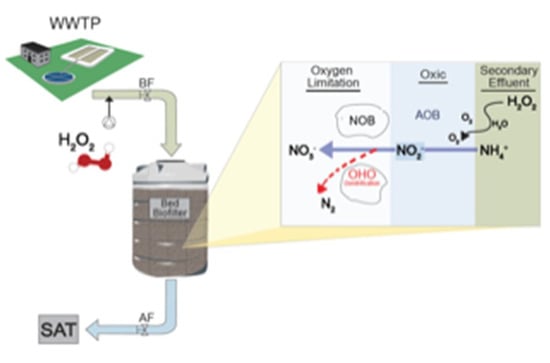
Stimulating Nitrogen Biokinetics with the Addition of Hydrogen Peroxide to Secondary Effluent Biofiltration
Abstract
1. Introduction
2. Experiments
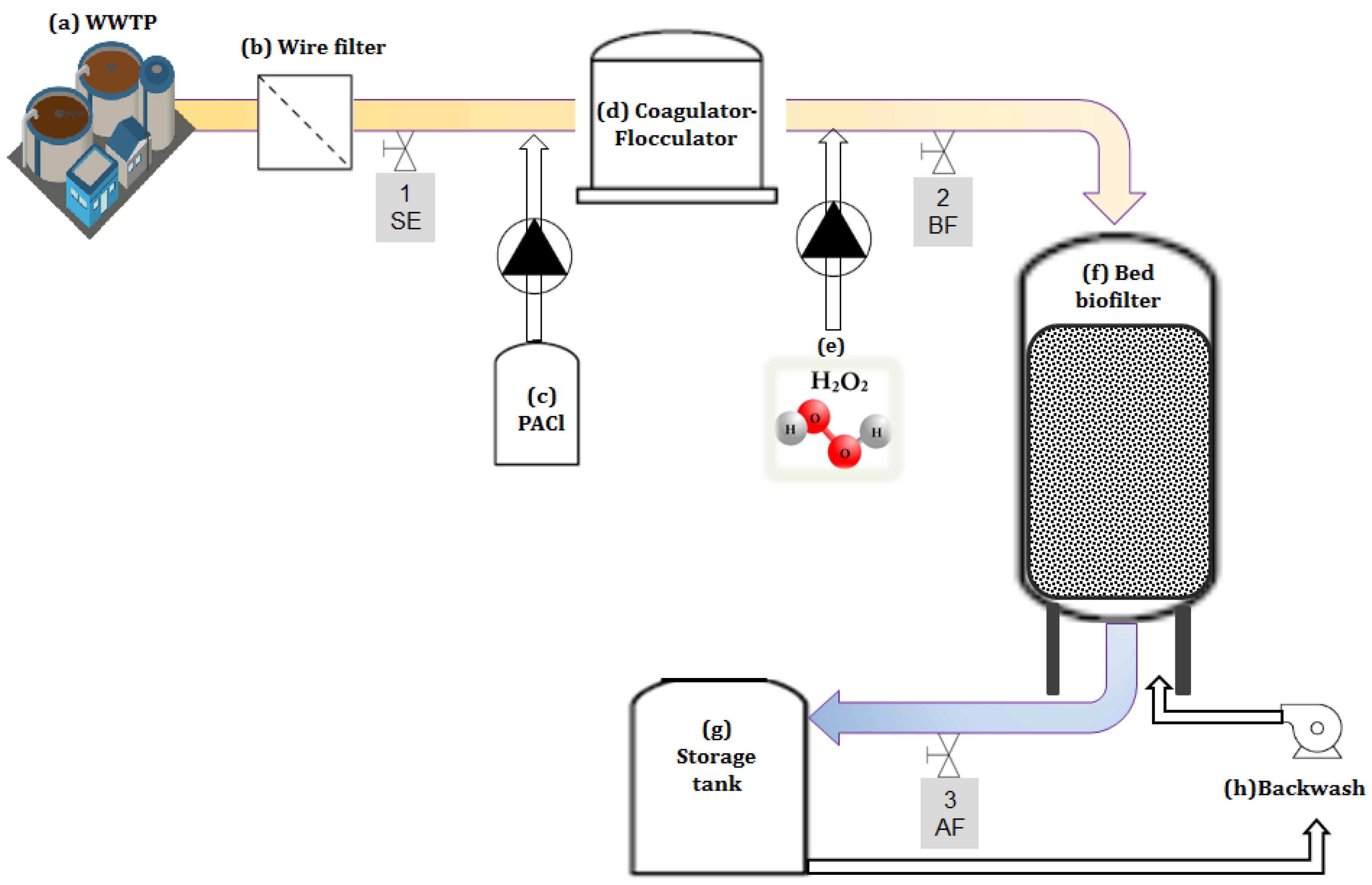
2.1. Pretreatment System Setup
- -
- -
- Coagulation/flocculation was carried out by injecting polyaluminum chloride (PACl, 18% Al2O3) by peristaltic pump (Figure 1c) to achieve a final concentration of ~2.7–3.6 mg/L as PACl. Flocculation was performed in a modified flocculator, which consisted of a pressure filter (Figure 1d) with ~15 min hydraulic retention time (HRT). PACl was chosen for this study as it has been shown to be preferable for flocculation and is widely used [35,36].
- -
- Following flocculation, H2O2 was added (10%) to the inlet of the high-rate biofilter (Figure 1f) using a peristaltic pump (Figure 1e). The final concentration of 27 mg/L was achieved to provide a surplus of DO for full nitrification of 3.5 mg/L-N NH4+ and 0.5 mg/L-N NO2−. The biofilter tank had a surface area of 1.13 m2, 1.2 m diameter, 1.1 m3 media volume, and 36% free headspace (additional characteristics are specified in Zucker et al., [34]).
2.2. System Specification and Operational Parameters
2.3. Solute Sampling and Analyses
2.4. Particle Sampling and Analysis
2.5. Media Sampling and Metagenome Analysis
3. Results
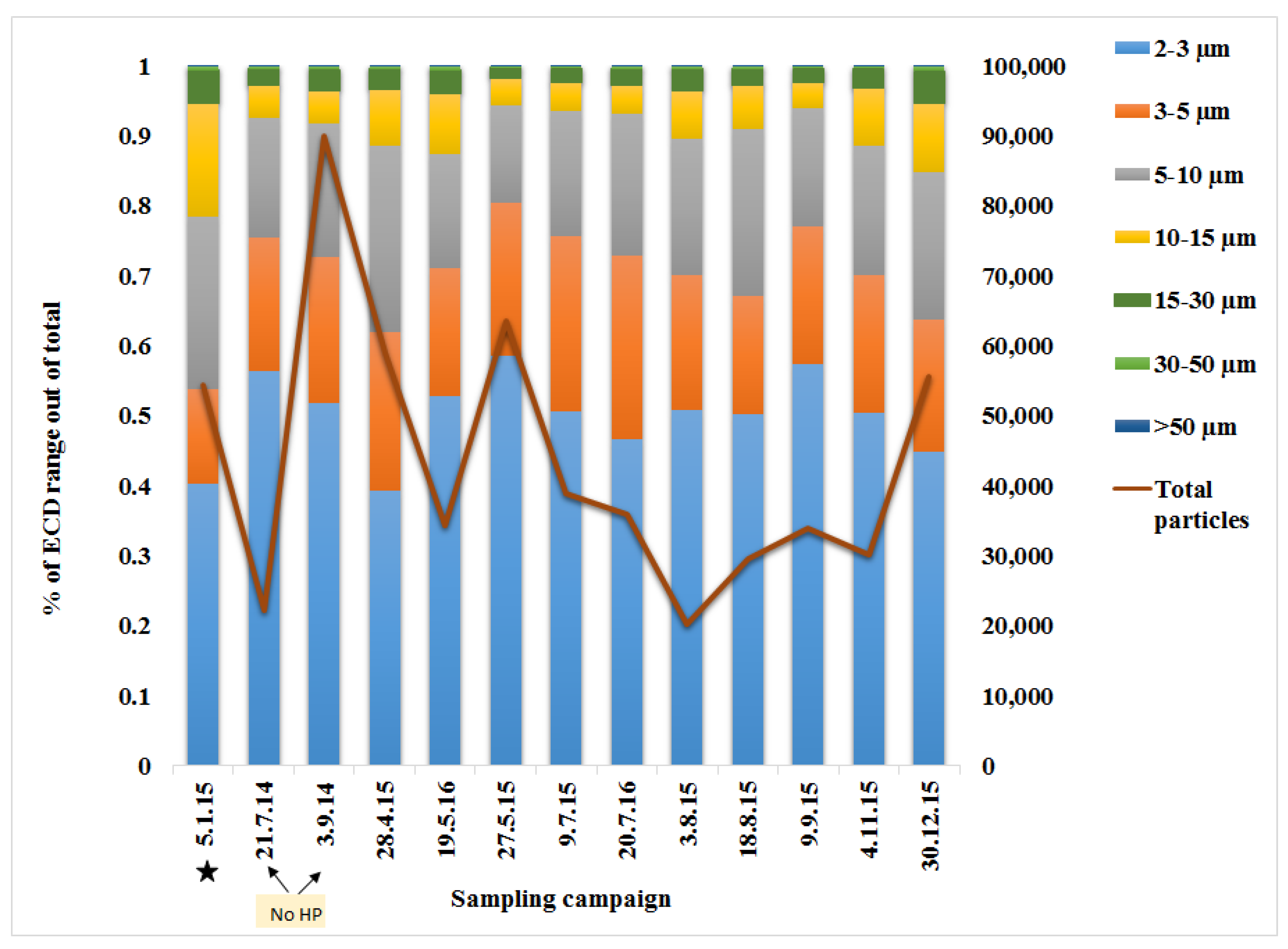
3.1. Particle Distribution
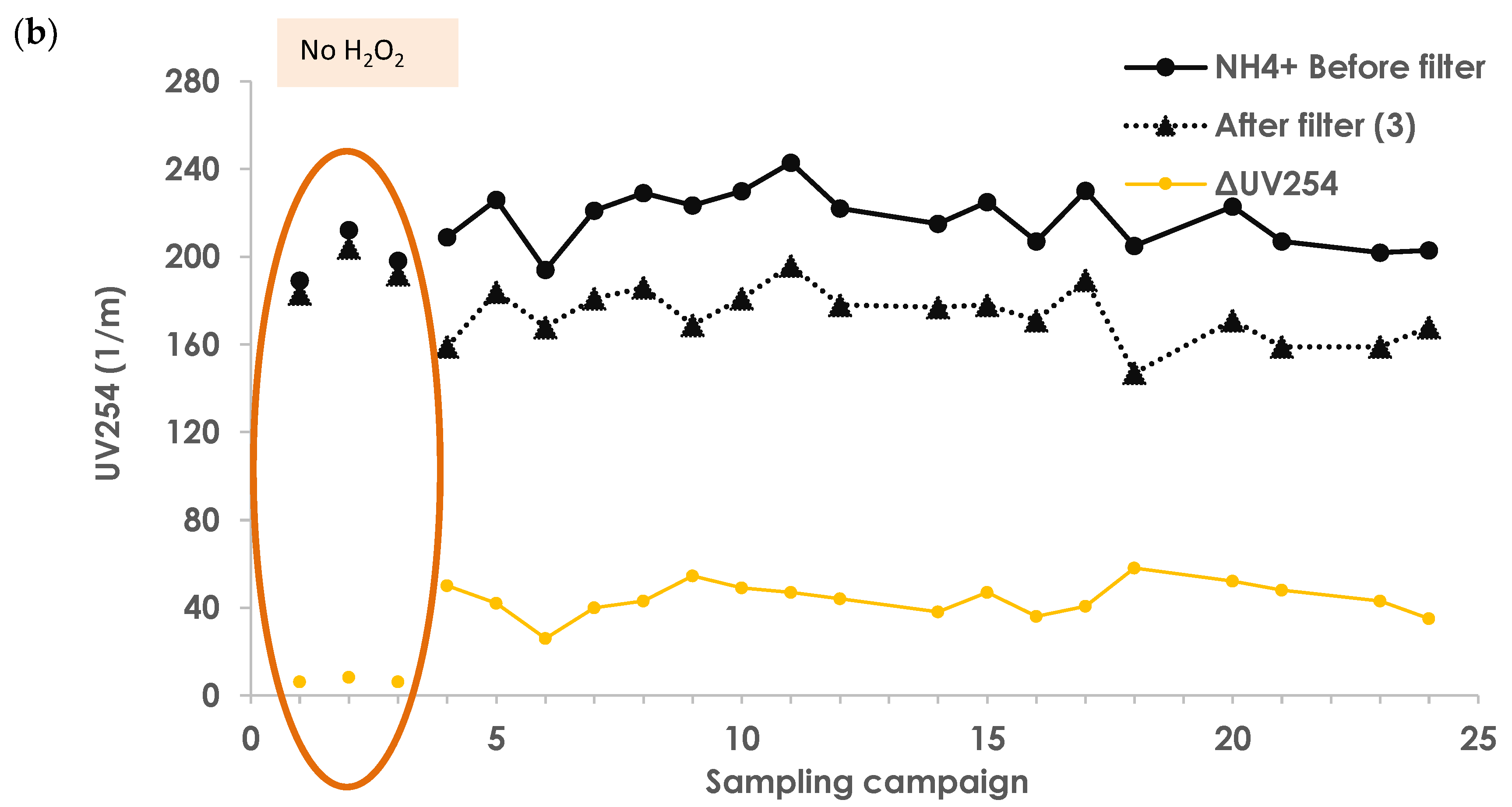
3.2. H2O2 Decomposition
3.3. Reference Measurements—No Addition of H2O2
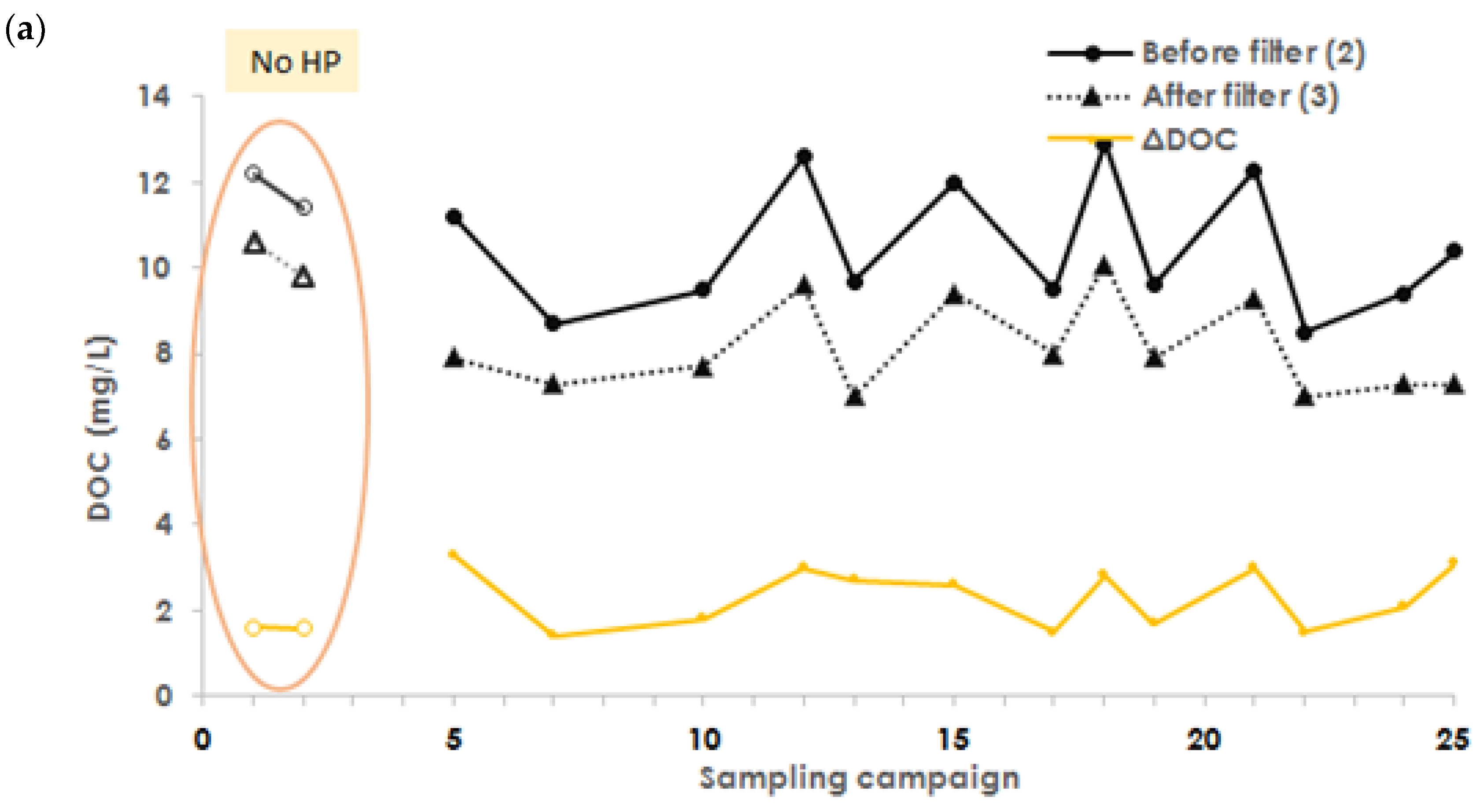
3.4. Organic Carbon Removal with H2O2 Addition
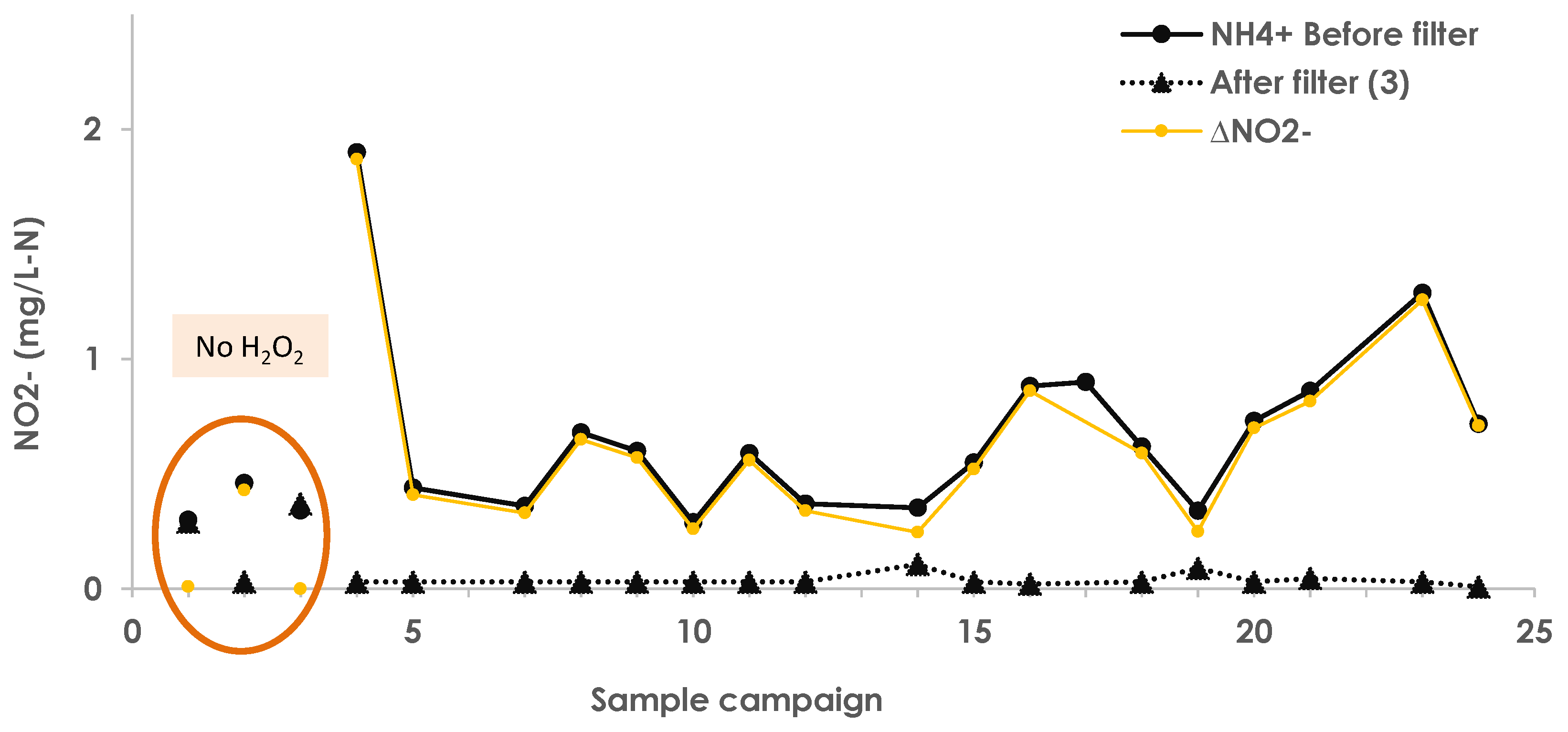
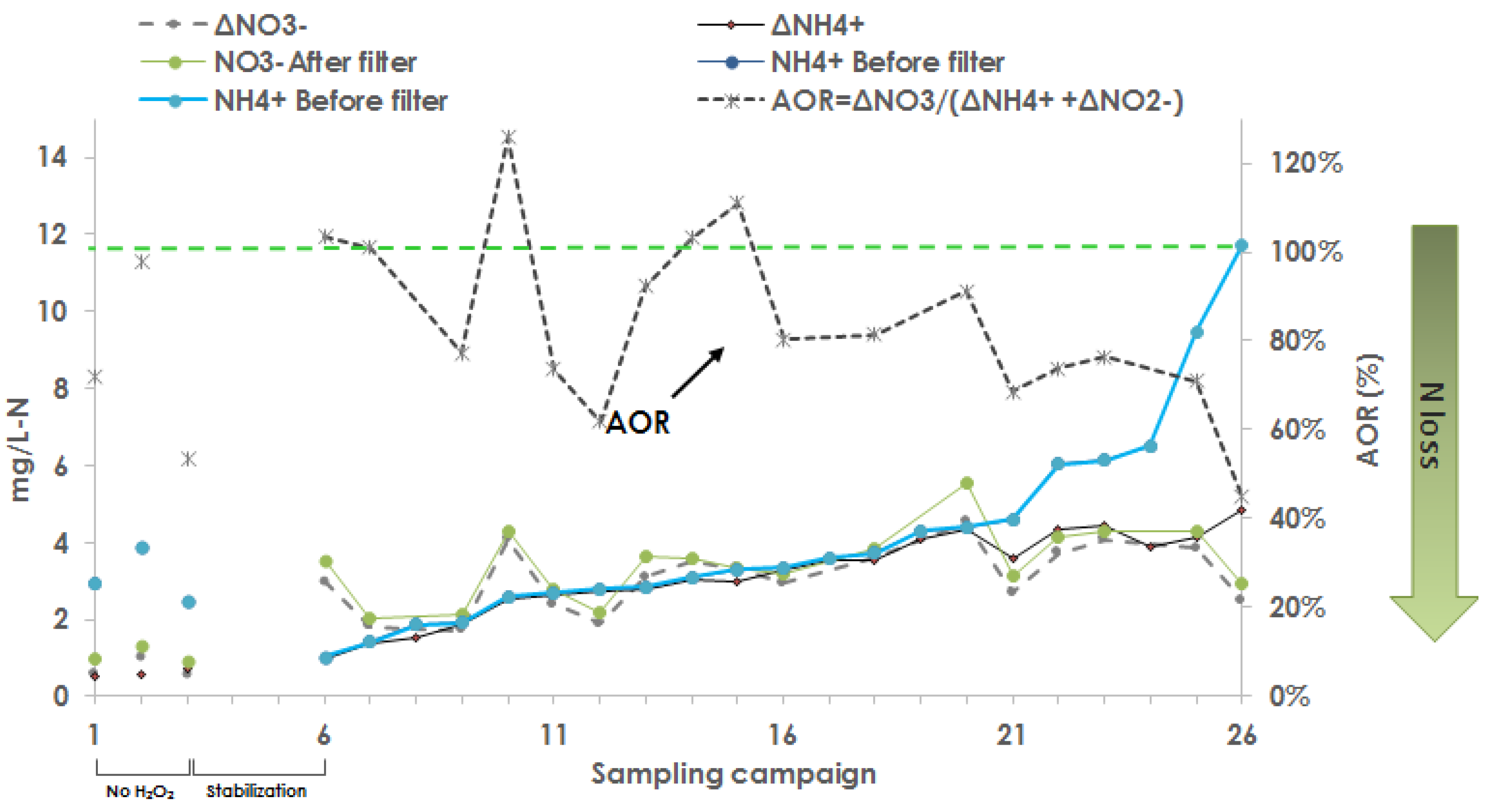
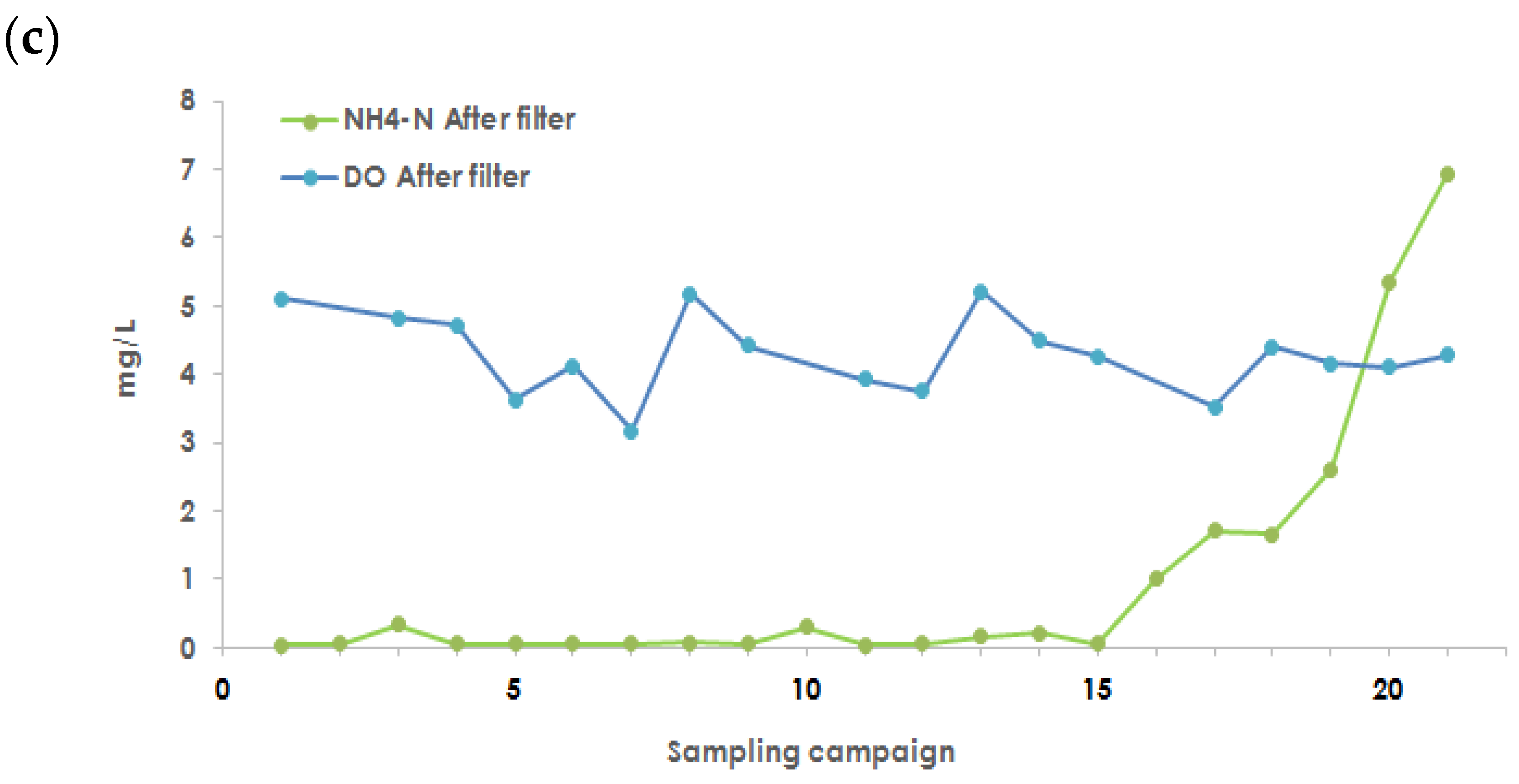
3.5. NH4+ and NO3− Transformation, Variation, and Nitrogen Mass Balance
- When AOR > 100%, ONT + Nit >> NX
- When AOR < 100%, significant NX
- When AOR ≈ 100%, Nit >> ONT, NX
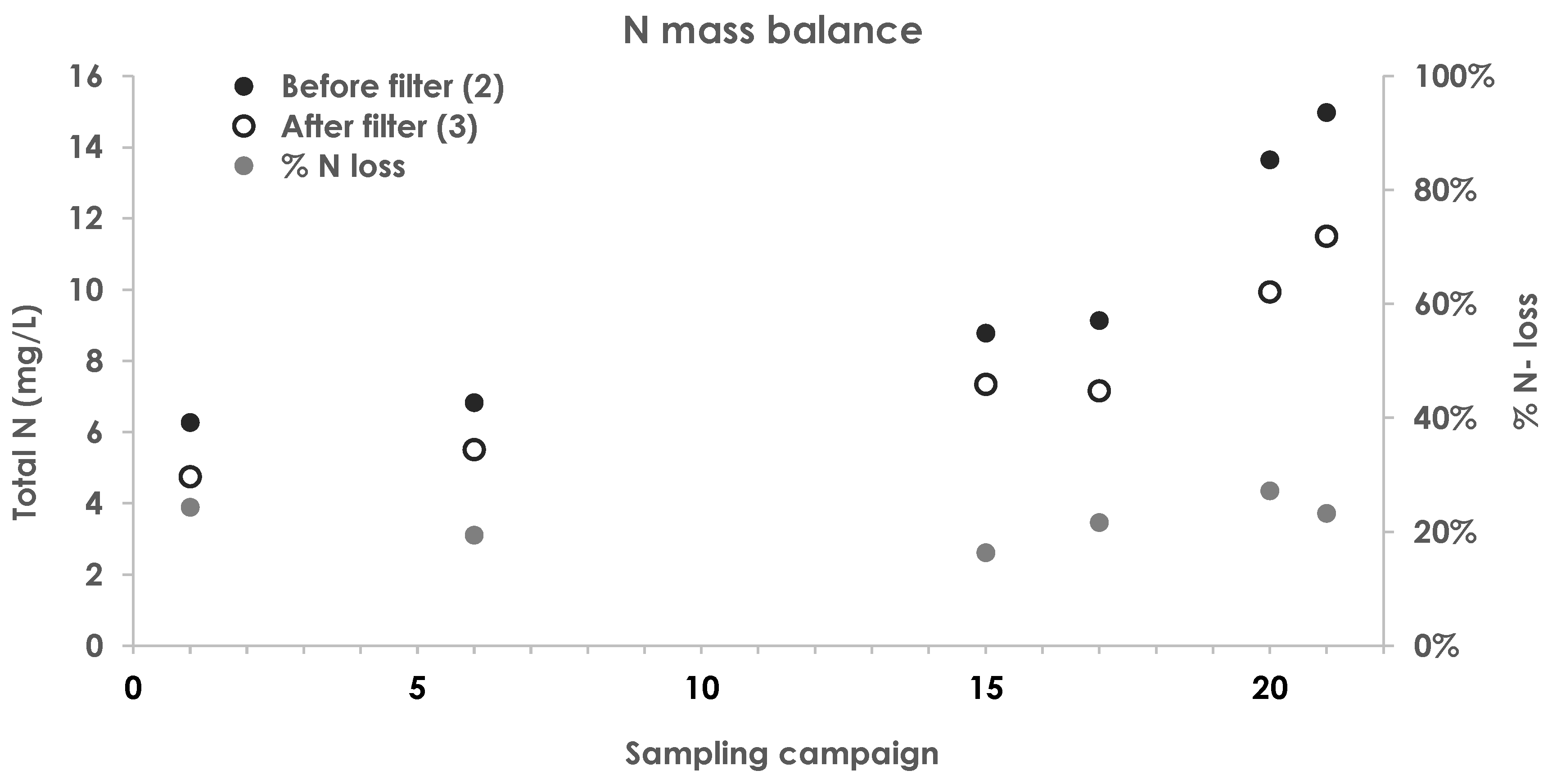
3.6. Mass Balance
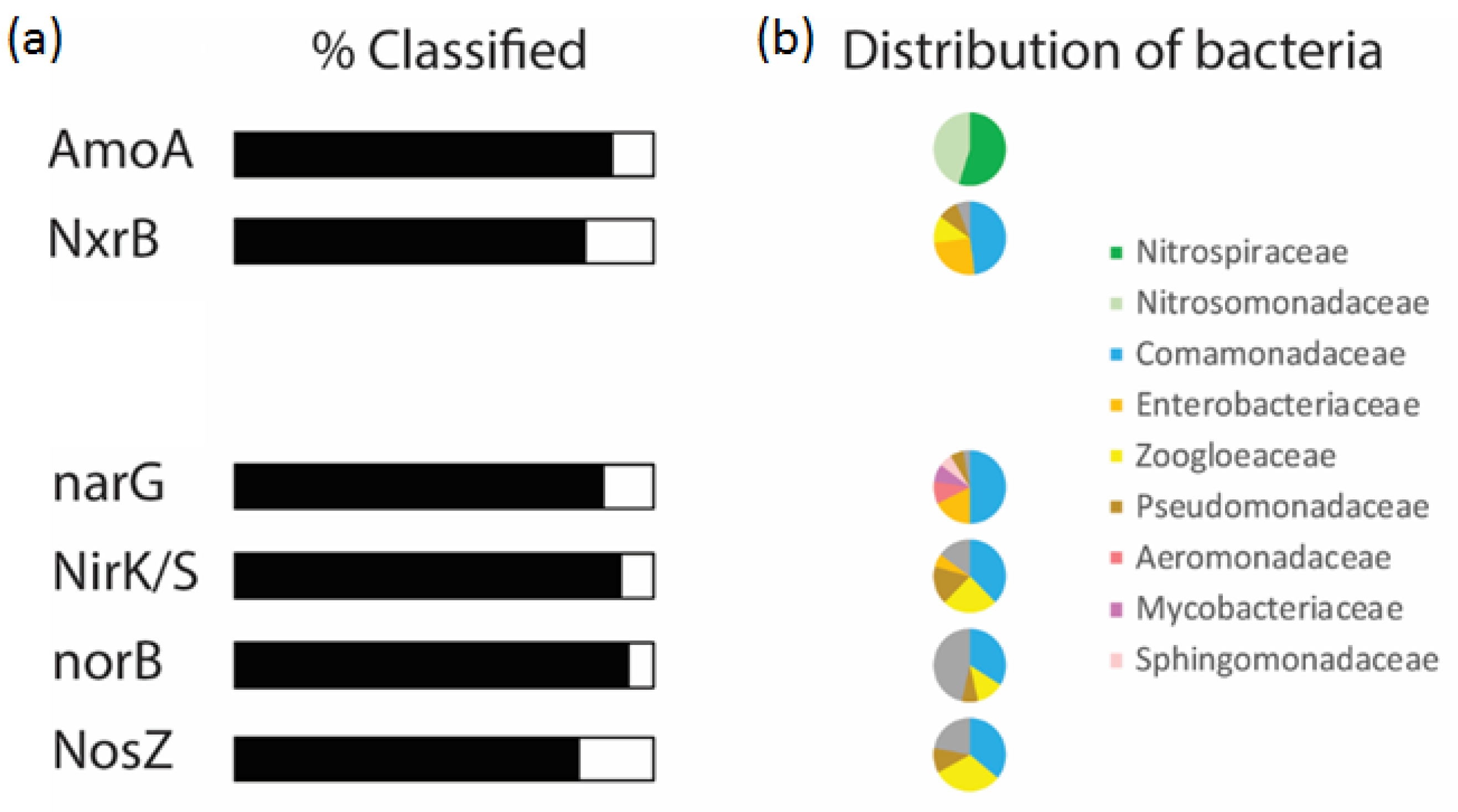
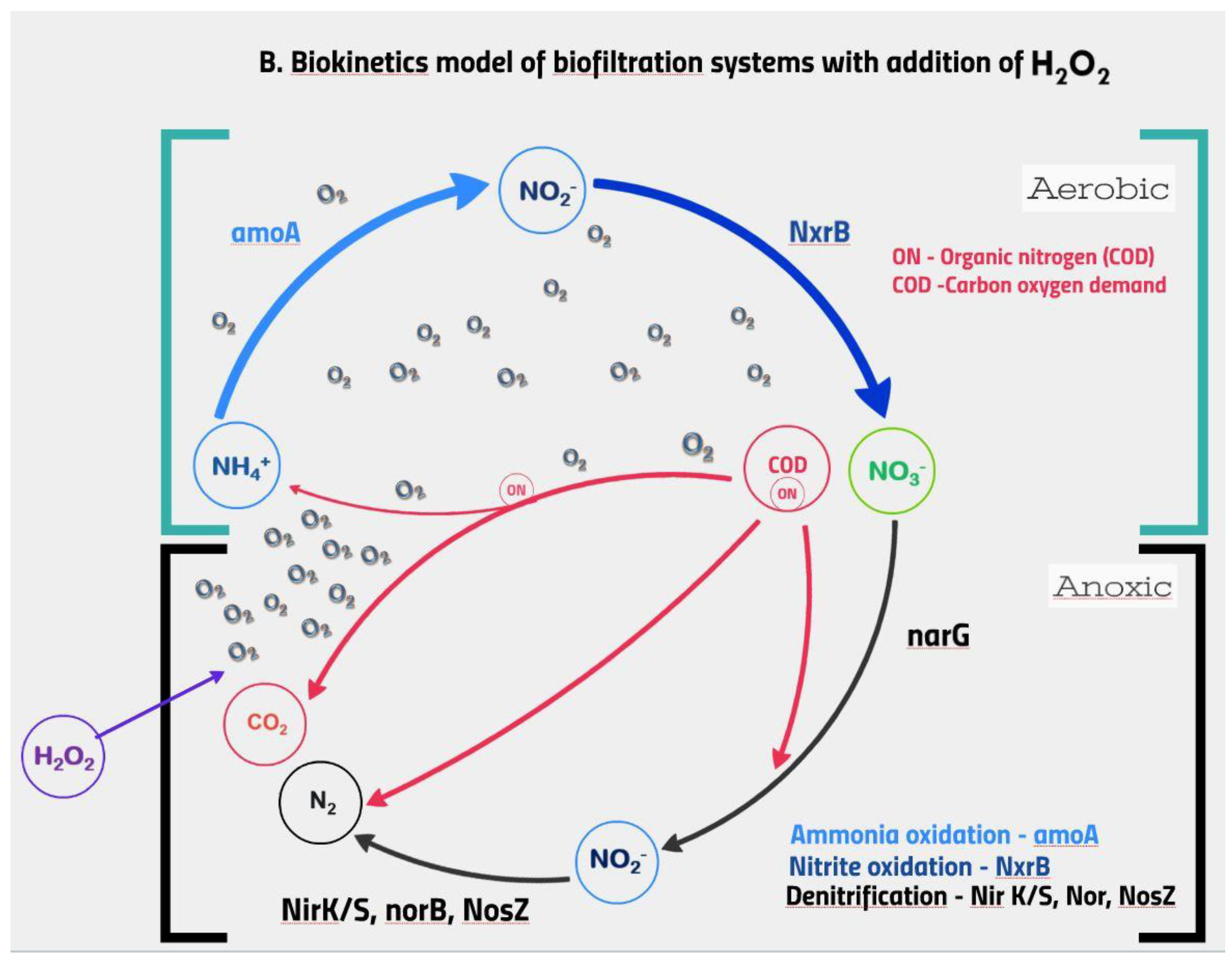
3.7. Functional Potential of the Biomass
4. Discussion
5. Conclusions
- -
- The addition of H2O2 to stimulate aerobic activity within a bed filtration process, practiced under the low range of the high rate bed filtration (5–6 m/h), enabled a combined effect of particle filtration and nitrification.The H2O2 was fully degraded, limiting the nitrification rate.
- -
- The trends in the concentration of NH4+, NO2−, and NO3− with addition of H2O2 demonstrated significant nitirification activity at the bed filter.
- -
- Metagenome analysis results were in line with the performance obtained and reflected significant potential for the simultanious nitrification denitrification activity of the attached biomass.
- -
- -
- In cases of residual NH4+ concentration (<5 mg N-NH4+/L) in secondary WWTP effluents, the presented technology shows potential for either managing ammonia concentration in the effluent or for reducing oxygen demand in following processes, such as SAT or direct reuse, under local regulation.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Theoretical Calculation of and Oxygen Nitrogen Mass Balance
| OHO Yield = 0.6 | # Campaign | N Loss (mg-N/L) | OHO Assimilation (mg-N/L) | N Loss for Denitrification (mg-N/L) | Denitrification Credit (COD mg/L Utilized for Denitrification) |
| 0 | 1.52 | 1.04 | 0.48 | 1.3 | |
| 6 | 1.32 | 0.82 | 0.50 | 1.4 | |
| 15 | 1.43 | 1.04 | 0.39 | 1.1 | |
| 17 | 1.97 | 1.26 | 0.71 | 2.0 | |
| 20 | 3.71 | 1.64 | 2.07 | 5.8 | |
| 21 | 3.47 | 1.12 | 2.36 | 6.6 | |
| Average H2O2 | 2.24 | 1.15 | 1.09 | 3.0 | |
| STD dev. H2O2 | 0.98 | 0.25 | 0.81 | 2.27 | |
| 28 | 0.32 | 0.30 | 0.02 | 0.1 | |
| 29 | 0.25 | 0.15 | 0.10 | 0.3 | |
| 30 | 0.18 | 0.30 | 0.00 | 0.0 | |
| Average Control | 0.25 | 0.25 | 0.04 | 0.1 | |
| STD dev Control | 0.06 | 0.07 | 0.04 | 0.12 | |
| OHO Yield = 0.4 | # Campaign | N Loss (mg-N/L) | OHO Assimilation (mg-N/L) | N Loss for Deni (mg-N/L) | Denitrification Credit (COD mg/L Utilized for Deni) |
| 0 | 1.52 | 0.69 | 0.83 | 2.3 | |
| 6 | 1.32 | 0.74 | 0.58 | 1.6 | |
| 15 | 1.43 | 0.69 | 0.74 | 2.1 | |
| 17 | 1.97 | 0.84 | 1.13 | 3.2 | |
| 20 | 3.71 | 1.09 | 2.62 | 7.3 | |
| 21 | 3.47 | 0.74 | 2.73 | 7.6 | |
| Average H2O2 | 2.24 | 0.80 | 1.44 | 4.0 | |
| STD H2O2 | 0.98 | 0.14 | 0.89 | 2.50 | |
| 28 | 0.32 | 0.20 | 0.12 | 0.3 | |
| 29 | 0.25 | 0.10 | 0.15 | 0.4 | |
| 30 | 0.18 | 0.20 | 0.00 | 0.0 | |
| Average Control | 0.25 | 0.17 | 0.09 | 0.3 | |
| STD dev. Control | 0.06 | 0.05 | 0.07 | 0.18 |
| # Campaign | O2 Inlet (mg-/L) | O2 Outlet (mg-/L) | O2 from H2O2 | O2 Balance (Consumed) | Delta NH4 | Potential Oxygen Consumed for Nitrification |
|---|---|---|---|---|---|---|
| 1 | 3.79 | 5.1 | 12.7 | 11.39 | 0.98 | 6.92 |
| 2 | 12.7 | 12.70 | 1.35 | 6.53 | ||
| 3 | 5.53 | 4.83 | 12.7 | 13.40 | 1.54 | 6.37 |
| 4 | 5 | 4.72 | 12.7 | 12.98 | 1.85 | 4.53 |
| 5 | 3.87 | 3.62 | 12.7 | 12.95 | 2.55 | 1.30 |
| 6 | 3.8 | 4.12 | 12.7 | 12.40 | 2.63 | 0.37 |
| 7 | 3.43 | 3.18 | 12.7 | 12.95 | 2.75 | 0.38 |
| 8 | 6.30 | 5.17 | 12.7 | 13.83 | 2.77 | 1.16 |
| 9 | 5.38 | 4.42 | 12.7 | 13.66 | 3.05 | −0.28 |
| 10 | 12.7 | 12.70 | 3.00 | −1.01 | ||
| 11 | 4.69 | 3.92 | 12.7 | 13.47 | 3.30 | −1.61 |
| 12 | 3.92 | 3.75 | 12.7 | 12.87 | 3.55 | −3.35 |
| 13 | 5.83 | 5.2 | 12.7 | 13.32 | 3.56 | −2.93 |
| 14 | 4.1 | 4.49 | 12.7 | 12.26 | 4.08 | −6.39 |
| 15 | 6.15 | 4.27 | 12.7 | 14.58 | 4.35 | −5.30 |
| 16 | 5.13 | 3.53 | 12.7 | 14.30 | 3.60 | −2.15 |
| 17 | 4.50 | 4.41 | 12.7 | 12.79 | 4.32 | −6.94 |
| 18 | 3.83 | 4.16 | 12.7 | 12.37 | 4.46 | −8.00 |
| 19 | 5.67 | 4.10 | 12.7 | 14.27 | 3.90 | −3.55 |
| 20 | 5.48 | 4.28 | 12.7 | 13.90 | 4.12 | −4.94 |
Appendix A.2. The Backwash Cycle
Appendix A.3. Water Parameters Methodology
Appendix A.4. H2O2 Decomposition
Appendix A.5. Secondary Effluent Particle Analysis
Appendix A.6. Particles Distribution
Appendix A.7. Media Sampling and Metagenome Analysis
Appendix A.8. Functional Molecular Analysis of the Biomass
Appendix A.9. Biofilter Performance—Particle Analysis
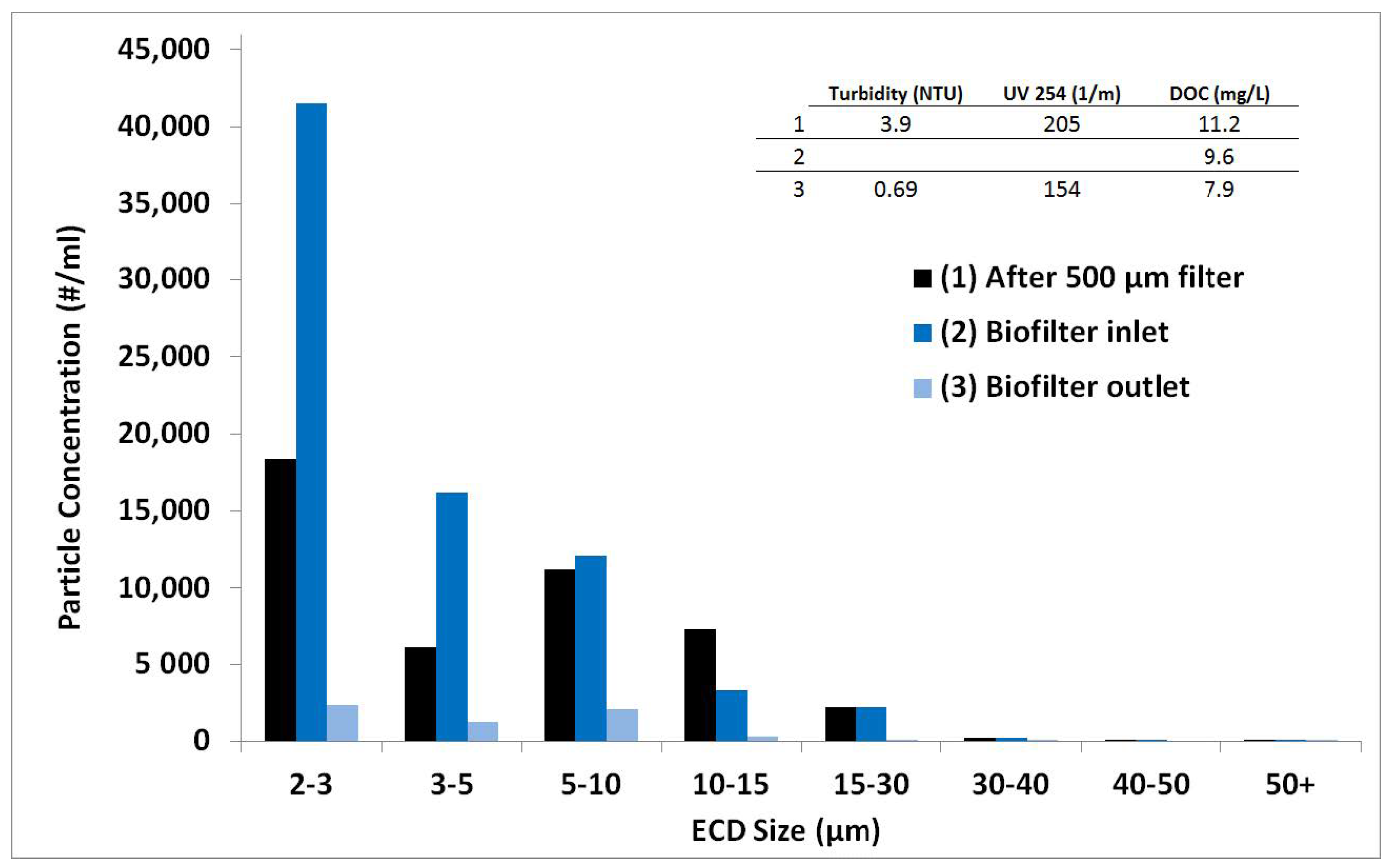
Appendix A.10. Organic Carbon Performance
Appendix A.11. Nitrite Removal and Concentration
References
- Bixio, D.; De Hayder, B.; Cikurel, H.; Muston, M.; Miska, V.; Joksimovic, D.; Schäfer, A.I.; Ravazzini, A.; Aharoni, A.; Savic, D.; et al. Municipal wastewater reclamation: Where do we stand? An overview of treatment technology and management practice. Water Sci. Technol. Water Supply 2005, 5, 77–86. [Google Scholar] [CrossRef]
- Siegel, S.M. Let There Be Water: Israel’s Solution for a Water-Starved World; Thomas Dunne Books/St. Martin’s Press: New York, NY, USA, 2015; ISBN 978-1-250-07395-2. [Google Scholar]
- Emerging Technologies for Wastewater Treatment and In-Plant Wet Weather Management; U.S Environmental Protection Agency (EPA): Washington DC, USA, 2013; Volume 832.
- Tchobanoglous, F.B.G. Wastewater Engineering and Reuse, 4th ed.; McGraw-Hill: New York, NY, USA, 2003; ISBN 978-1-259-01079-8. [Google Scholar]
- Van Den Akker, B.; Holmes, M.; Cromar, N.; Fallowfield, H. Application of high rate nitrifying trickling filters for potable water treatment. Water Res. 2008, 42, 4514–4524. [Google Scholar] [CrossRef]
- Miska-Markusch, V. Effluent Filtration for more than Particle Removal; Leibniz Universität Hannover: Hannover, Germany, 2009. [Google Scholar]
- Müller, J.; Drewes, J.E.; Hübner, U. Sequential biofiltration—A novel approach for enhanced biological removal of trace organic chemicals from wastewater treatment plant effluent. Water Res. 2017, 127, 127–138. [Google Scholar] [CrossRef]
- Alidina, M.; Li, D.; Drewes, J.E. Investigating the role for adaptation of the microbial community to transform trace organic chemicals during managed aquifer recharge. Water Res. 2014, 56, 172–180. [Google Scholar] [CrossRef]
- Palomo, A.; Fowler, S.J.; Gülay, A.; Rasmussen, S.; Sicheritz-Ponten, T.; Smets, B.F. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 2016, 10, 2569–2581. [Google Scholar] [CrossRef]
- Pinto, A.J.; Marcus, D.N.; Ijaz, Z.; Bautista-de los Santos, Q.M.; Dick, G.J.; Raskin, L. Metagenomic Evidence for the Presence of Comammox Nitrospira-Like Bacteria in a Drinking Water System. mSphere 2015, 1, e00054-15. [Google Scholar] [CrossRef]
- Salerno, C.; Berardi, G.; Laera, G.; Pollice, A. Functional Response of MBR Microbial Consortia to Substrate Stress as Revealed by Metaproteomics. Microb. Ecol. 2019, 78, 873–884. [Google Scholar] [CrossRef]
- Salerno, C.; Benndorf, D.; Kluge, S.; Palese, L.L. Metaproteomics Applied to Activated Sludge for Industrial Wastewater Treatment Revealed a Dominant Methylotrophic Metabolism of Hyphomicrobium zavarzinii. Microb. Ecol. 2016, 72, 9–13. [Google Scholar] [CrossRef]
- Mendoza-Espinoza, L.; Stephenson, T. A Review of Biological Aerated Filters (BAFs) for Wastewater Treatment. Environ. Eng. Sci. 1999, 16, 201–216. [Google Scholar] [CrossRef]
- Rassamee, V.; Sattayatewa, C.; Pagilla, K.; Chandran, K. Effect of oxic and anoxic conditions on nitrous oxide emissions from nitrification and denitrification processes. Biotechnol. Bioeng. 2011, 108, 2036–2045. [Google Scholar] [CrossRef]
- Yoo, H.; Ahn, K.; Lee, H.J.; Lee, K.; Kwak, Y.; Song, K. Nitrogen removal from synthetic wastewater by simultaneous nitrification and denitrification (SND) via nitrite in an intermittently-aerated reactor. Water Res. 1999, 33, 145–154. [Google Scholar] [CrossRef]
- Bassin, J.P.; Abbas, B.; Vilela, C.L.S.; Kleerebezem, R.; Muyzer, G.; Rosado, A.S.; van Loosdrecht, M.C.M.; Dezotti, M. Tracking the dynamics of heterotrophs and nitrifiers in moving-bed biofilm reactors operated at different COD/N ratios. Bioresour. Technol. 2015, 192, 131–141. [Google Scholar] [CrossRef]
- de Vet, W.W.J.M.; Kleerebezem, R.; van der Wielen, P.W.J.J.; Rietveld, L.C.; van Loosdrecht, M.C.M. Assessment of nitrification in groundwater filters for drinking water production by qPCR and activity measurement. Water Res. 2011, 45, 4008–4018. [Google Scholar] [CrossRef]
- Yu, X.; Qi, Z.; Zhang, X.; Yu, P.; Liu, B.; Zhang, L.; Fu, L. Nitrogen loss and oxygen paradox in full-scale biofiltration for drinking water treatment. Water Res. 2007, 41, 1455–1464. [Google Scholar] [CrossRef]
- Jiang, B.; Hu, W.; Pei, H.; Chen, P.; Liu, Q. The influence of aeration on nitrification and the nitrifier distribution in an upflow biological aerated filter for tertiary treatment of municipal sewage. Desalin. Water Treat. 2010, 24, 308–320. [Google Scholar] [CrossRef]
- Albuquerque, A.; Makinia, J.; Pagilla, K. Impact of aeration conditions on the removal of low concentrations of nitrogen in a tertiary partially aerated biological filter. Ecol. Eng. 2012, 44, 44–52. [Google Scholar] [CrossRef]
- Matsumoto, S.; Terada, A.; Tsuneda, S. Modeling of membrane-aerated biofilm: Effects of C/N ratio, biofilm thickness and surface loading of oxygen on feasibility of simultaneous nitrification and denitrification. Biochem. Eng. J. 2007, 37, 98–107. [Google Scholar] [CrossRef]
- Brindle, K.; Stephenson, T.; Semmens, M.J. Nitrification and oxygen utilisation in a membrane aeration bioreactor. J. Memb. Sci. 1998, 144, 197–209. [Google Scholar] [CrossRef]
- Pellicer-Nàcher, C.; Smets, B.F. Structure, composition, and strength of nitrifying membrane-aerated biofilms. Water Res. 2014, 57, 151–161. [Google Scholar] [CrossRef]
- Fiorenza, S.; Ward, C.H. Microbial adaptation to hydrogen peroxide and biodegradation of aromatic hydrocarbons. J. Ind. Microbiol. Biotechnol. 1997, 18, 140–151. [Google Scholar] [CrossRef]
- Zappi, M.; White, K.; Hwang, H.-M.; Bajpai, R.; Qasim, M. The Fate of Hydrogen Peroxide as an Oxygen Source for Bioremediation Activities within Saturated Aquifer Systems. J. Air Waste Manag. Assoc. 2000, 50, 1818–1830. [Google Scholar] [CrossRef]
- Chance, B.; Herbert, D. The enzyme-substrate compounds of bacterial catalase and peroxides. Biochem. J. 1950, 46, 402–414. [Google Scholar] [CrossRef]
- Northrop, J.H. Experimental Conditions and General Results. Effect of Varying the Concentration of Catalase. J. Gen. Physiology 1924, 5, 373–387. [Google Scholar]
- Sooch, B.S.; Kauldhar, B.S.; Puri, M. Recent insights into microbial catalases: Isolation, production and purification. Biotechnol. Adv. 2014, 32, 1429–1447. [Google Scholar] [CrossRef]
- Flores, M.J.; Brandi, R.J.; Cassano, A.E.; Labas, M.D. Chemical disinfection with H2O2—The proposal of a reaction kinetic model. Chem. Eng. J. 2012, 198–199, 388–396. [Google Scholar] [CrossRef]
- Lakretz, A.; Ron, E.Z.; Mamane, H. Biofilm control in water by a UV-based advanced oxidation process. Biofouling 2011, 27, 295–307. [Google Scholar] [CrossRef]
- Tusseau-Vuillemin, M.H.; Lagarde, F.; Chauviere, C.; Heduit, A. Hydrogen peroxide (H2O2) as a source of dissolved oxygen in COD-degradation respirometric experiments. Water Res. 2002, 36, 793–798. [Google Scholar] [CrossRef]
- Lakretz, A.; Ron, E.Z.; Harif, T.; Mamane, H. Biofilm control in water by advanced oxidation process (AOP) pre-treatment: Effect of natural organic matter (NOM). Water Sci. Technol. 2011, 64, 1876–1884. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, D.; Li, Y.; Yu, L.; Gin, K.Y.H.; Chen, J.P.; Reinhard, M. Effects of monochloramine and hydrogen peroxide on the bacterial community shifts in biologically treated wastewater. Chemosphere 2017, 189, 399–406. [Google Scholar] [CrossRef]
- Zucker, I.; Mamane, H.; Cikurel, H.; Jekel, M.; Hubner, U.; Avisar, D. A hybrid process of biofiltration of secondary effluent followed by ozonation and short soil aquifer treatment for water reuse. Water Res. 2015, 84, 315–322. [Google Scholar] [CrossRef]
- Cikurel, H.; Rebhun, M.; Amirtharajah, A.; Adin, A. Wastewater effluent reuse by in-line flocculation filtration process. Water Sci. Technol. 1996, 33, 203–211. [Google Scholar] [CrossRef]
- Mamane, H.; Kohn, C.; Adin, A. Characterizing Shape of Effluent Particles by Image Analysis. Sep. Sci. Technol. 2008, 43, 1737–1753. [Google Scholar] [CrossRef]
- Li, D.; Sharp, J.O.; Saikaly, P.E.; Ali, S.; Alidina, M.; Alarawi, M.S.; Keller, S.; Hoppe-Jones, C.; Drewes, J.E. Dissolved organic carbon influences microbial community composition and diversity in managed aquifer recharge systems. Appl. Environ. Microbiol. 2012, 78, 6819–6828. [Google Scholar] [CrossRef] [PubMed]
- Lauderdale, C.; Chadik, P.; Kirisits, M.J.; Brown, A.J. Engineered biofiltration: Enhanced biofilter performance through nutrient and peroxide addition. J. Am. Water Work. Assoc. 2012, 104, E298–E309. [Google Scholar] [CrossRef]
- Lakretz, A.; Elifantz, H.; Kviatkovski, I.; Eshel, G.; Mamane, H. Automatic microfiber filtration (AMF) of surface water: Impact on water quality and biofouling evolution. Water Res. 2014, 48, 592–604. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Shon, H.K.; Vigneswaran, S.; Snyder, S.A. Effluent Organic Matter (EfOM) in Wastewater: Constituents, Effects, and Treatment. Crit. Rev. Environ. Sci. Technol. 2006, 36, 327–374. [Google Scholar] [CrossRef]
- Petala, M.; Tsiridis, V.; Samaras, P.; Zouboulis, A.; Sakellaropoulos, G.P. Wastewater reclamation by advanced treatment of secondary effluents. Desalination 2006, 195, 109–118. [Google Scholar] [CrossRef]
- Grady, C.P.L.; Daigger, G.T.; Love, N.G.; Filipe, C.D.M. Biological Wastewater Treatment; IWA Publishing—Co-Publication: London, UK, 2011; ISBN 9781843393429. [Google Scholar]
- Friedman, L.; Mamane, H.; Avisar, D.; Chandran, K. The role of influent organic carbon-to-nitrogen (COD/N) ratio in removal rates and shaping microbial ecology in soil aquifer treatment (SAT). Water Res. 2018, 146, 197–205. [Google Scholar] [CrossRef]
- Cua, L.S.; Stein, L.Y. Effects of nitrite on ammonia-oxidizing activity and gene regulation in three ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 2011, 319, 169–175. [Google Scholar] [CrossRef]
- Chen, J.; Strous, M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lücker, S.; Wagner, M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Kits, K.D.; Sedlacek, C.J.; Lebedeva, E.V.; Han, P.; Bulaev, A.; Pjevac, P.; Daebeler, A.; Romano, S.; Albertsen, M.; Stein, L.Y.; et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 2017, 549, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Jetten, M.S.M. New pathways for ammonia conversion in soil and aquatic systems. Plant Soil 2001, 230, 9–19. [Google Scholar] [CrossRef]
- Stein, L.Y. Heterotrophic nitrification and nitrifier denitrification. In Nitrification; American Society of Microbiology: Washington, DC, USA, 2011. [Google Scholar]
- Ge, S.; Wang, S.; Yang, X.; Qiu, S.; Li, B.; Peng, Y. Detection of nitrifiers and evaluation of partial nitrification for wastewater treatment: A review. Chemosphere 2015, 140, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Park, M.R.; Park, H.; Chandran, K. Molecular and kinetic characterization of planktonic Nitrospira spp. selectively enriched from activated sludge. Environ. Sci. Technol. 2017, 51, 2720–2728. [Google Scholar] [CrossRef] [PubMed]
- Siahrostami, S.; Verdaguer-Casadevall, A.; Karamad, M.; Deiana, D.; Malacrida, P.; Wickman, B.; Escudero-Escribano, M.; Paoli, E.A.; Frydendal, R.; Hansen, T.W.; et al. Enabling direct H2O2 production through rational electrocatalyst design. Nat. Mater. 2013, 12, 1137–1143. [Google Scholar] [CrossRef]
- Yang, S.; Verdaguer-casadevall, A.; Arnarson, L.; Silvioli, L.; Viktor, C.; Frydendal, R.; Rossmeisl, J.; Chorkendor, I.; Stephens, I.E.L. Toward the Decentralized Electrochemical Production of H2O2: A Focus on the Catalysis. ACS Catal. 2018, 8, 4064–4081. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Manimaran, S.; Shen, Y.; Perez-Rogers, J.F.; Byrd, A.L.; Castro-Nallar, E.; Crandall, K.A.; Johnson, W.E. PathoScope 2.0: A complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2014, 2, 33. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Friedman, L.; Mamane, H.; Chandran, K.; Jekel, M.; Cikurel, H.; Hübner, U.; Elgart, M.; Dagan, S.; Santo-Domingo, J.; Avisar, D. Stimulating Nitrogen Biokinetics with the Addition of Hydrogen Peroxide to Secondary Effluent Biofiltration. Clean Technol. 2020, 2, 53-73. https://doi.org/10.3390/cleantechnol2010005
Friedman L, Mamane H, Chandran K, Jekel M, Cikurel H, Hübner U, Elgart M, Dagan S, Santo-Domingo J, Avisar D. Stimulating Nitrogen Biokinetics with the Addition of Hydrogen Peroxide to Secondary Effluent Biofiltration. Clean Technologies. 2020; 2(1):53-73. https://doi.org/10.3390/cleantechnol2010005
Chicago/Turabian StyleFriedman, Liron, Hadas Mamane, Kartik Chandran, Martin Jekel, Haim Cikurel, Uwe Hübner, Michael Elgart, Shlomi Dagan, Jorge Santo-Domingo, and Dror Avisar. 2020. "Stimulating Nitrogen Biokinetics with the Addition of Hydrogen Peroxide to Secondary Effluent Biofiltration" Clean Technologies 2, no. 1: 53-73. https://doi.org/10.3390/cleantechnol2010005
APA StyleFriedman, L., Mamane, H., Chandran, K., Jekel, M., Cikurel, H., Hübner, U., Elgart, M., Dagan, S., Santo-Domingo, J., & Avisar, D. (2020). Stimulating Nitrogen Biokinetics with the Addition of Hydrogen Peroxide to Secondary Effluent Biofiltration. Clean Technologies, 2(1), 53-73. https://doi.org/10.3390/cleantechnol2010005