholar] [CrossRef]
- Wu, J.; Wei, Z.; Cheng, P.; Qian, C.; Xu, F.; Yang, Y.; Wang, A.; Chen, W.; Sun, Z.; Lu, Y. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics 2020, 10, 10665–10679. [Google Scholar] [CrossRef]
- Ammara, A.; Sobia, A.; Nureen, Z.; Sohail, A.; Abid, S.; Aziz, T.; Nahaa, M.A.; Rewaa, S.J.; Ahellah, M.J.; Nouf, S.A.A.; et al. Revolutionizing the effect of Azadirachta indica extracts on edema induced changes in C-reactive protein and interleukin-6 in albino rats: In silico and in vivo approach. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 5951–5963. [Google Scholar] [CrossRef]
- Naz, R.; Saqib, F.; Awadallah, S.; Wahid, M.; Latif, M.F.; Iqbal, I.; Mubarak, M.S. Food Polyphenols and Type II Diabetes Mellitus: Pharmacology and Mechanisms. Molecules 2023, 28, 3996. [Google Scholar] [CrossRef] [PubMed]
- Egbuna, C.; Awuchi, C.G.; Kushwaha, G.; Rudrapal, M.; Patrick-Iwuanyanwu, K.C.; Singh, O.; Odoh, U.E.; Khan, J.; Jeevanandam, J.; Kumarasamy, S.; et al. Bioactive Compounds Effective Against Type 2 Diabetes Mellitus: A Systematic Review. Curr. Top. Med. Chem. 2021, 21, 1067–1095. [Google Scholar] [CrossRef] [PubMed]
- Chawansuntati, K.; Hongjaisee, S.; Sirita, K.; Kingkaew, K.; Rattanathammethee, K.; Kumrapich, B.; Ounjaijean, S.; Kongkaew, A.; Lumjuan, N. Effects of quercetin and extracts from Phyllanthus emblica, Morus alba, and Ginkgo biloba on platelet recovery in a rat model of chemotherapy-induced thrombocytopenia. Heliyon 2024, 10, e25013. [Google Scholar] [CrossRef]
- Alomar, M.Y. Physiological and histopathological study on the influence of Ocimum basilicum leaves extract on thioacetamide-induced nephrotoxicity in male rats. Saudi J. Biol. Sci. 2020, 27, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Vijayakumar, S.; Kothandaraman, S.; Palani, M. Anti-diabetic activity of quercetin extracted from Phyllanthus emblica L. fruit: In silico and in vivo approaches. J. Pharm. Anal. 2018, 8, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Denniston, K.; Chopra, D. Therapeutic Uses of Triphala in Ayurvedic Medicine. J. Altern. Complement. Med. 2017, 23, 607–614. [Google Scholar] [CrossRef]
- Olennikov, D.N.; Kashchenko, N.I.; Chirikova, N.K. In Vitro Bioaccessibility, Human Gut Microbiota Metabolites and Hepatoprotective Potential of Chebulic Ellagitannins: A Case of Padma Hepaten(R) Formulation. Nutrients 2015, 7, 8456–8477. [Google Scholar] [CrossRef]
- Peng, X.; Zhang, Z.; Zhang, N.; Liu, L.; Li, S.; Wei, H. In vitro catabolism of quercetin by human fecal bacteria and the antioxidant capacity of its catabolites. Food Nutr. Res. 2014, 58, 23406. [Google Scholar] [CrossRef]
- Zhang, Z.; Peng, X.; Li, S.; Zhang, N.; Wang, Y.; Wei, H. Isolation and identification of quercetin degrading bacteria from human fecal microbes. PLoS ONE 2014, 9, e90531. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Chen, C.; Lu, J.; Yu, J.; Xu, X.; Peng, Y.; Zhang, S.; Jiang, S.; Guo, J.; et al. Targeting the gut microbial metabolic pathway with small molecules decreases uremic toxin production. Gut Microbes 2020, 12, 1823800. [Google Scholar] [CrossRef]
- Pivari, F.; Mingione, A.; Piazzini, G.; Ceccarani, C.; Ottaviano, E.; Brasacchio, C.; Dei Cas, M.; Vischi, M.; Cozzolino, M.G.; Fogagnolo, P.; et al. Curcumin Supplementation (Meriva((R))) Modulates Inflammation, Lipid Peroxidation and Gut Microbiota Composition in Chronic Kidney Disease. Nutrients 2022, 14, 231. [Google Scholar] [CrossRef]
- Jabczyk, M.; Nowak, J.; Hudzik, B.; Zubelewicz-Szkodzinska, B. Curcumin and Its Potential Impact on Microbiota. Nutrients 2021, 13, 2004. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, U.; Rubab, M.; Daliri, E.B.; Chelliah, R.; Javed, A.; Oh, D.H. Curcumin, Quercetin, Catechins and Metabolic Diseases: The Role of Gut Microbiota. Nutrients 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Akinrinde, A.S.; Adekanmbi, A.O.; Olojo, F.O. Nigella sativa oil protects against cadmium-induced intestinal toxicity via promotion of anti-inflammatory mechanisms, mucin expression and microbiota integrity. Avicenna J. Phytomed. 2022, 12, 241–256. [Google Scholar] [CrossRef]
- Lin, L.; Tang, R.; Liu, Y.; Li, Z.; Li, H.; Yang, H. Research on the anti-aging mechanisms of Panax ginseng extract in mice: A gut microbiome and metabolomics approach. Front. Pharmacol. 2024, 15, 1415844. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Duan, Z.; Deng, J.; Zhang, Z.; Fu, R.; Zhu, C.; Fan, D. Ginsenoside Rh4 inhibits colorectal cancer via the modulation of gut microbiota-mediated bile acid metabolism. J. Adv. Res. 2024, in press. [CrossRef]
- Liu, Y.; Bai, X.; Wu, H.; Duan, Z.; Zhu, C.; Fu, R.; Fan, D. Ginsenoside CK Alleviates DSS-Induced IBD in Mice by Regulating Tryptophan Metabolism and Activating Aryl Hydrocarbon Receptor via Gut Microbiota Modulation. J. Agric. Food Chem. 2024, 72, 9867–9879. [Google Scholar] [CrossRef]
- Zhang, K.X.; Zhu, Y.; Song, S.X.; Bu, Q.Y.; You, X.Y.; Zou, H.; Zhao, G.P. Ginsenoside Rb1, Compound K and 20(S)-Protopanaxadiol Attenuate High-Fat Diet-Induced Hyperlipidemia in Rats via Modulation of Gut Microbiota and Bile Acid Metabolism. Molecules 2024, 29, 1108. [Google Scholar] [CrossRef]
- Liao, C.P.; Liu, X.C.; Dong, S.Q.; An, M.; Zhao, L.; Zhang, A.J.; Liu, J.F.; Hou, W.B.; Fan, H.R.; Liu, C.X. Investigation of the metabolites of five major constituents from Berberis amurensis in normal and pseudo germ-free rats. Chin. J. Nat. Med. 2021, 19, 758–771. [Google Scholar] [CrossRef]
- Du, H.; Xu, T.; Yi, H.; Xu, X.; Zhao, C.; Ge, Y.; Zhang, C.; Fan, G. Effect of Gut Microbiota on the Metabolism of Chemical Constituents of Berberis kansuensis Extract Based on UHPLC-Orbitrap-MS Technique. Planta Med. 2022, 88, 933–949. [Google Scholar] [CrossRef]
- Wu, C.; Zhao, Y.; Zhang, Y.; Yang, Y.; Su, W.; Yang, Y.; Sun, L.; Zhang, F.; Yu, J.; Wang, Y.; et al. Gut microbiota specifically mediates the anti-hypercholesterolemic effect of berberine (BBR) and facilitates to predict BBR’s cholesterol-decreasing efficacy in patients. J. Adv. Res. 2022, 37, 197–208. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Y.; Ren, H.; Wang, S.; Zhong, H.; Zhao, X.; Ma, J.; Gu, X.; Xue, Y.; Huang, S.; et al. Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the PREMOTE study). Nat. Commun. 2020, 11, 5015. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Pourang, A.; Dhaliwal, S.; Kohn, J.N.; Uchitel, S.; Singh, H.; Mills, P.J.; Peterson, S.N.; Sivamani, R.K. Modulatory Effects of Triphala and Manjistha Dietary Supplementation on Human Gut Microbiota: A Double-Blind, Randomized, Placebo-Controlled Pilot Study. J. Altern. Complement. Med. 2020, 26, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Iablokov, S.N.; Uchitel, S.; Chopra, D.; Perez-Santiago, J.; Rodionov, D.A.; Peterson, S.N. Community Metabolic Interactions, Vitamin Production and Prebiotic Potential of Medicinal Herbs Used for Immunomodulation. Front. Genet. 2021, 12, 584197. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Perez-Santiago, J.; Iablokov, S.N.; Rodionov, D.A.; Peterson, S.N. Alteration of Community Metabolism by Prebiotics and Medicinal Herbs. Microorganisms 2023, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.T.; Vaughn, A.R.; Sharma, V.; Chopra, D.; Mills, P.J.; Peterson, S.N.; Sivamani, R.K. Effects of Turmeric and Curcumin Dietary Supplementation on Human Gut Microbiota: A Double-Blind, Randomized, Placebo-Controlled Pilot Study. J. Evid. -Based Integr. Med. 2018, 23, 2515690X18790725. [Google Scholar] [CrossRef]
- Dong, J.; Liang, Q.; Niu, Y.; Jiang, S.; Zhou, L.; Wang, J.; Ma, C.; Kang, W. Effects of Nigella sativa seed polysaccharides on type 2 diabetic mice and gut microbiota. Int. J. Biol. Macromol. 2020, 159, 725–738. [Google Scholar] [CrossRef]
- Liang, Q.; Dong, J.; Wang, S.; Shao, W.; Ahmed, A.F.; Zhang, Y.; Kang, W. Immunomodulatory effects of Nigella sativa seed polysaccharides by gut microbial and proteomic technologies. Int. J. Biol. Macromol. 2021, 184, 483–496. [Google Scholar] [CrossRef]
- Wang, T.; Han, J.; Dai, H.; Sun, J.; Ren, J.; Wang, W.; Qiao, S.; Liu, C.; Sun, L.; Liu, S.; et al. Polysaccharides from Lyophyllum decastes reduce obesity by altering gut microbiota and increasing energy expenditure. Carbohydr. Polym. 2022, 295, 119862. [Google Scholar] [CrossRef]
- Peterson, C.T.; Iablokov, S.N.; Rodionov, D.A.; Peterson, S.N. Personalized Response of Parkinson’s Disease Gut Microbiota to Nootropic Medicinal Herbs In Vitro: A Proof of Concept. Microorganisms 2023, 11, 1979. [Google Scholar] [CrossRef]
- Gojda, J.; Cahova, M. Gut Microbiota as the Link between Elevated BCAA Serum Levels and Insulin Resistance. Biomolecules 2021, 11, 1414. [Google Scholar] [CrossRef]
- Guo, C.J.; Allen, B.M.; Hiam, K.J.; Dodd, D.; Van Treuren, W.; Higginbottom, S.; Nagashima, K.; Fischer, C.R.; Sonnenburg, J.L.; Spitzer, M.H.; et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science 2019, 366, eaav1282. [Google Scholar] [CrossRef]
- Needham, B.D.; Funabashi, M.; Adame, M.D.; Wang, Z.; Boktor, J.C.; Haney, J.; Wu, W.L.; Rabut, C.; Ladinsky, M.S.; Hwang, S.J.; et al. A gut-derived metabolite alters brain activity and anxiety behaviour in mice. Nature 2022, 602, 647–653. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898.e3811. [Google Scholar] [CrossRef]
- Li, J.W.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef]
- Damale, M.G.; Harke, S.N.; Kalam Khan, F.A.; Shinde, D.B.; Sangshetti, J.N. Recent advances in multidimensional QSAR (4D-6D): A critical review. Mini-Rev. Med. Chem. 2014, 14, 35–55. [Google Scholar] [CrossRef]
- Saikia, S.; Bordoloi, M. Molecular Docking: Challenges, Advances and its Use in Drug Discovery Perspective. Curr. Drug Targets 2019, 20, 501–521. [Google Scholar] [CrossRef]
- Xu, Z.; Park, T.J.; Cao, H. Advances in mining and expressing microbial biosynthetic gene clusters. Crit. Rev. Microbiol. 2023, 49, 18–37. [Google Scholar] [CrossRef]
- Ro, D.K.; Paradise, E.M.; Ouellet, M.; Fisher, K.J.; Newman, K.L.; Ndungu, J.M.; Ho, K.A.; Eachus, R.A.; Ham, T.S.; Kirby, J.; et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 2006, 440, 940–943. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef]
- Santero, E.; Floriano, B.; Govantes, F. Harnessing the power of microbial metabolism. Crit. Rev. Microbiol. 2016, 31, 63–69. [Google Scholar] [CrossRef]
- Ren, H.; Shi, C.; Zhao, H. Computational Tools for Discovering and Engineering Natural Product Biosynthetic Pathways. iScience 2020, 23, 100795. [Google Scholar] [CrossRef]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castano-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef]
- Caboche, S.; Pupin, M.; Leclere, V.; Fontaine, A.; Jacques, P.; Kucherov, G. NORINE: A database of nonribosomal peptides. Nucleic Acids Res. 2008, 36, D326–D331. [Google Scholar] [CrossRef]
- Ho, S.H.; Ye, X.; Hasunuma, T.; Chang, J.S.; Kondo, A. Perspectives on engineering strategies for improving biofuel production from microalgae—A critical review. Biotechnol. Adv. 2014, 32, 1448–1459. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Qi, Y.K.; Tang, X.; Wei, N.N.; Pang, C.J.; Du, S.S.; Wang, K. Discovery, synthesis, and optimization of teixobactin, a novel antibiotic without detectable bacterial resistance. J. Pept. Sci. 2022, 28, e3428. [Google Scholar] [CrossRef]
- Feng, X.; Cao, S.; Qiu, F.; Zhang, B. Traditional application and modern pharmacological research of Artemisia annua L. Pharmacol. Ther. 2020, 216, 107650. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Duhrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Liu, C.; Du, M.X.; Abuduaini, R.; Yu, H.Y.; Li, D.H.; Wang, Y.J.; Zhou, N.; Jiang, M.Z.; Niu, P.X.; Han, S.S.; et al. Enlightening the taxonomy darkness of human gut microbiomes with a cultured biobank. Microbiome 2021, 9, 119. [Google Scholar] [CrossRef]
- Chang, S.C.; Kao, C.Y.; Lin, L.C.; Hidrosollo, J.H.; Lu, J.J. Lugdunin production and activity in Staphylococcus lugdunensis isolates are associated with its genotypes. Microbiol. Spectr. 2023, 11, e0129823. [Google Scholar] [CrossRef]
- Sato, Y.; Atarashi, K.; Plichta, D.R.; Arai, Y.; Sasajima, S.; Kearney, S.M.; Suda, W.; Takeshita, K.; Sasaki, T.; Okamoto, S.; et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 2021, 599, 458–464. [Google Scholar] [CrossRef]
- Diakite, A.; Dubourg, G.; Dione, N.; Afouda, P.; Bellali, S.; Ngom, I.I.; Valles, C.; Tall, M.L.; Lagier, J.C.; Raoult, D. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci. Rep. 2020, 10, 9674. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, P.; Ma, Y.; Wang, J.; Chen, Y. Artificial intelligence accelerates the mining of bioactive small molecules from human microbiome. Clin. Transl. Med. 2022, 12, e1011. [Google Scholar] [CrossRef]
- Cohen, L.J.; Kang, H.S.; Chu, J.; Huang, Y.H.; Gordon, E.A.; Reddy, B.V.; Ternei, M.A.; Craig, J.W.; Brady, S.F. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. USA 2015, 112, E4825–E4834. [Google Scholar] [CrossRef]
- Stuart, K.A.; Welsh, K.; Walker, M.C.; Edrada-Ebel, R. Metabolomic tools used in marine natural product drug discovery. Expert Opin. Drug Discov. 2020, 15, 499–522. [Google Scholar] [CrossRef]
- Gaudencio, S.P.; Bayram, E.; Lukic Bilela, L.; Cueto, M.; Diaz-Marrero, A.R.; Haznedaroglu, B.Z.; Jimenez, C.; Mandalakis, M.; Pereira, F.; Reyes, F.; et al. Advanced Methods for Natural Products Discovery: Bioactivity Screening, Dereplication, Metabolomics Profiling, Genomic Sequencing, Databases and Informatic Tools, and Structure Elucidation. Mar. Drugs 2023, 21, 308. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. J. Vis. Exp. 2022, 183, e61715. [Google Scholar] [CrossRef]
- Ullah, A.; Sun, Q.; Li, J.; Li, J.; Khatun, P.; Kou, G.; Lyu, Q. Bioactive Compounds in Citrus reticulata Peel Are Potential Candidates for Alleviating Physical Fatigue through a Triad Approach of Network Pharmacology, Molecular Docking, and Molecular Dynamics Modeling. Nutrients 2024, 16, 1934. [Google Scholar] [CrossRef]
- Qiao, S.; Liu, C.; Sun, L.; Wang, T.; Dai, H.; Wang, K.; Bao, L.; Li, H.; Wang, W.; Liu, S.J.; et al. Gut Parabacteroides merdae protects against cardiovascular damage by enhancing branched-chain amino acid catabolism. Nat. Metab. 2022, 4, 1271–1286. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, L.; Ma, Q.; Zhang, X.; Chen, M.; Liu, F.; Zhang, T.; Jia, W.; Zhu, L.; Qi, W.; et al. Astragalus Polysaccharide Modulates the Gut Microbiota and Metabolites of Patients with Type 2 Diabetes in an In Vitro Fermentation Model. Nutrients 2024, 16, 1698. [Google Scholar] [CrossRef]
- Sun, W.L.; Hua, S.; Li, X.Y.; Shen, L.; Wu, H.; Ji, H.F. Microbially produced vitamin B12 contributes to the lipid-lowering effect of silymarin. Nat. Commun. 2023, 14, 477. [Google Scholar] [CrossRef]
- McCoubrey, L.E.; Elbadawi, M.; Orlu, M.; Gaisford, S.; Basit, A.W. Harnessing machine learning for development of microbiome therapeutics. Gut Microbes 2021, 13, 1872323. [Google Scholar] [CrossRef]
- Kouraki, A.; Nogal, A.; Nocun, W.; Louca, P.; Vijay, A.; Wong, K.; Michelotti, G.A.; Menni, C.; Valdes, A.M. Machine Learning Metabolomics Profiling of Dietary Interventions from a Six-Week Randomised Trial. Metabolites 2024, 14, 311. [Google Scholar] [CrossRef]
- Zhang, Y.; Bhosle, A.; Bae, S.; McIver, L.J.; Pishchany, G.; Accorsi, E.K.; Thompson, K.N.; Arze, C.; Wang, Y.; Subramanian, A.; et al. Discovery of bioactive microbial gene products in inflammatory bowel disease. Nature 2022, 606, 754–760. [Google Scholar] [CrossRef]
- Sengupta, S.; Pabbaraja, S.; Mehta, G. Natural products from the human microbiome: An emergent frontier in organic synthesis and drug discovery. Org. Biomol. Chem. 2024, 22, 4006–4030. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, P.; Liu, F.; Pan, S. Regulations of Citrus Pectin Oligosaccharide on Cholesterol Metabolism: Insights from Integrative Analysis of Gut Microbiota and Metabolites. Nutrients 2024, 16, 2002. [Google Scholar] [CrossRef]
- Ji, Y.; Xiao, Y.; Li, S.; Fan, Y.; Cai, Y.; Yang, B.; Chen, H.; Hu, S. Protective effect and mechanism of Xiaoyu Xiezhuo decoction on ischemia-reperfusion induced acute kidney injury based on gut-kidney crosstalk. Ren. Fail. 2024, 46, 2365982. [Google Scholar] [CrossRef]
- Mullowney, M.W.; Duncan, K.R.; Elsayed, S.S.; Garg, N.; van der Hooft, J.J.J.; Martin, N.I.; Meijer, D.; Terlouw, B.R.; Biermann, F.; Blin, K.; et al. Artificial intelligence for natural product drug discovery. Nat. Rev. Drug Discov. 2023, 22, 895–916. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Medema, M.H.; Cimermancic, P.; Sali, A.; Takano, E.; Fischbach, M.A. A systematic computational analysis of biosynthetic gene cluster evolution: Lessons for engineering biosynthesis. PLoS Comput. Biol. 2014, 10, e1004016. [Google Scholar] [CrossRef]
- Piel, J.; Hui, D.; Wen, G.; Butzke, D.; Platzer, M.; Fusetani, N.; Matsunaga, S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 2004, 101, 16222–16227. [Google Scholar] [CrossRef]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Ruckert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).