Synthetic Bacterial Consortium Induces Dynamic Shifts in Fungal Community and Alters Microbial Network Topology in Barley Soil Under Field Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Barley Variety
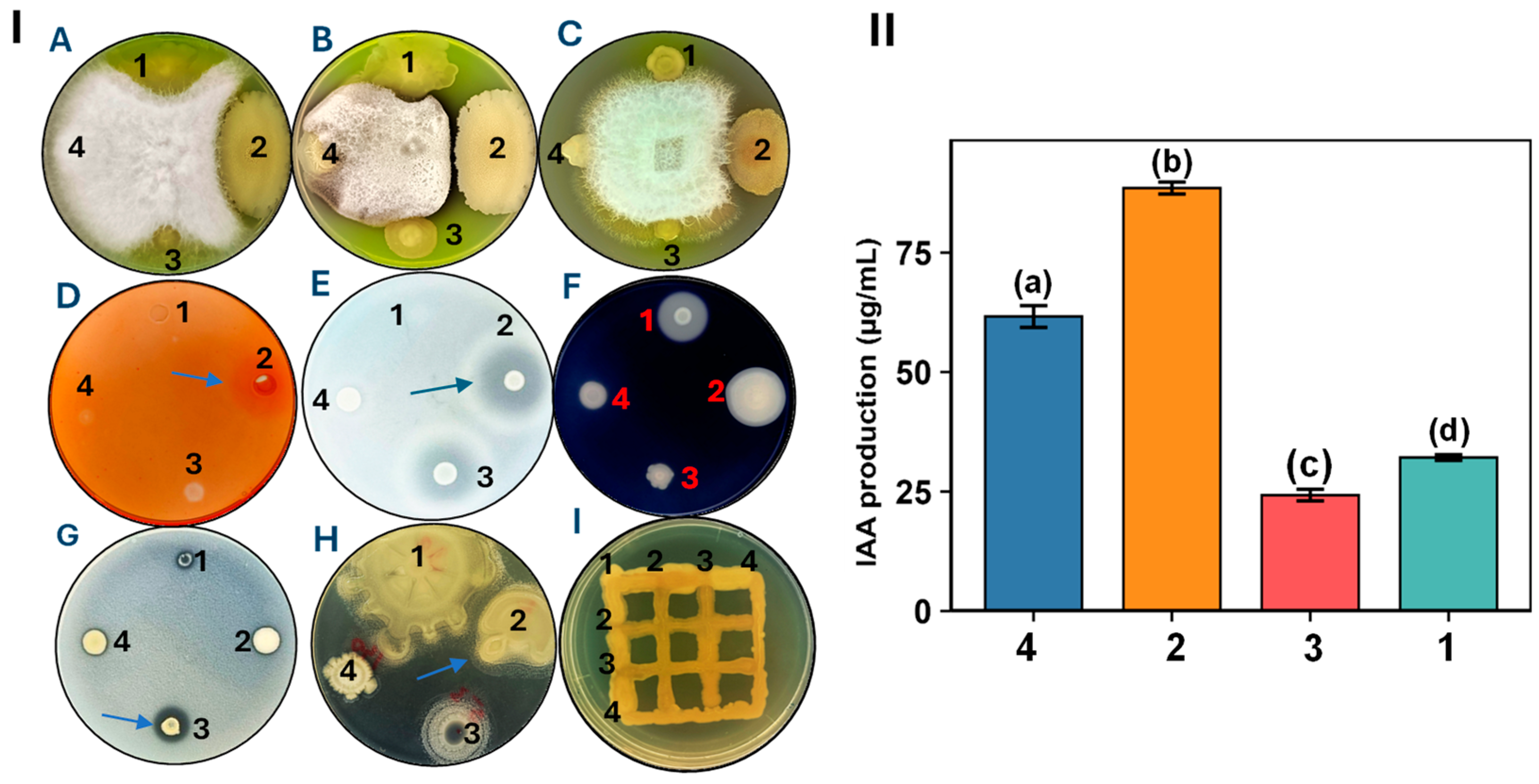
2.2. Plant Growth-Promoting Characteristics of Selected Bacterial Strains
2.3. Experimental Setup
2.3.1. Basal Fertilizer and Fertigation Management
2.3.2. Bacterial Inoculum Preparation and Crop Management
2.4. Soil Sample Collection and Property Analysis
Soil Chemical Analyses
2.5. DNA Isolation and Metabarcoding Analysis
2.5.1. DNA Isolation
2.5.2. Metabarcoding Sequencing and Library Preparation
2.5.3. Bioinformatics Analysis
2.6. Statistical Analysis
2.6.1. Fold Changes in Fungal Community Analysis
2.6.2. Fungal Community Networks and Comparison
3. Results
3.1. Biological Characterization of Selected Microbial Strains
3.2. Dynamic Shifts in Fungal Community Composition
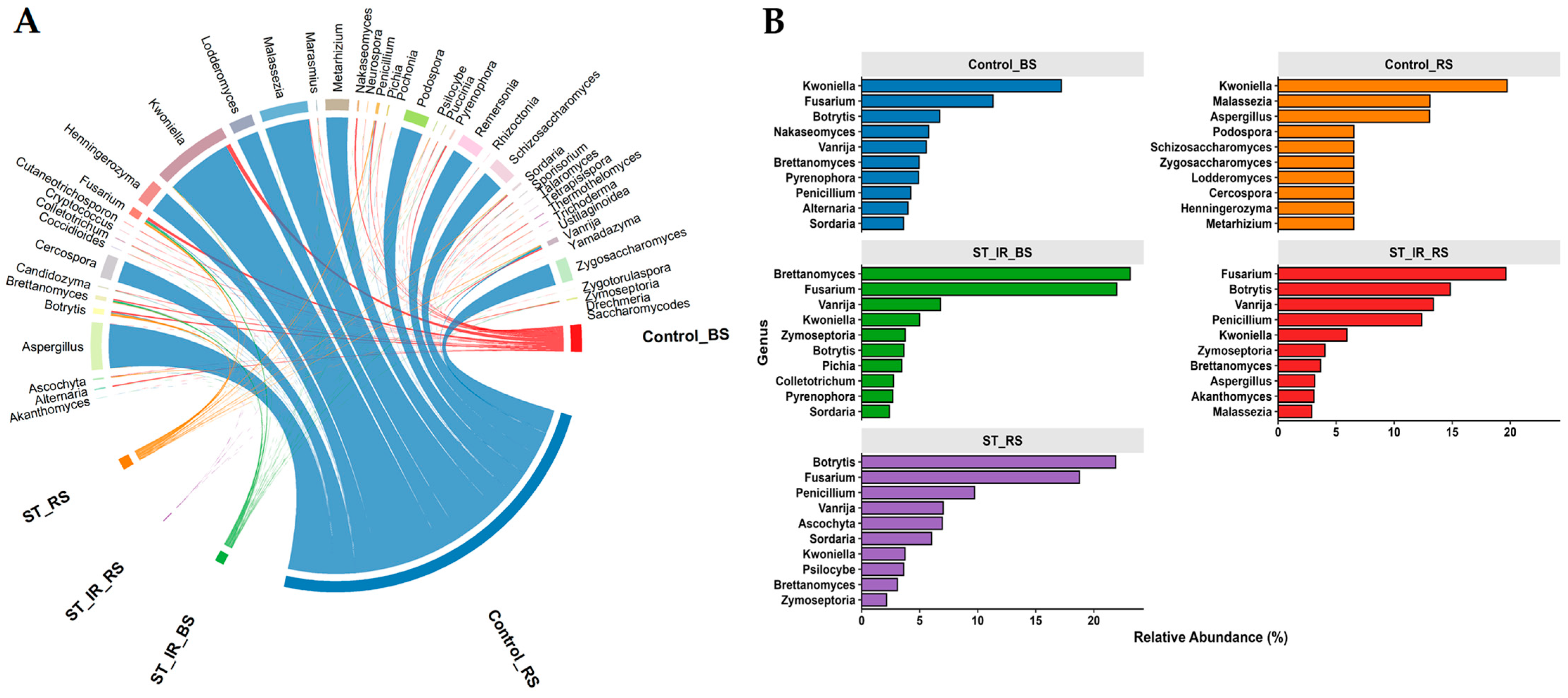
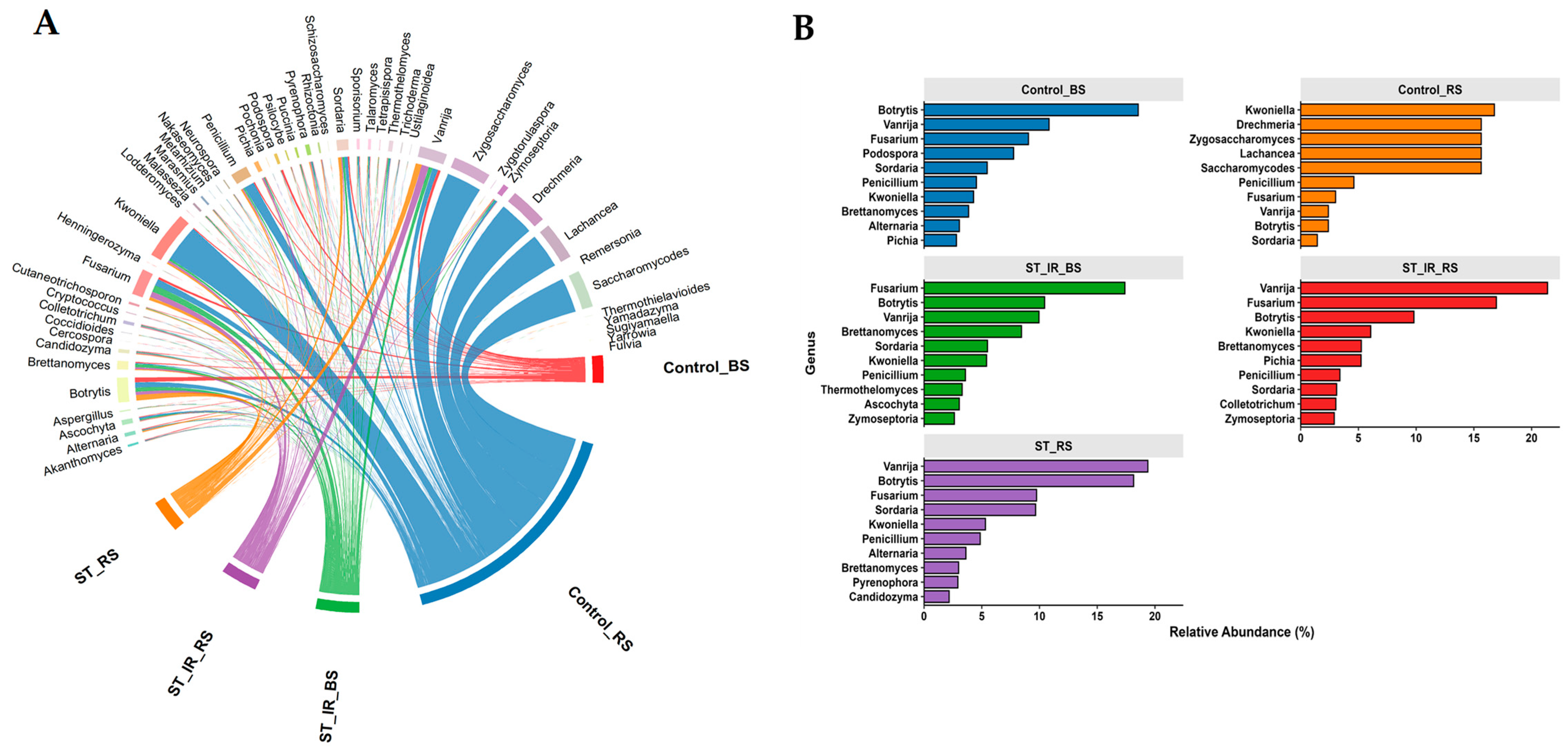
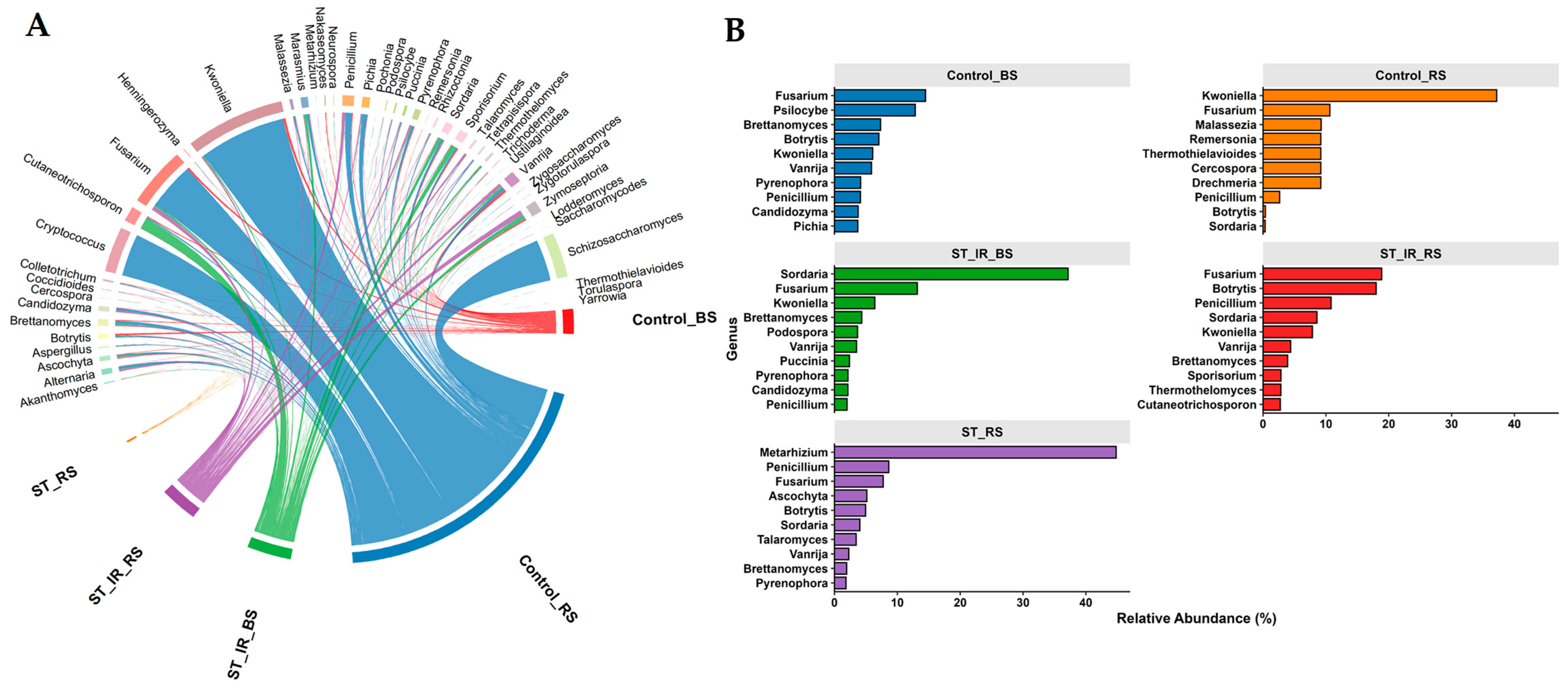
3.3. Fungal Community Composition and Unique Taxa Across Experimental Groups
3.4. Fungal Subcommunity Analysis
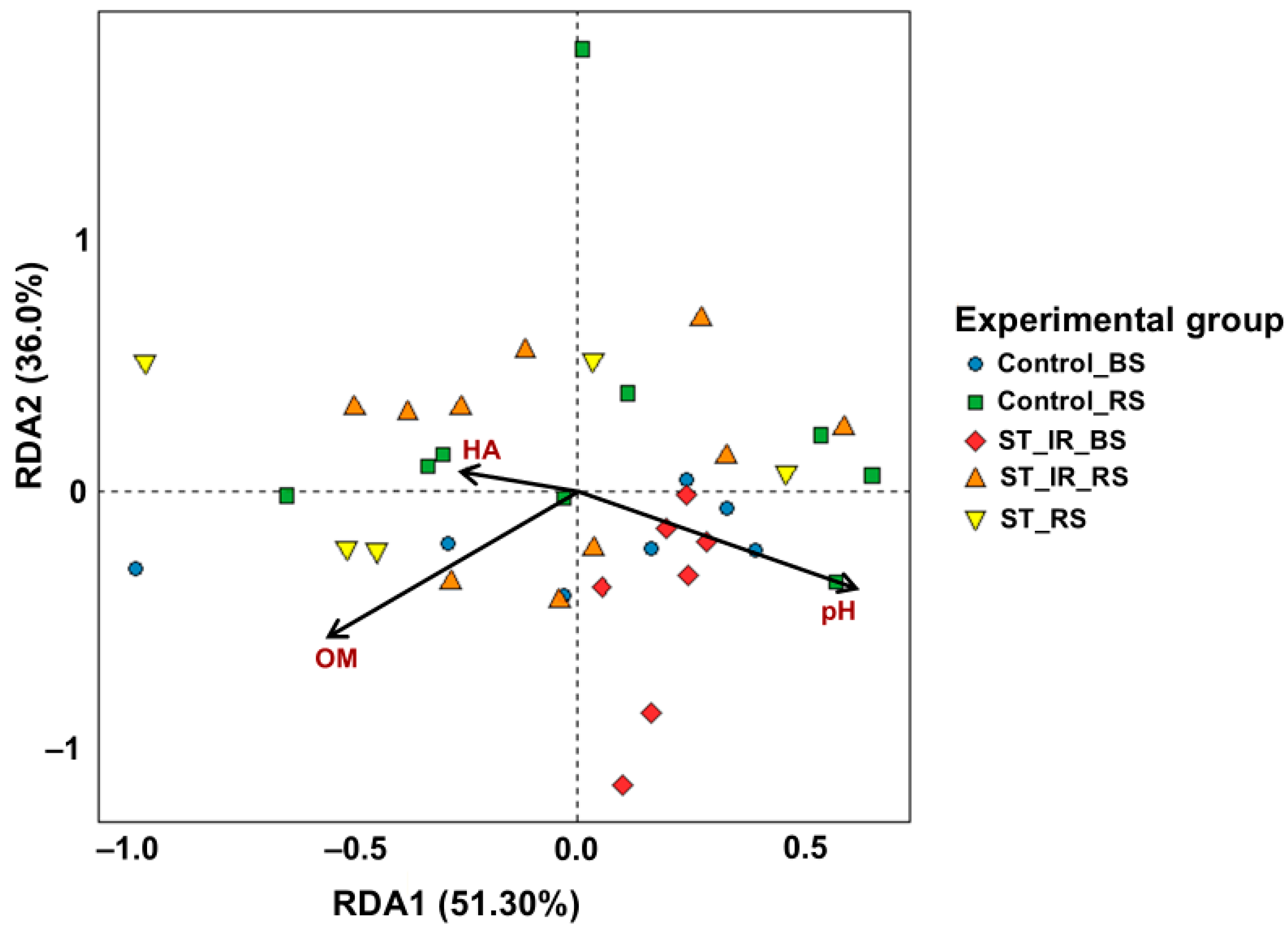
3.5. Correlation Between Soil Properties and Microbial Community
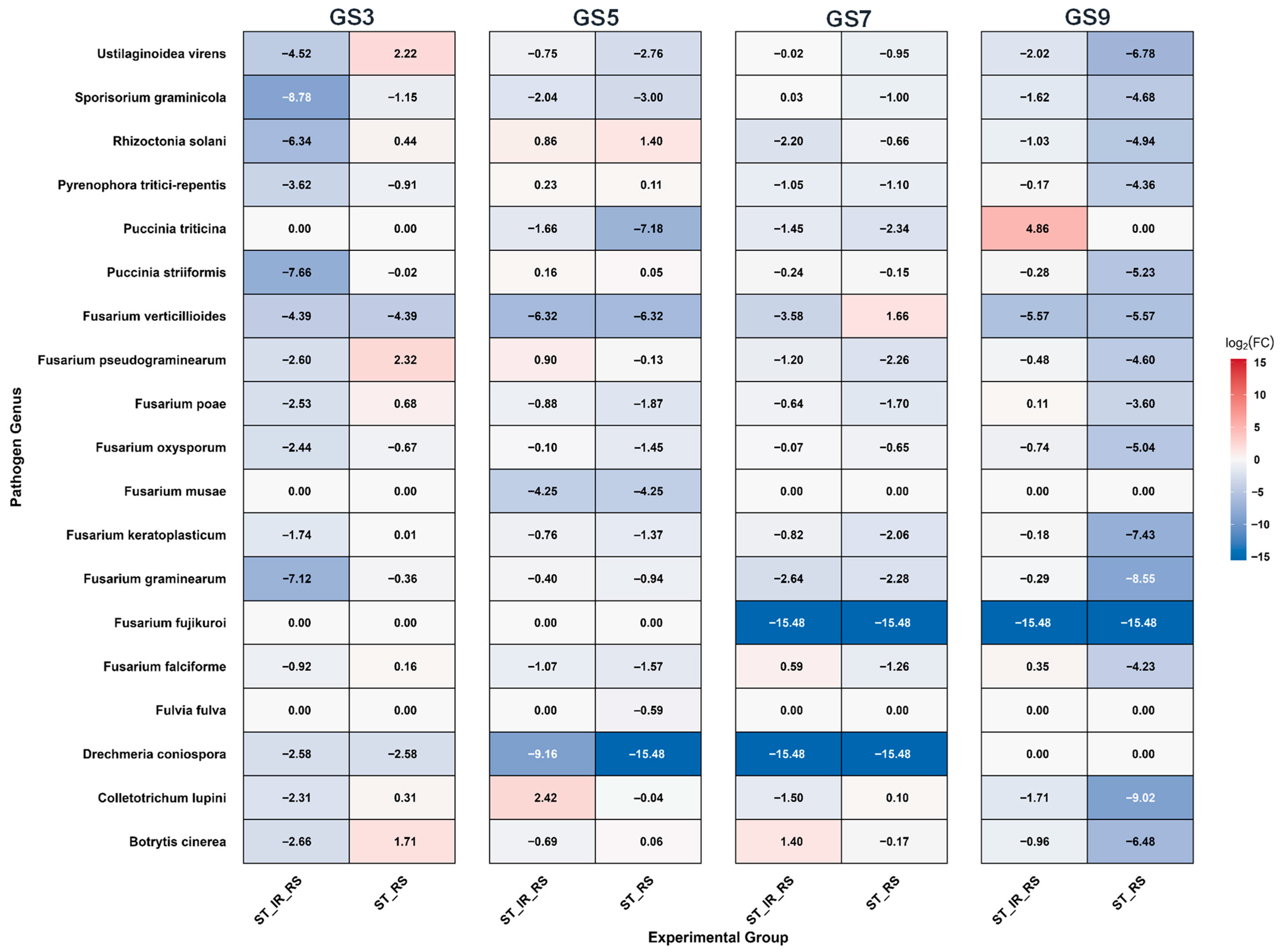
3.6. Fold Change Dynamics of Phytopathogens in the Rhizosphere of Barley Under Different Treatments
3.7. Alpha Diversity Analysis of Fungal Communities in Barley Under Treatments of Microbial Consortium
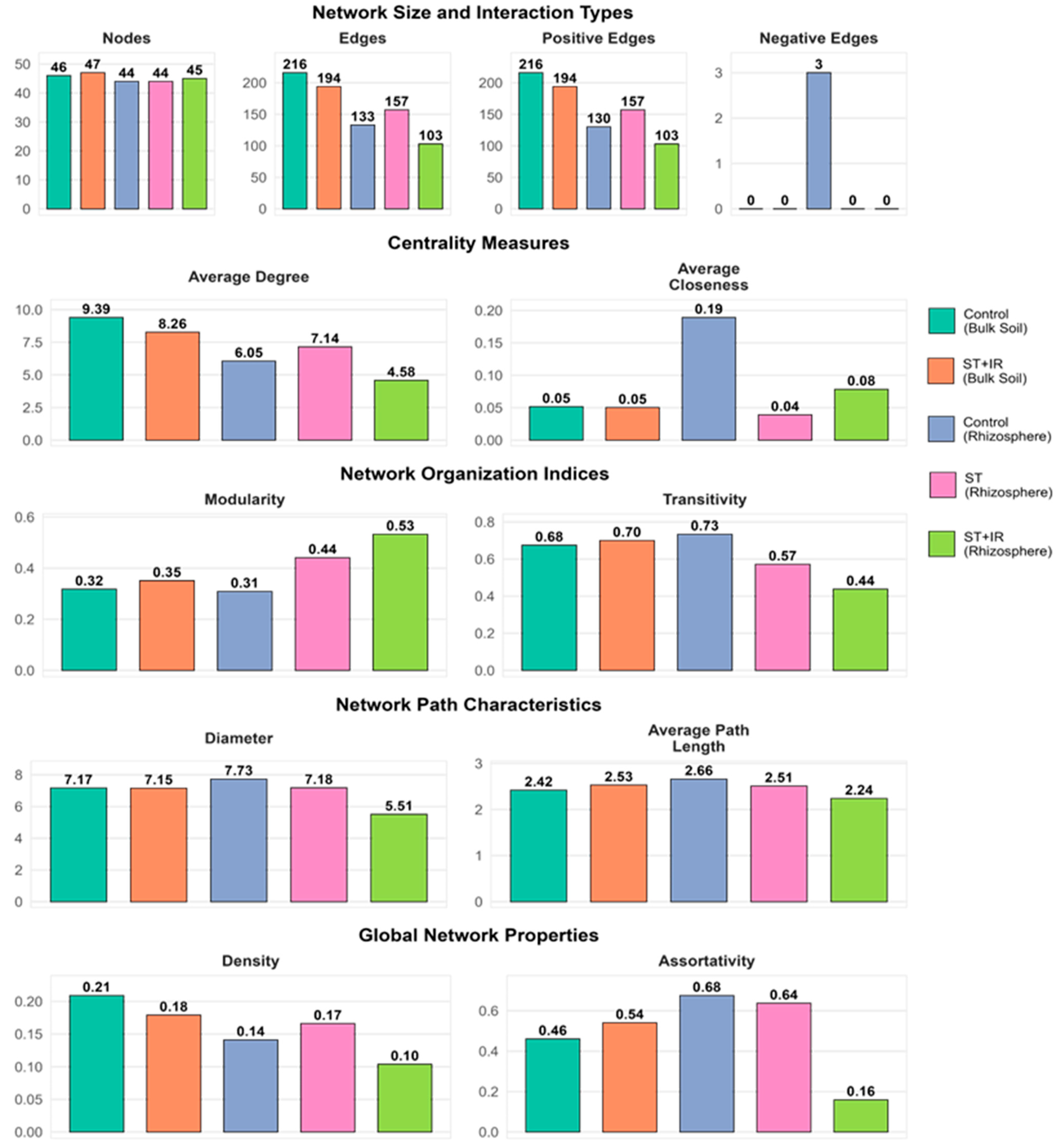
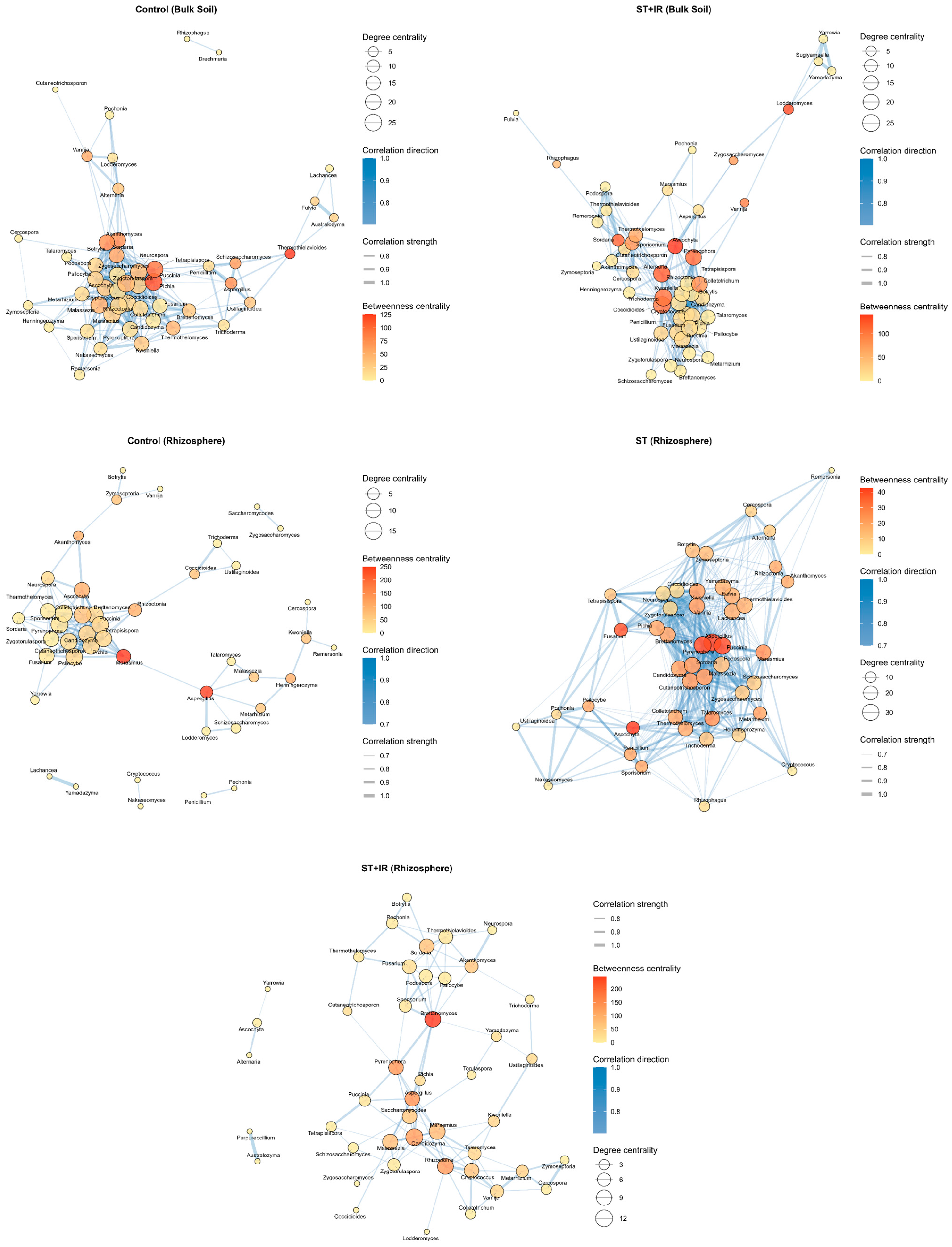
3.8. Fungal Community Interaction Networks in Soil Under Treatments at Different Growth Stages
Key Topological Features in Network Co-Occurrence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Devlet, A. Modern agriculture, and challenges. Front. Life Sci. Relat. Technol. 2021, 2, 21–29. [Google Scholar] [CrossRef]
- Tahat, M.M.; Alananbeh, K.M.; Othman, Y.A.; Leskovar, D.I. Soil health and sustainable agriculture. Sustainability 2020, 12, 4859. [Google Scholar] [CrossRef]
- Cárceles Rodríguez, B.; Durán-Zuazo, V.H.; Soriano Rodríguez, M.; García-Tejero, I.F.; Gálvez Ruiz, B.; Cuadros Tavira, S. Conservation Agriculture as a Sustainable System for Soil Health: A Review. Soil Syst. 2022, 6, 87. [Google Scholar] [CrossRef]
- Condron, L.; Stark, C.; O’Callaghan, M.; Clinton, P.; Huang, Z. The role of microbial communities in the formation and decomposition of soil organic matter. In Soil Microbiology and Sustainable Crop Production; Springer: Dordrecht, The Netherlands, 2010; pp. 81–118. [Google Scholar]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, K.C.; Razdan, N.; Saharan, K. Rhizospheric Microbiome: Bio-Based Emerging Strategies for Sustainable Agriculture Development and Future Perspectives. Microbiol. Res. 2022, 254, 126901. [Google Scholar] [CrossRef]
- Santoyo, G. How plants recruit their microbiome? New insights into beneficial interactions. J. Adv. Res. 2022, 40, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Ficke, A.; Aubertot, J.N.; Hollier, C. Crop losses due to diseases and their implications for global food production and food security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Hakim, S.; Naqqash, T.; Nawaz, M.S.; Laraib, I.; Siddique, M.J.; Zia, R.; Mirza, M.S.; Imran, A. Rhizosphere engineering with plant growth-promoting microorganisms for agriculture and ecological sustainability. Front. Sustain. Food Syst. 2021, 5, 617157. [Google Scholar] [CrossRef]
- Thepbandit, W.; Athinuwat, D. Rhizosphere microorganisms supply availability of soil nutrients and induce plant defense. Microorganisms 2024, 12, 558. [Google Scholar] [CrossRef]
- Mahmud, K.; Missaoui, A.; Lee, K.; Ghimire, B.; Presley, H.W.; Makaju, S. Rhizosphere microbiome manipulation for sustainable crop production. Curr. Plant Biol. 2021, 27, 100210. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Kumar, N.; Shandilya, C.; Mohapatra, S.; Bhayana, S.; Varma, A. Revisiting plant–microbe interactions and microbial consortia application for enhancing sustainable agriculture: A review. Front. Microbiol. 2020, 11, 560406. [Google Scholar] [CrossRef]
- Weller, D.M.; Raaijmakers, J.M.; Gardener, B.B.M.; Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [PubMed]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease suppressive soils: New insights from the soil microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Godoy, O. Coexistence theory as a tool to understand biological invasions in species interaction networks: Implications for the study of novel ecosystems. Funct. Ecol. 2019, 33, 1190–1201. [Google Scholar] [CrossRef]
- Qian, X.; Li, X.; Li, H.; Zhang, D. Floral fungal-bacterial community structure and co-occurrence patterns in four sympatric island plant species. Fungal Biol. 2021, 125, 49–61. [Google Scholar] [CrossRef]
- Shayanthan, A.; Ordoñez, P.A.C.; Oresnik, I.J. The role of synthetic microbial communities (SynCom) in sustainable agriculture. Front. Agron. 2022, 4, 896307. [Google Scholar] [CrossRef]
- Santoyo, G.; Guzmán-Guzmán, P.; Parra-Cota, F.I.; Santos-Villalobos, S.D.L.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Plant growth stimulation by microbial consortia. Agronomy 2021, 11, 219. [Google Scholar] [CrossRef]
- Khan, S.T. Consortia-based microbial inoculants for sustaining agricultural activities. Appl. Soil Ecol. 2022, 176, 104503. [Google Scholar] [CrossRef]
- Trabelsi, D.; Mhamdi, R. Microbial inoculants and their impact on soil microbial communities: A review. BioMed Res. Int. 2013, 2013, 863240. [Google Scholar] [CrossRef]
- Čaušević, S.; Dubey, M.; Morales, M.; Salazar, G.; Sentchilo, V.; Carraro, N.; Ruscheweyh, H.-J.; Sunagawa, S.; van der Meer, J.R. Niche availability and competitive loss by facilitation control the proliferation of bacterial strains intended for soil microbiome interventions. Nat. Commun. 2024, 15, 2557. [Google Scholar] [CrossRef]
- Khare, E.; Arora, N.K. Effects of soil environment on field efficacy of microbial inoculants. In Plant Microbes Symbiosis: Applied Facets; Springer: New Delhi, India, 2014; pp. 353–381. [Google Scholar] [CrossRef]
- Shen, Z.Z.; Penton, C.R.; Lv, N.; Xue, C.; Yuan, X.; Ruan, Y.; Li, R. Banana Fusarium wilt disease incidence is influenced by shifts of soil microbial communities under different monoculture spans. Microb. Ecol. 2017, 75, 739–750. [Google Scholar] [CrossRef]
- Xiong, W.; Li, R.; Ren, Y.; Liu, C.; Zhao, Q.; Wu, H.; Jousset, A.; Shen, Q. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol. Biochem. 2017, 107, 198–207. [Google Scholar] [CrossRef]
- Xu, T.; Jiang, W.; Qin, D.; Liu, T.; Zhang, J.; Chen, W.; Gao, L. Characterization of the microbial communities in wheat tissues and rhizosphere soil caused by dwarf bunt of wheat. Sci. Rep. 2021, 11, 5773. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Xia, Y.; Fan, C.; Kou, J.; Wu, F.; Li, W.; Pan, K. Control of Fusarium wilt by wheat straw is associated with microbial network changes in watermelon rhizosphere. Sci. Rep. 2020, 10, 12736. [Google Scholar] [CrossRef]
- Orland, C.; Emilson, E.J.; Basiliko, N.; Mykytczuk, N.C.; Gunn, J.M.; Tanentzap, A.J. Microbiome functioning depends on individual and interactive effects of the environment and community structure. ISME J. 2019, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Eng, A.; Borenstein, E. Taxa-function robustness in microbial communities. Microbiome 2018, 6, 45. [Google Scholar] [CrossRef]
- Tian, L.; Wang, X.-W.; Wu, A.-K.; Fan, Y.; Friedman, J.; Dahlin, A.; Waldor, M.K.; Weinstock, G.M.; Weiss, S.T. Deciphering functional redundancy in the human microbiome. Nat. Commun. 2020, 11, 6217. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N. Friends or foes—Microbial interactions in nature. Biology 2021, 10, 496. [Google Scholar] [CrossRef]
- Srinivasan, S.; Jnana, A.; Murali, T.S. Modeling microbial community networks: Methods and tools for studying microbial interactions. Microb. Ecol. 2024, 87, 56. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Afordoanyi, D.M.; Diabankana, R.G.C.; Krupin, E.; Bikchantaev, I.; Taylan, A.; Validov, S. Inhibitory effects of Bacillus licheniformis WJ53A and homofermentative lactic acid bacteria on clostridial growth in corn silage. Pak. J. Agric. Sci. 2023, 60, 53–63. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Frolov, M.; Islamov, B.; Shulga, E.; Filimonova, M.N.; Afordoanyi, D.M.; Validov, S. Identification and Aggressiveness of Fusarium Species Associated with Onion Bulb (Allium cepa L.) during Storage. J. Fungi 2024, 10, 161. [Google Scholar] [CrossRef]
- Validov, S.Z.; Kamilova, F.D.; Lugtenberg, B.J. Monitoring of pathogenic and non-pathogenic Fusarium oxysporum strains during tomato plant infection. Microb. Biotechnol. 2011, 4, 82–88. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Afordoanyi, D.M.; Safin, R.I.; Nizamov, R.M.; Karimova, L.Z.; Validov, S.Z. Antifungal properties, abiotic stress resistance, and biocontrol ability of Bacillus mojavensis PS17. Curr. Microbiol. 2021, 78, 3124–3132. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Shulga, E.U.; Validov, S.Z.; Afordoanyi, D.M. Genetic characteristics and enzymatic activities of Bacillus velezensis KS04AU as a stable biocontrol agent against phytopathogens. Int. J. Plant Biol. 2022, 13, 201–222. [Google Scholar] [CrossRef]
- Bae, H.D.; Yanke, L.J.; Cheng, K.J.; Selinger, L.B. A novel staining method for detecting phytase activity. J. Microbiol. Methods 1999, 39, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Santiago, C.D.; Yagi, S.; Ijima, M.; Nashimoto, T.; Sawada, M.; Ikeda, S.; Asano, K.; Orikasa, Y.; Ohwada, T. Bacterial compatibility in combined inoculations enhances the growth of potato seedlings. Microbes Environ. 2017, 32, 14–23. [Google Scholar] [CrossRef]
- Ehmann, A. The Van Urk-Salkowski reagent—A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. A 1977, 132, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Riley, D.; Barber, S.A. Bicarbonate accumulation and pH changes at the soybean (Glycine max (L.) Merr.) root-soil interface. Soil Sci. Soc. Am. J. 1969, 33, 905–908. [Google Scholar] [CrossRef]
- Thomas, G.W. Soil pH and soil acidity. In Methods of Soil Analysis: Part 3 Chemical Methods; SSSA: Madison, WI, USA, 1996; pp. 475–490. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An Examination of the Degtjareff Method for Determining Soil Organic Matter, and a Proposed Modification of the Chromic Acid Titration Method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Leelamanie, D.A.L.; Liyanage, T.D.P.; Rajarathna, I.M.L.V. A Comparison of Weight Loss and C Analysis Methods in Determining Organic Matter Content in Sri Lankan Soils. Trop. Agric. Res. Ext. 2015, 18, 3. [Google Scholar] [CrossRef]
- Abakumov, E.V.; Rodina, O.A.; Eskov, A.K. Humification and Humic Acid Composition of Suspended Soil in Oligotrophous Environments in South Vietnam. Appl. Environ. Soil Sci. 2018, 2018, 1026237. [Google Scholar] [CrossRef]
- Jarukas, L.; Ivanauskas, L.; Kasparaviciene, G.; Baranauskaite, J.; Marksa, M.; Bernatoniene, J. Determination of Organic Compounds, Fulvic Acid, Humic Acid, and Humin in Peat and Sapropel Alkaline Extracts. Molecules 2021, 26, 2995. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, R.H.; Hildebrand, F.; et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Tedersoo, L.; Tooming-Klunderud, A.; Anslan, S. PacBio metabarcoding of fungi and other eukaryotes: Errors, biases and perspectives. New Phytol. 2018, 217, 1370–1385. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Buetas, E.; Jordán-López, M.; López-Roldán, A.; D’Auria, G.; Martínez-Priego, L.; De Marco, G.; Carda-Diéguez, M. Full-length 16S rRNA gene sequencing by PacBio improves taxonomic resolution in human microbiome samples. BMC Genom. 2024, 25, 310. [Google Scholar] [CrossRef]
- Shen, W.; Sipos, B.; Zhao, L. SeqKit2: A Swiss Army Knife for Sequence and Alignment Processing. iMeta 2024, 3, e191. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics 2021, 37, 4572–4574. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2024, 52, D791–D797. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Lynch, M.D.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Jones, S.E.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J.A. Conditionally rare taxa disproportionately contribute to changes in microbial diversity. mBio 2014, 5, e01371-14. [Google Scholar] [CrossRef]
- Wu, L.; Ning, D.; Zhang, B.; Li, Y.; Sanabria, J. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019, 4, 1183–1195. [Google Scholar] [CrossRef]
- Dai, T.; Wen, D.; Bates, C.T.; Wu, L.; Guo, X.; Liu, S.; Su, Y.; Lei, J.; Zhou, J. Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 2022, 13, 175. [Google Scholar] [CrossRef]
- Aertsen, W.; Kint, V.; Van Orshoven, J.; Özkan, K.; Muys, B. Comparison and ranking of different modelling techniques for prediction of site index in Mediterranean mountain forests. Ecol. Model. 2010, 217, 1119–1130. [Google Scholar] [CrossRef]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian barley industry. Australas. Plant Pathol. 2010, 39, 85–96. [Google Scholar] [CrossRef]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Röttjers, L.; Faust, K. From hairballs to hypotheses—Biological insights from microbial networks. FEMS Microbiol. Rev. 2018, 42, 761–780. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Ge, Z.; Li, S.; Bol, R.; Zhu, P.; Peng, C.; An, T.; Cheng, N.; Liu, X.; Li, T.; Xu, Z.; et al. Differential long-term fertilization alters residue-derived labile organic carbon fractions and microbial community during straw residue decomposition. Soil Till. Res. 2021, 213, 105120. [Google Scholar] [CrossRef]
- Zamkovaya, T.; Foster, J.S.; de Crécy-Lagard, V.; Conesa, A. A network approach to elucidate and prioritize microbial dark matter in microbial communities. ISME J. 2021, 15, 228–244. [Google Scholar] [CrossRef] [PubMed]
- Raag, V.A.; Waltman, L.; van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef] [PubMed]
- Kusstatscher, P.; Wicaksono, W.A.; Thenappan, D.P.; Adam, E.; Müller, H.; Berg, G. Microbiome management by biological and chemical treatments in maize is linked to plant health. Microorganisms 2020, 8, 1506. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.M.; Johnsson, L.; Gerhardson, B. Suppression of wheat-seedling diseases caused by Fusarium culmorum and Microdochium nivale using bacterial seed treatment. Plant Pathol. 2003, 52, 219–227. [Google Scholar] [CrossRef]
- Todorović, I.; Moënne-Loccoz, Y.; Raičević, V.; Jovičić-Petrović, J.; Muller, D. Microbial diversity in soils suppressive to Fusarium diseases. Front. Plant Sci. 2023, 14, 1228749. [Google Scholar] [CrossRef] [PubMed]
- Solórzano, R.; Ramírez Maguiña, H.A.; Johnson, L.; Ureta Sierra, C.; Cruz, J. Current progress in microbial biocontrol of banana Fusarium wilt: A systematic review. Agronomy 2025, 15, 619. [Google Scholar] [CrossRef]
- Müller, T.; Behrendt, U.; Ruppel, S.; von der Waydbrink, G.; Müller, M.E. Fluorescent pseudomonads in the phyllosphere of wheat: Potential antagonists against fungal phytopathogens. Curr. Microbiol. 2016, 72, 383–389. [Google Scholar] [CrossRef]
- Sundaramoorthy, S.; Raguchander, T.; Ragupathi, N.; Samiyappan, R. Combinatorial effect of endophytic and plant growth promoting rhizobacteria against wilt disease of Capsicum annum L. caused by Fusarium solani. Biol. Control 2012, 60, 59–68. [Google Scholar] [CrossRef]
- Kumari, P.; Khanna, V. Seed bacterization stimulated resistance in chickpea against Fusarium oxysporum f. sp. ciceris. Indian Phytopathol. 2019, 72, 689–697. [Google Scholar] [CrossRef]
- Ankati, S.; Srinivas, V.; Pratyusha, S.; Gopalakrishnan, S. Streptomyces consortia-mediated plant defense against Fusarium wilt and plant growth-promotion in chickpea. Microb. Pathog. 2021, 157, 104961. [Google Scholar] [CrossRef]
- Devi, N.O.; Tombisana Devi, R.K.; Debbarma, M.; Hajong, M.; Thokchom, S. Effect of endophytic Bacillus and arbuscular mycorrhiza fungi (AMF) against Fusarium wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici. Egypt. J. Biol. Pest. Control 2022, 32, 1–14. [Google Scholar] [CrossRef]
- Thakkar, A.; Saraf, M. Development of microbial consortia as a biocontrol agent for effective management of fungal diseases in Glycine max L. Arch. Phytopathol. Plant Prot. 2015, 48, 459–474. [Google Scholar] [CrossRef]
- Wong, C.K.F.; Saidi, N.B.; Vadamalai, G.; Teh, C.Y.; Zulperi, D. Effect of bioformulations on the biocontrol efficacy, microbial viability, and storage stability of a consortium of biocontrol agents against Fusarium wilt of banana. J. Appl. Microbiol. 2019, 127, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.; Leeman, M.; van Oorschot, M.; van der Sluis, I.; Schippers, B.; Bakker, P. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 1995, 85, 1075–1081. Phytopathology 1995, 85, 1075–1081. [Google Scholar] [CrossRef]
- Birr, T.; Hasler, M.; Verreet, J.A.; Klink, H. Composition and predominance of Fusarium species causing Fusarium head blight in winter wheat grain depending on cultivar susceptibility and meteorological factors. Microorganisms 2020, 8, 617. [Google Scholar] [CrossRef] [PubMed]
- Ghoul, M.; Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Tjamos, E.C.; Tsitsigiannis, D.I.; Tjamos, S.E.; Antoniou, P.P.; Katinakis, P. Selection and screening of endorhizosphere bacteria from solarized soils as biocontrol agents against Verticillium dahliae of solanaceous hosts. Eur. J. Plant Pathol. 2004, 110, 35–44. [Google Scholar] [CrossRef]
- Abdel-Kader, M.M.; El-Mougy, N.S.; Khalil, M.S.A.; El-Gamal, N.G.; Attia, M. Soil drenching and foliar spray with bioagents for reducing wheat leaf diseases under natural field conditions. J. Plant Dis. Prot. 2023, 130, 279–291. [Google Scholar] [CrossRef]
- Hol, W.H.G.; De Boer, W.; Termorshuizen, A.J.; Meyer, K.M.; Schneider, J.H.M.; Van Dam, N.M.; Van Veen, J.A.; Van Der Putten, W.H. Reduction of rare soil microbes modifies plant–herbivore interactions. Ecol. Lett. 2010, 13, 292–301. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-L.; Ding, J.; Zhu, D.; Hu, H.-W.; Delgado-Baquerizo, M.; Ma, Y.-B.; He, J.-Z.; Zhu, Y.-G. Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol. Biochem. 2020, 141, 107686. [Google Scholar] [CrossRef]
- Deng, X.; Zhang, N.; Li, Y.; Zhu, C.; Qu, B.; Liu, H.; Li, R.; Bai, Y.; Shen, Q.; Salles, J.F. Bio-organic soil amendment promotes the suppression of Ralstonia solanacearum by inducing changes in the functionality and composition of rhizosphere bacterial communities. New Phytol. 2022, 235, 1558–1574. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X. Functional molecular ecological networks. mBio 2010, 1, e00169-10. [Google Scholar] [CrossRef]
- Kurm, V.; Van der Putten, W.H.; Hol, W.G. Cultivation-success of rare soil bacteria is not influenced by incubation time and growth medium. PLoS ONE 2019, 14, e0210073. [Google Scholar] [CrossRef]
- Albuquerque, U.P.; Nascimento, A.L.B.D.; da Silva Chaves, L.; Feitosa, I.S.; de Moura, J.M.B.; Gonçalves, P.H.S.; da Silva, R.H.; da Silva, T.C.; Júnior, W.S.F. A brief introduction to niche construction theory for ecologists and conservationists. Biol. Conserv. 2019, 237, 50–56. [Google Scholar] [CrossRef]
- Newsham, K.K.; Goodall-Copestake, W.P.; Ochyra, R.; Váňa, J. Mycothalli of the hepatic Barbilophozia hatcheri in Antarctica: Distribution and identities of mycobionts. Fungal Ecol. 2014, 11, 91–99. [Google Scholar] [CrossRef]
- Xu, Q.; Li, L.; Guo, J.; Guo, H.; Liu, M.; Guo, S.; Kuzyakov, Y.; Ling, N.; Shen, Q. Active microbial population dynamics and life strategies drive the enhanced carbon use efficiency in high-organic matter soils. mBio 2024, 15, e00177-24. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef]
- Guan, H.; Fan, J.; Zhang, H.; Harris, W. Comparison of drivers of soil microbial communities developed in karst ecosystems with shallow and deep soil depths. Agronomy 2021, 11, 173. [Google Scholar] [CrossRef]
- Duan, N.; Li, L.; Liang, X.; Fine, A.; Zhuang, J.; Radosevich, M.; Schaeffer, S.M. Variation in bacterial community structure under long-term fertilization, tillage, and cover cropping in continuous cotton production. Front. Microbiol. 2022, 13, 847005. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dong, Y.; Wang, H.-B.; Xue, D.-H.; Zhang, H.-F.; Li, Z.-J.; Wang, H.; Bo, H.-J.; Zhang, W.-J.; Zhang, B.-H.; et al. Long-term legume cultivation affects the soil bacterial community via altering the soil pore structure in coal mine reclamation agroecosystems. Plant Cell Environ. 2025, 48, 1–15. [Google Scholar] [CrossRef]
- Luo, X.; Liu, K.; Shen, Y.; Yao, G.; Yang, W.; Mortimer, P.E.; Gui, H. Fungal community composition and diversity vary with soil horizons in a subtropical forest. Front. Microbiol. 2021, 12, 650440. [Google Scholar] [CrossRef]
- Daraz, U.; Erhunmwunse, A.S.; Dubeux Jr, J.C.; Mackowiak, C.; Liao, H.L.; Wang, X.B. Soil fungal community structure and function response to rhizoma perennial peanut cultivars. BMC Plant Biol. 2024, 24, 582. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef]
- Panke-Buisse, K.; Poole, A.C.; Goodrich, J.K.; Ley, R.E.; Kao-Kniffin, J. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 2015, 9, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Sato, H. Root-associated fungi shared between arbuscular mycorrhizal and ectomycorrhizal conifers in a temperate forest. Front. Microbiol. 2018, 9, 433. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Hansen, J.C.; Schillinger, W.F.; Sullivan, T.S.; Paulitz, T.C. Common and unique rhizosphere microbial communities of wheat and canola in a semiarid Mediterranean environment. Appl. Soil Ecol. 2019, 144, 170–181. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Ondreičková, K.; Piliarová, M.; Klčová, L.; Žofajová, A.; Gubiš, J.; Horník, M.; Gubišová, M.; Hudcovicová, M.; Kraic, J. The impact of sewage sludge on the fungal communities in the rhizosphere and roots of barley and on barley yield. Open Life Sci. 2021, 16, 210–221. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Eldridge, D.J.; Liu, Y.-R.; Sokoya, B.; Wang, J.-T.; Hu, H.-W.; He, J.-Z.; Bastida, F.; Moreno, J.L.; Bamigboye, A.R.; et al. Global homogenization of the structure and function in the soil microbiome of urban greenspaces. Sci. Adv. 2021, 7, eabg5809. [Google Scholar] [CrossRef]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.-C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef]
- Barua, P.; You, M.P.; Bayliss, K.; Lanoiselet, V.; Barbetti, M.J. A rapid and miniaturized system using Alamar blue to assess fungal spore viability: Implications for biosecurity. Eur. J. Plant Pathol. 2017, 148, 139–150. [Google Scholar] [CrossRef]
- Barua, P.; You, M.P.; Bayliss, K.L.; Lanoiselet, V.; Barbetti, M.J. Extended survival of Puccinia graminis f. sp. tritici urediniospores: Implications for biosecurity and on-farm management. Plant Pathol. 2018, 67, 799–809. [Google Scholar] [CrossRef]
- Sohaliya, N.; Naik, A.; Patel, D. Puccinia. In Compendium of Phytopathogenic Microbes in Agro-Ecology; Springer: Cham, Switzerland, 2025; pp. 609–632. [Google Scholar]
- Chao, H.; Cai, A.; Heimburger, B.; Wu, Y.; Zhao, D.; Sun, M.; Hu, F. Keystone taxa enhance the stability of soil bacterial communities and multifunctionality under steelworks disturbance. J. Environ. Manag. 2024, 356, 120664. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q. Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef]
- Kaisermann, A.; de Vries, F.T.; Griffiths, R.I.; Bardgett, R.D. Legacy effects of drought on plant–soil feedbacks and plant–plant interactions. New Phytol. 2017, 215, 1413–1424. [Google Scholar] [CrossRef]
- Wachowska, U.; Sulyok, M.; Wiwart, M.; Suchowilska, E.; Kandler, W.; Krska, R. The application of antagonistic yeasts and bacteria: An assessment of in vivo and under field conditions pattern of Fusarium mycotoxins in winter wheat grain. Food Control 2022, 138, 109039. [Google Scholar] [CrossRef]
- Stark, J.M.; Firestone, M.K. Mechanisms for soil moisture effects on activity of nitrifying bacteria. Appl. Environ. Microbiol. 1995, 61, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Brockett, B.F.; Prescott, C.E.; Grayston, S.J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 2012, 44, 9–20. [Google Scholar] [CrossRef]
- Torres, N.; Yu, R.; Kurtural, S.K. Inoculation with mycorrhizal fungi and irrigation management shape the bacterial and fungal communities and networks in vineyard soils. Microorganisms 2021, 9, 1273. [Google Scholar] [CrossRef]
- He, X.; Zhang, Q.; Li, B.; Jin, Y.; Jiang, L.; Wu, R. Network mapping of root–microbe interactions in Arabidopsis thaliana. npj Biofilms Microbiomes 2021, 7, 72. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Pétriacq, P.; López, A.; Luna, E. Fruit decay to diseases: Can induced resistance and priming help? Plants 2018, 7, 77. [Google Scholar] [CrossRef] [PubMed]





| Growth Stages (GS) | Sample Collections | ||||
|---|---|---|---|---|---|
| Bulk Soil (BS) | Rhizosphere Soil (RS) | ||||
| Untreated (CT_BS) | Seed Treatment and Rhizosphere Drenching (ST_IR_BS) | Untreated (CT_RS) | Seed Treatment (ST_RS) | Seed Treatment and Rhizosphere Drenching (ST_IR_RS) | |
| Tillering (GS3) | ✔ | ✔ | ✔ | ✔ | ✔ |
| Booting (GS5) | ✔ | ✔ | ✔ | ✔ | ✔ |
| Flowering (GS7) | ✔ | ✔ | ✔ | ✔ | ✔ |
| Dough development (GS9) | ✔ | ✔ | ✔ | ✔ | ✔ |
| Network Properties | Control BS | ST_IR_BS | Control RS | ST_RS | ST_IR_RS |
|---|---|---|---|---|---|
| Nodes | 46 | 47 | 44 | 44 | 45 |
| Edges | 216 | 194 | 133 | 157 | 103 |
| Positive edges | 216 | 194 | 130 | 157 | 103 |
| Negative edges | 0 | 0 | 3 | 0 | 0 |
| Average degree | 9.39 ± 0.04 | 8.25 ± 0.08 | 6.04 ± 0.03 | 7.14 ± 0.02 | 4.58 ± 0.01 |
| Average closeness | 0.05 ± 0.07 | 0.05 ± 0.01 | 0.19 ± 0.02 | 0.04 ± 0.01 | 0.08 ± 0.02 |
| Modularity | 0.32 | 0.35 | 0.31 | 0.44 | 0.53 |
| Transitivity | 0.68 | 0.70 | 0.74 | 0.57 | 0.44 |
| Diameter | 7.17 | 7.15 | 7.73 | 7.18 | 5.51 |
| Average path length | 2.42 ± 0.13 | 2.53 ± 0.10 | 2.66 ± 0.08 | 2.51 ± 0.07 | 2.24 ± 0.18 |
| Density | 0.21 | 0.18 | 0.14 | 0.17 | 0.10 |
| Assortativity | 0.46 | 0.54 | 0.67 | 0.64 | 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diabankana, R.G.C.; Komissarov, E.N.; Afordoanyi, D.M.; Islamov, B.; Sukhanov, A.Y.; Shulga, E.; Filimonova, M.N.; Saparmyradov, K.; Trachtmann, N.V.; Validov, S.Z. Synthetic Bacterial Consortium Induces Dynamic Shifts in Fungal Community and Alters Microbial Network Topology in Barley Soil Under Field Conditions. Soil Syst. 2025, 9, 116. https://doi.org/10.3390/soilsystems9040116
Diabankana RGC, Komissarov EN, Afordoanyi DM, Islamov B, Sukhanov AY, Shulga E, Filimonova MN, Saparmyradov K, Trachtmann NV, Validov SZ. Synthetic Bacterial Consortium Induces Dynamic Shifts in Fungal Community and Alters Microbial Network Topology in Barley Soil Under Field Conditions. Soil Systems. 2025; 9(4):116. https://doi.org/10.3390/soilsystems9040116
Chicago/Turabian StyleDiabankana, Roderic Gilles Claret, Ernest Nailevich Komissarov, Daniel Mawuena Afordoanyi, Bakhtiyar Islamov, Artemiy Yurievich Sukhanov, Elena Shulga, Maria Nikolaevna Filimonova, Keremli Saparmyradov, Natalia V. Trachtmann, and Shamil Z. Validov. 2025. "Synthetic Bacterial Consortium Induces Dynamic Shifts in Fungal Community and Alters Microbial Network Topology in Barley Soil Under Field Conditions" Soil Systems 9, no. 4: 116. https://doi.org/10.3390/soilsystems9040116
APA StyleDiabankana, R. G. C., Komissarov, E. N., Afordoanyi, D. M., Islamov, B., Sukhanov, A. Y., Shulga, E., Filimonova, M. N., Saparmyradov, K., Trachtmann, N. V., & Validov, S. Z. (2025). Synthetic Bacterial Consortium Induces Dynamic Shifts in Fungal Community and Alters Microbial Network Topology in Barley Soil Under Field Conditions. Soil Systems, 9(4), 116. https://doi.org/10.3390/soilsystems9040116







