A New Era in the Discovery of Biological Control Bacteria: Omics-Driven Bioprospecting
Abstract
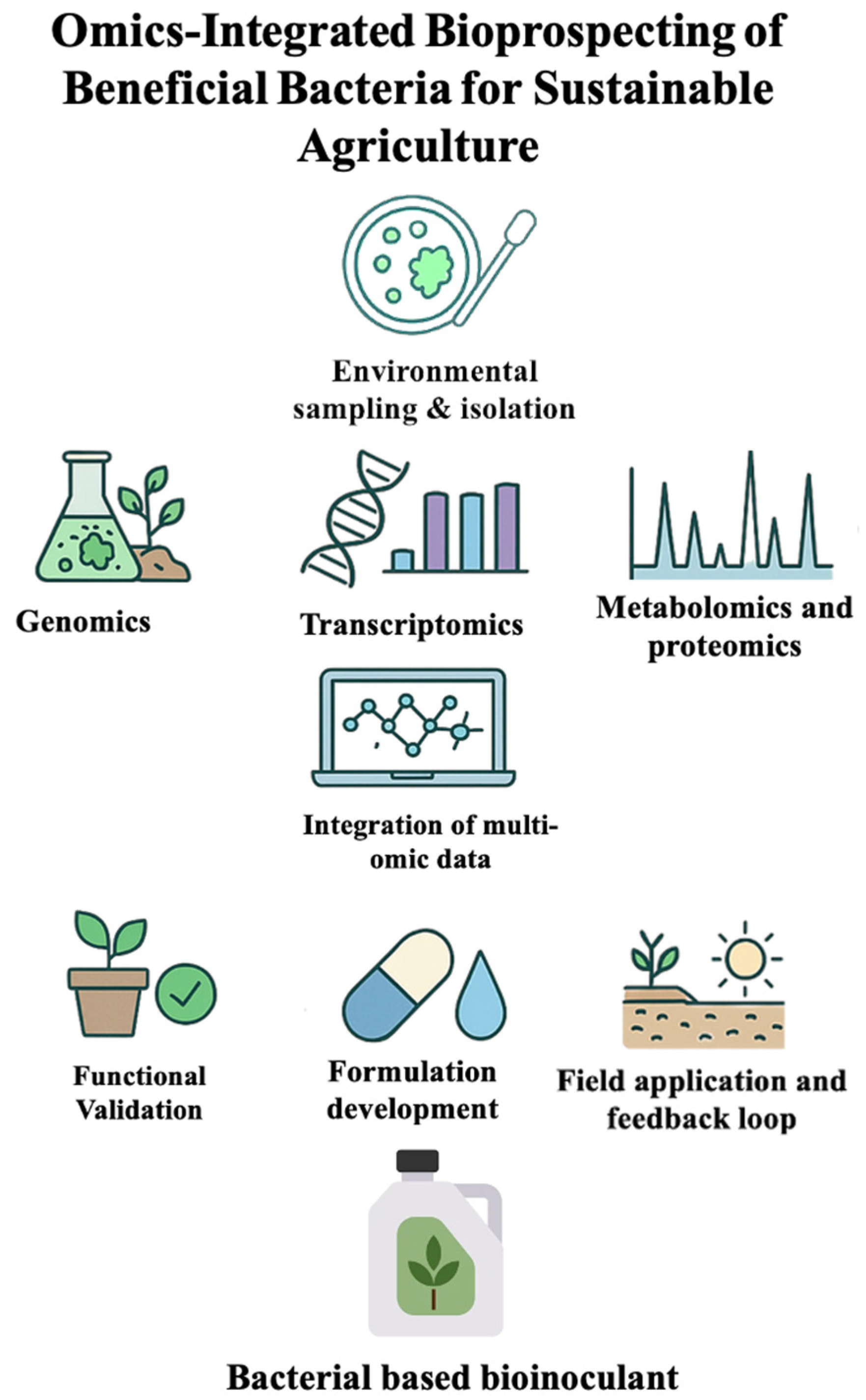
1. Introduction
2. Genomics: Unlocking the Genetic Potential of Biological Control Bacteria
3. Metagenomics: Exploring Uncultured Microbial Reservoirs
4. Transcriptomics: Deciphering Molecular Interactions
5. Metabolomics: Profiling Bioactive Compounds for Enhanced Biocontrol
6. Integrating Multi-Omics Data for Precision Biocontrol Strategies: Case Study
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). New Standards to Curb the Global Spread of Plant Pests and Diseases; Food and Agriculture Organization of the United Nations: Roma, Italy, 2025. [Google Scholar]
- Han, P.; Rodriguez-Saona, C.; Zalucki, M.P.; Liu, S.S.; Desneux, N. A theoretical framework to improve the adoption of green Integrated Pest Management tactics. Commun. Biol. 2024, 7, 337. [Google Scholar] [CrossRef]
- Shattuck, A.; Werner, M.; Mempel, F.; Dunivin, Z.; Galt, R. Global pesticide use and trade database (GloPUT): New estimates show pesticide use trends in low-income countries substantially underestimated. Glob. Environ. Chang. 2023, 81, 102693. [Google Scholar] [CrossRef]
- Córdova-Albores, L.C.; Zelaya-Molina, L.X.; Ávila-Alistac, N.; Valenzuela-Ruíz, V.; Cortés-Martínez, N.E.; Parra-Cota, F.I.; Burgos-Canul, Y.Y.; Chávez-Díaz, I.F.; Fajardo-Franco, M.L.; de los Santos-Villalobos, S. Omics sciences potential on bioprospecting of biological control microbial agents: The case of the Mexican agro-biotechnology. Rev. Mex. Fitopatol. 2020, 39, 147–184. [Google Scholar] [CrossRef]
- Sforza, R.F. Biological Control: Global Impacts, Challenges and Future Directions of Pest Management; CSIRO Publishing: Canberra, Australia, 2021. [Google Scholar]
- Valenzuela Ruiz, V.; Gándara-Ledezma, A.; Villarreal-Delgado, M.F.; Villa-Rodríguez, E.D.; Parra-Cota, F.I.; Santoyo, G.; Gómez-Godínez, L.J.; Cira-Chávez, L.A.; de los Santos Villalobos, S. Regulation, Biosynthesis, and Extraction of Bacillus-Derived Lipopeptides and Its Implications in Biological Control of Phytopathogens. Stresses 2024, 4, 107–132. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, Q.; Li, Y.; Deng, Z.; Song, G.; Wang, H.; Yan, M.; Wang, X. Integrated transcriptome and metabolome analyses shed light on the defense mechanisms in tomato plants after (E)-2-hexenal fumigation. Genomics 2023, 115, 110592. [Google Scholar] [CrossRef]
- Singh, A.; Yadav, V.K.; Gautam, H.; Rathod, L.; Chundawat, R.S.; Singh, G.; Verma, R.K.; Sahoo, D.K.; Patel, A. The role of plant growth promoting rhizobacteria in strengthening plant resistance to fluoride toxicity: A review. Front. Microbiol. 2023, 14, 1271034. [Google Scholar] [CrossRef]
- Ding, X.; Duan, S.; Ding, X.; Liu, R.; Xu, F.-J. Versatile Antibacterial Materials: An Emerging Arsenal for Combatting Bacterial Pathogens. Adv. Funct. Mater. 2018, 28, 1802140. [Google Scholar] [CrossRef]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant Pathol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Ait Barka, E. Biological Control of Plant Pathogens: A Global Perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef]
- Kumar, S.; Nikunj, C.; Rathore, A.; Nigam, R.; Shivam; M, A.; Nilesh, R.; Yuvarani, R. Role of Biocontrol Agents in Suppressing Plant Pathogen: A Compressive Analysis. J. Sci. Res. Rep. 2024, 30, 1004–1015. [Google Scholar] [CrossRef]
- de Faria, M.R.; Costa, L.S.A.S.; Chiaramonte, J.B.; Bettiol, W.; Mendes, R. The rhizosphere microbiome: Functions, dynamics, and role in plant protection. Trop. Plant Pathol. 2021, 46, 13–25. [Google Scholar] [CrossRef]
- Chaudhary, R.; Nawaz, A.; Khattak, Z.; Butt, M.A.; Fouillaud, M.; Dufossé, L.; Munir, M.; Haq, I.u.; Mukhtar, H. Microbial bio-control agents: A comprehensive analysis on sustainable pest management in agriculture. J. Agric. Food Res. 2024, 18, 101421. [Google Scholar] [CrossRef]
- Tripathi, A.N.; Meena, B.R.; Pandey, K.K.; Singh, J. Microbial Bioagents in Agriculture: Current Status and Prospects. In New Frontiers in Stress Management for Durable Agriculture; Springer: Singapore, 2020. [Google Scholar]
- He, D.-C.; He, M.-H.; Amalin, D.M.; Liu, W.; Alvindia, D.G.; Zhan, J. Biological Control of Plant Diseases: An Evolutionary and Eco-Economic Consideration. Pathogens 2021, 10, 1311. [Google Scholar] [CrossRef] [PubMed]
- Karaoğlan, B.; Alkassab, A.T.; Borges, S.; Fisher, T.; Link-Vrabie, C.; McVey, E.; Ortego, L.; Nuti, M. Microbial pesticides: Challenges and future perspectives for non-target organism testing. Environ. Sci. Eur. 2024, 36, 205. [Google Scholar] [CrossRef]
- Tavares, M.; Kozak, M.; Balola, A.; Sá-Correia, I. Burkholderia cepacia Complex Bacteria: A Feared Contamination Risk in Water-Based Pharmaceutical Products. Clin. Microbiol. Rev. 2020, 33, 10–1128. [Google Scholar] [CrossRef]
- Abreo, E.; Altier, N. Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the links between them. Sci. Rep. 2019, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M.; Altier, N. Fluorescent pseudomonads as biocontrol agents for sustainable agricultural systems. Res. Microbiol. 2010, 161, 464–471. [Google Scholar] [CrossRef]
- Wend, K.; Zorrilla, L.; Freimoser, F.M.; Gallet, A. Microbial pesticides—Challenges and future perspectives for testing and safety assessment with respect to human health. Environ. Health 2024, 23, 49. [Google Scholar] [CrossRef]
- Lensch, A.; Lindfors, H.A.; Duwenig, E.; Fleischmann, T.; Hjort, C.; Kärenlampi, S.O.; McMurtry, L.; Melton, E.-D.; Andersen, M.R.; Skinner, R.; et al. Safety aspects of microorganisms deliberately released into the environment. EFB Bioecon. J. 2024, 4, 100061. [Google Scholar] [CrossRef]
- Beattie, A.J.; Hay, M.; Magnusson, B.; de Nys, R.; Smeathers, J.; Vincent, J.F.V. Ecology and bioprospecting. Austral. Ecol. 2011, 36, 341–356. [Google Scholar] [CrossRef]
- Montes-Montes, G.; González-Escobedo, R.; Muñoz-Castellanos, L.N.; Avila-Quezada, G.D.; Ramírez-Sánchez, O.; Borrego-Loya, A.; Ortiz-Aguirre, I.; Muñoz-Ramírez, Z.Y. Whole Genome Analysis of Streptomyces spp. Strains Isolated from the Rhizosphere of Vitis vinifera L. Reveals Their Role in Nitrogen and Phosphorus Metabolism. Nitrogen 2024, 5, 301–314. [Google Scholar] [CrossRef]
- Helué Morales, P.; Valenzuela Ruiz, V.; Ortega Urquieta, M.E.; Martínez Vidales, A.D.; Félix Pablos, C.M.; Chávez Luzania, R.A.; Parra Cota, F.I.; de los Santos Villalobos, S. Taxonomía bacteriana basada en índices relacionados al genoma completo. La Soc. Académica 2024, 58, 39–50. [Google Scholar]
- Martinez Vidales, A.D.; Cervantes Enríquez, E.P.; Ruiz Castrejón, A.; Romero Silva, J.H.; Ortega Urquieta, M.E.; Parra Cota, F.I.; de los Santos Villalobos, S. Phylogenomic diversity and genome mining of type Bacillus species: Searching for genes associated with biological control of phytopathogens. Rev. Mex. Fitopatol. 2024, 42, 21. [Google Scholar] [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 006300. [Google Scholar] [CrossRef] [PubMed]
- Helué Morales-Sandoval, P.; Valenzuela-Ruíz, V.; Santoyo, G.; Hyder, S.; Mitra, D.; Zelaya-Molina, L.X.; Ávila-Alistac, N.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Draft genome of a biological control agent against Bipolaris sorokiniana, the causal phytopathogen of spot blotch in wheat (Triticum turgidum L. subsp. durum): Bacillus inaquosorum TSO22. Open Agric. 2024, 9, 20220309. [Google Scholar] [CrossRef]
- Du, Y.; Wang, T.; Lv, C.; Yan, B.; Wan, X.; Wang, S.; Kang, C.; Guo, L.; Huang, L. Whole Genome Sequencing Reveals Novel Insights about the Biocontrol Potential of Burkholderia ambifaria CF3 on Atractylodes lancea. Microorganisms 2024, 12, 1043. [Google Scholar] [CrossRef]
- Shiekh Suliman, N.; Talaei-Hassanloui, R.; Abachi, H.; Zarei, S.; Osdaghi, E. Taxonomic refinement of Bacillus thuringiensis. Front. Microbiol. 2025, 16, 1518307. [Google Scholar] [CrossRef]
- Ercole, T.G.; Kava, V.M.; Petters-Vandresen, D.A.L.; Gomes, M.E.N.; Aluizio, R.; Ribeiro, R.A.; Hungria, M.; Galli, L.V. Unlocking the growth-promoting and antagonistic power: A comprehensive whole genome study on Bacillus velezensis strains. Gene 2024, 927, 148669. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, T.; Wang, Z.; Wang, X.; Wang, H.; Li, Y.; Zheng, W.; Wei, S.; Leng, Y.; Li, J.; et al. Whole-genome sequencing and secondary metabolite exploration of the novel Bacillus velezensis BN with broad-spectrum antagonistic activity against fungal plant pathogens. Front. Microbiol. 2025, 15, 1498653. [Google Scholar] [CrossRef] [PubMed]
- Mian, S.; Machado, A.C.Z.; Hoshino, R.T.; Mosela, M.; Higashi, A.Y.; Shimizu, G.D.; Teixeira, G.M.; Nogueira, A.F.; Giacomin, R.M.; Ribeiro, L.A.B.; et al. Complete genome sequence of Bacillus velezensis strain Ag109, a biocontrol agent against plant-parasitic nematodes and Sclerotinia sclerotiorum. BMC Microbiol. 2024, 24, 194. [Google Scholar] [CrossRef]
- Du, J.; Huang, B.; Huang, J.; Long, Q.; Zhang, C.; Guo, Z.; Wang, Y.; Chen, W.; Tan, S.; Liu, Q. Pan-Genome Analysis and Secondary Metabolic Pathway Mining of Biocontrol Bacterium Brevibacillus brevis. Agronomy 2024, 14, 1024. [Google Scholar] [CrossRef]
- Kukreti, A.; Kotasthane, A.S.; Tandon, A.L.; Nekkanti, A.; Prasannakumar, M.K.; Devanna, P.; Aravindaram, K.; Sreedevi, K.; Sushil, S.N.; Manjunatha, C. Hybrid de novo whole genome assembly of lipopeptide producing novel Bacillus thuringiensis strain NBAIR BtAr exhibiting antagonistic activity against Sclerotium rolfsii. Microb. Pathog. 2024, 195, 106867. [Google Scholar] [CrossRef]
- Valencia-Marin, M.F.; Chávez-Avila, S.; Sepúlveda, E.; Delgado-Ramírez, C.S.; Meza-Contreras, J.J.; Orozco-Mosqueda, M.d.C.; De Los Santos-Villalobos, S.; Babalola, O.O.; Hernández-Martinez, R.; Santoyo, G. Stress-tolerant Bacillus strains for enhancing tomato growth and biocontrol of Fusarium oxysporum under saline conditions: Functional and genomic characterization. World J. Microbiol. Biotechnol. 2025, 41, 96. [Google Scholar] [CrossRef]
- Ayala-Zepeda, M.; Parra-Cota, F.I.; Chinchilla-Soto, C.; De La Cruz-Torres, E.; Ibba, M.I.; Estrada-Alvarado, M.I.; de los Santos-Villalobos, S. 15N-Nitrogen Use Efficiency, Productivity, and Quality of Durum Wheat Integrating Nitrogen Management and an Indigenous Bacterial Inoculant in a Single Growing Season. Appl. Sci. 2025, 15, 1429. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Moreno-Ulloa, A.; Castro-Longoria, E.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Integrated omics approaches for deciphering antifungal metabolites produced by a novel Bacillus species, B. cabrialesii TE3T, against the spot blotch disease of wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2021, 251, 126826. [Google Scholar] [CrossRef]
- Valenzuela Ruiz, V.; Santoyo, G.; Gómez Godínez, L.J.; Cira Chávez, L.A.; Parra Cota, F.I.; de los Santos Villalobos, S. Complete genome sequencing of Bacillus cabrialesii TE3T: A plant growth-promoting and biological control agent isolated from wheat (Triticum turgidum subsp. durum) in the Yaqui Valley. Curr. Res. Microb. Sci. 2023, 4, 100193. [Google Scholar] [CrossRef]
- Valenzuela Ruiz, V.; Parra Cota, F.I.; Santoyo, G.; Estrada Alvarado, M.I.; Cira Chávez, L.A.; Castro Longoria, E.; de los Santos Villalobos, S. Potential biological control Mechanisms of Bacillus paralicheniformis TRQ65 against phytopathogenic fungi. Rev. Mex. Fitopatol. 2024, 42, 6. [Google Scholar] [CrossRef]
- Dahar, G.Y.; Wang, H.W.; Rajer, F.U.; Jin, P.; Xu, P.; Abro, M.A.; Qureshi, A.S.; Karim, A.; Miao, W. Comparative genomic analysis of Bacillus atrophaeus HAB-5 reveals genes associated with antimicrobial and plant growth-promoting activities. Front. Microbiol. 2024, 15, 1384691. [Google Scholar] [CrossRef] [PubMed]
- Settu, V.; Annaiyan, S.; Mannu, J. Revealing the genetic arsenal of Bacillus firmus TNAU1: Unleashing nematicidal and plant growth promotion traits. Physiol. Mol. Plant Pathol. 2024, 129, 102177. [Google Scholar] [CrossRef]
- González-León, Y.; De la Vega-Camarillo, E.; Ramírez-Vargas, R.; Anducho-Reyes, M.A.; Mercado-Flores, Y. Whole genome analysis of Bacillus velezensis 160, biological control agent of corn head smut. Microbiol. Spectr. 2024, 12, e03264-23. [Google Scholar] [CrossRef] [PubMed]
- Yim, B.; Heider, M.A.; Bloem, E.; Vetterlein, D.; Behr, J.H.; Babin, D.; Smalla, K. Exploring the potential of seed inoculation with microbial consortia to mitigate drought stress in maize plants under greenhouse conditions. Plant Soil 2025. [Google Scholar] [CrossRef]
- Wang, Y.; Dall’Agnol, R.F.; Bertani, I.; Bez, C.; Venturi, V. Identification of synthetic consortia from a set of plant--beneficial bacteria. Microb. Biotechnol. 2024, 17, e14330. [Google Scholar] [CrossRef] [PubMed]
- Cangioli, L.; Tabacchioni, S.; Visca, A.; Fiore, A.; Aprea, G.; Ambrosino, P.; Ercole, E.; Sørensen, S.; Mengoni, A.; Bevivino, A. Genome Insights into Beneficial Microbial Strains Composing SIMBA Microbial Consortia Applied as Biofertilizers for Maize, Wheat and Tomato. Microorganisms 2024, 12, 2562. [Google Scholar] [CrossRef]
- Nam, N.; Do, H.; Loan Trinh, K.; Lee, N. Metagenomics: An Effective Approach for Exploring Microbial Diversity and Functions. Foods 2023, 12, 2140. [Google Scholar] [CrossRef]
- Berini, F.; Casciello, C.; Marcone, G.L.; Marinelli, F. Metagenomics: Novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 2017, 364, fnx211. [Google Scholar] [CrossRef]
- Nwachukwu, B.C.; Babalola, O.O. Metagenomics: A Tool for Exploring Key Microbiome With the Potentials for Improving Sustainable Agriculture. Front. Sustain. Food Syst. 2022, 6, 886987. [Google Scholar] [CrossRef]
- Xiao, J.; Guo, X.; Qiao, X.; Zhang, X.; Chen, X.; Zhang, D. Activity of Fengycin and Iturin A Isolated From Bacillus subtilis Z-14 on Gaeumannomyces graminis Var. tritici and Soil Microbial Diversity. Front. Microbiol. 2021, 12, 682437. [Google Scholar] [CrossRef]
- Han, V.-C.; Yu, N.H.; Yoon, H.; Ahn, N.-H.; Son, Y.K.; Lee, B.-H.; Kim, J.-C. Identification, Characterization, and Efficacy Evaluation of Bacillus velezensis for Shot-Hole Disease Biocontrol in Flowering Cherry. Plant Pathol. J. 2022, 38, 115–130. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Omotayo, O.P.; Igiehon, O.N.; Babalola, O.O. Microbial Genes of Agricultural Importance in Maize Rhizosphere Unveiled Through Shotgun Metagenomics. Span. J. Soil Sci. 2022, 12, 10427. [Google Scholar] [CrossRef]
- Malard, L.A.; Guisan, A. Into the microbial niche. Trends Ecol. Evol. 2023, 38, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bennke, C.; Beier, S.; Bunse, C.; Quince, C.; Ininbergs, K.; Riemann, L.; Ekman, M.; Jürgens, K.; Labrenz, M.; et al. Ecosystem-wide metagenomic binning enables prediction of ecological niches from genomes. Commun. Biol. 2020, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Rosabal Ayan, L.; Macías Coutiño, P.; Maza González, M.; López Vázquez, R.; Guevara Hernández, F. Microorganismos del suelo y sus usos potenciales en la agricultura frente al escenario del cambio climático. Magna Sci. UCEVA 2021, 1, 104–119. [Google Scholar] [CrossRef]
- Bodilis, J.; Calbrix, R.; Guérillon, J.; Mérieau, A.; Pawlak, B.; Orange, N.; Barray, S. Phylogenetic Relationships Between Environmental and Clinical Isolates of Pseudomonas fluorescens and Related Species Deduced from 16S rRNA Gene and OprF Protein Sequences. Syst. Appl. Microbiol. 2004, 27, 93–108. [Google Scholar] [CrossRef]
- Zhong, C.; Hu, G.; Hu, C.; Xu, C.; Zhang, Z.; Ning, K. Comparative genomics analysis reveals genetic characteristics and nitrogen fixation profile of Bradyrhizobium. iScience 2024, 27, 108948. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Boynton, P.J. Evidence for microbial local adaptation in nature. Mol. Ecol. 2017, 26, 1860–1876. [Google Scholar] [CrossRef]
- Bleuven, C.; Landry, C.R. Molecular and cellular bases of adaptation to a changing environment in microorganisms. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161458. [Google Scholar] [CrossRef]
- Kukkar, D.; Sharma, P.K.; Kim, K.-H. Recent advances in metagenomic analysis of different ecological niches for enhanced biodegradation of recalcitrant lignocellulosic biomass. Environ. Res. 2022, 215, 114369. [Google Scholar] [CrossRef]
- Taş, N.; de Jong, A.E.E.; Li, Y.; Trubl, G.; Xue, Y.; Dove, N.C. Metagenomic tools in microbial ecology research. Curr. Opin. Biotechnol. 2021, 67, 184–191. [Google Scholar] [CrossRef]
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science 2013, 342, 621–624. [Google Scholar] [CrossRef]
- Hultman, J.; Waldrop, M.P.; Mackelprang, R.; David, M.M.; McFarland, J.; Blazewicz, S.J.; Harden, J.; Turetsky, M.R.; McGuire, A.D.; Shah, M.B.; et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 2015, 521, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Kannan, V.; Sureendar, R. Synergistic effect of beneficial rhizosphere microflora in biocontrol and plant growth promotion. J. Basic Microbiol. 2009, 49, 158–164. [Google Scholar] [CrossRef]
- Niu, B.; Wang, W.; Yuan, Z.; Sederoff, R.R.; Sederoff, H.; Chiang, V.L.; Borriss, R. Microbial Interactions Within Multiple-Strain Biological Control Agents Impact Soil-Borne Plant Disease. Front. Microbiol. 2020, 11, 585404. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, D.; Li, F.; Dong, Y.; Jin, Z.; Liao, Y.; Li, X.; Peng, S.; Delgado-Baquerizo, M.; Li, X. Superiority of native soil core microbiomes in supporting plant growth. Nat. Commun. 2024, 15, 6599. [Google Scholar] [CrossRef]
- Hartmann, M.; Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 2022, 4, 4–18. [Google Scholar] [CrossRef]
- Schmitz, L.; Yan, Z.; Schneijderberg, M.; de Roij, M.; Pijnenburg, R.; Zheng, Q.; Franken, C.; Dechesne, A.; Trindade, L.M.; van Velzen, R.; et al. Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. ISME J. 2022, 16, 1907–1920. [Google Scholar] [CrossRef]
- Jiang, M.; Delgado-Baquerizo, M.; Yuan, M.M.; Ding, J.; Yergeau, E.; Zhou, J.; Crowther, T.W.; Liang, Y. Home-based microbial solution to boost crop growth in low-fertility soil. New Phytol. 2023, 239, 752–765. [Google Scholar] [CrossRef]
- Saiyam, D.; Dubey, A.; Malla, M.A.; Kumar, A. Lipopeptides from Bacillus: Unveiling biotechnological prospects—Sources, properties, and diverse applications. Braz. J. Microbiol. 2024, 55, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, Y.; Gu, W.; Liu, Z.; Wang, W.; Sun, Y.; Xiu, L.; Zhang, W.; Chen, W. Rhizosphere metabolite-mediated soil enhancement: Long-term biochar application optimizes continuous soybean production systems. Biochar 2025, 7, 95. [Google Scholar] [CrossRef]
- Tomita, S.; Kuroda, K.; Narihiro, T. A small step to discover candidate biological control agents from preexisting bioresources by using novel nonribosomal peptide synthetases hidden in activated sludge metagenomes. PLoS ONE 2023, 18, e0294843. [Google Scholar] [CrossRef]
- Diwan, D.; Rashid, M.M.; Vaishnav, A. Current understanding of plant-microbe interaction through the lenses of multi-omics approaches and their benefits in sustainable agriculture. Microbiol. Res. 2022, 265, 127180. [Google Scholar] [CrossRef]
- Ahmdikhah, A.; Safaeizadeh, M.; Tehranian, A.S. Responses of rice plant to multiple abiotic stresses revealed by transcriptome meta-analysis and identification of novel genetic factors. Sci. Rep. 2025, 15, 8248. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Mann, R.; Kaur, J.; Spangenberg, G.; Sawbridge, T. Transcriptome analyses of barley roots inoculated with novel Paenibacillus sp. and Erwinia gerundensis strains reveal beneficial early-stage plant–bacteria interactions. Plants 2021, 10, 1802. [Google Scholar] [CrossRef]
- Rosman, A.C.; Urquiaga, M.C.d.O.; Thiebaut, F.; Forero Ballesteros, H.G.; Gamosa de Oliveira, E.A.; Hemerly, A.S. Dual RNA-seq of maize and H. seropedicae ZAE94 association, in different doses of nitrate, reveals novel insights into Plant-PGPB-environment relationship. Front. Plant Sci. 2024, 15, 1346523. [Google Scholar] [CrossRef]
- Sahoo, A.; Yadav, G.; Mehta, T.; Meena, M.; Swapnil, P. Omics-driven insights into plant growth-promoting microorganisms for sustainable agriculture. Discov. Sustain. 2025, 6, 659. [Google Scholar] [CrossRef]
- Chang, P.E.; Wu, Y.-H.; Tai, C.-Y.; Lin, I.-H.; Wang, W.-D.; Tseng, T.-S.; Chuang, H.-W. Examining the transcriptomic and biochemical signatures of Bacillus subtilis strains: Impacts on plant growth and abiotic stress tolerance. Int. J. Mol. Sci. 2023, 24, 13720. [Google Scholar] [CrossRef] [PubMed]
- Khan, N. Molecular insights into ABA-mediated regulation of stress tolerance and development in plants. Int. J. Mol. Sci. 2025, 26, 7872. [Google Scholar] [CrossRef]
- Mavrodi, O.V.; McWilliams, J.R.; Peter, J.O.; Berim, A.; Hassan, K.A.; Elbourne, L.D.H.; LeTourneau, M.K.; Gang, D.R.; Paulsen, I.T.; Weller, D.M.; et al. Root exudates alter the expression of diverse metabolic, transport, regulatory, and stress response genes in rhizosphere Pseudomonas. Front. Microbiol. 2021, 12, 651282. [Google Scholar] [CrossRef]
- Ampntelnour, L.; Poulaki, E.G.; Dimitrakas, V.; Mavrommati, M.; Amourgis, G.G.; Tjamos, S.E. Enhancing Botrytis disease management in tomato plants: Insights from a Pseudomonas putida strain with biocontrol activity. J. Appl. Microbiol. 2024, 135, lxae094. [Google Scholar] [CrossRef]
- Doni, F.; Miranti, M.; Mispan, M.S.; Mohamed, Z.; Uphoff, N. Multi-omics approaches for deciphering the microbial modulation of plants’ genetic potentials: What’s known and what’s next? Rhizosphere 2022, 24, 100613. [Google Scholar] [CrossRef]
- Du, X.-Q.; Sun, T.-X.; Xu, W.-L.; Zhu, T.; Wang, Q.; Gu, P.-W.; Lu, J. Multi-omics analysis reveals the specific role of biocontrol reagents against tomato bacterial wilt. Front. Plant Sci. 2025, 16, 1620460. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, R.; Tang, Y.; Cheng, Z.; Qian, M.; Li, W.; Shao, Y. Transcriptome analysis reveals the inducing effect of Bacillus siamensis on disease resistance in postharvest mango fruit. Foods 2022, 11, 107. [Google Scholar] [CrossRef]
- Miggiels, P.; Wouters, B.; van Westen, G.J.P.; Dubbelman, A.-C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trends Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Chen, Y.; Li, E.-M.; Xu, L.-Y. Guide to metabolomics analysis: A bioinformatics workflow. Metabolites 2022, 12, 357. [Google Scholar] [CrossRef]
- Holbrook-Smith, D.; Trouillon, J.; Sauer, U. Metabolomics and microbial metabolism: Toward a systematic understanding. Annu. Rev. Biophys. 2024, 53, 41–64. [Google Scholar] [CrossRef]
- Etminani, F.; Harighi, B.; Mozafari, A.A. Effect of volatile compounds produced by endophytic bacteria on virulence traits of grapevine crown gall pathogen, Agrobacterium tumefaciens. Sci. Rep. 2022, 12, 10510. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Feng, Y.; Ma, J.; Zhang, Q.; Dong, Y.; Li, D.; Duan, X.; Zhou, L.; Li, Z.; Yang, Y.; et al. Genomic and metabolomic insights into the antimicrobial compounds and plant growth-promoting potential of Bacillus velezensis B115. Sci. Rep. 2025, 15, 10666. [Google Scholar] [CrossRef] [PubMed]
- Sokołowski, W.; Marek-Kozaczuk, M.; Sosnowski, P.; Sajnaga, E.; Jach, M.E.; Karaś, M.A. Profiling metabolites with antifungal activities from endophytic plant-beneficial strains of Pseudomonas chlororaphis isolated from Chamaecytisus albus (Hack.) Rothm. Molecules 2024, 29, 4370. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Aragon, B.; Parra-Cota, F.I.; Santoyo, G.; Arellano-Wattenbarger, G.L.; de los Santos-Villalobos, S. Plant-assisted selection: A promising alternative for in vivo identification of wheat (Triticum turgidum L. subsp. durum) growth promoting bacteria. Plant Soil 2019, 435, 367–384. [Google Scholar] [CrossRef]
- Kumawat, K.C.; Razdan, N.; Saharan, K. Rhizospheric microbiome: Bio-based emerging strategies for sustainable agriculture development and future perspectives. Microbiol. Res. 2022, 254, 126901. [Google Scholar] [CrossRef]
- Ayaz, M.; Li, C.-H.; Ali, Q.; Zhao, W.; Chi, Y.-K.; Shafiq, M.; Ali, F.; Yu, X.-Y.; Yu, Q.; Zhao, J.-T.; et al. Bacterial and fungal biocontrol agents for plant disease protection: Journey from lab to field, current status, challenges, and global perspectives. Molecules 2023, 28, 6735. [Google Scholar] [CrossRef] [PubMed]
- Sahil, R.; Pal, V.; Kharat, A.S.; Jain, M. A multi-omics meta-analysis of rhizosphere microbiome reveals growth-promoting marker bacteria at different stages of legume development. Plant Cell Environ. 2025, 3, 915–917. [Google Scholar] [CrossRef]
- Wankhade, A.; Wilkinson, E.; Britt, D.W.; Kaundal, A. A review of plant–microbe interactions in the rhizosphere and the role of root exudates in microbiome engineering. Appl. Sci. 2025, 15, 7127. [Google Scholar] [CrossRef]
- de los Santos Villalobos, S.; Robles, R.I.; Parra Cota, F.I.; Larsen, J.; Lozano, P.; Tiedje, J.M. Bacillus cabrialesii sp. nov., an endophytic plant growth promoting bacterium isolated from wheat (Triticum turgidum subsp. durum) in the Yaqui Valley, Mexico. Int. J. Syst. Evol. Microbiol. 2019, 69, 3939–3945. [Google Scholar] [CrossRef]
- Díaz-Rodríguez, A.M.; Salcedo Gastelum, L.A.; Félix Pablos, C.M.; Parra-Cota, F.I.; Santoyo, G.; Puente, M.L.; Bhattacharya, D.; Mukherjee, J.; de los Santos-Villalobos, S. The current and future role of microbial culture collections in food security worldwide. Front. Sustain. Food Syst. 2021, 4, 614739. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Lugo-Enríquez, C.; de los Santos-Villalobos, S.; Parra-Cota, F.I.; Figueroa-López, P. First report of Cochliobolus sativus causing spot blotch on durum wheat (Triticum durum) in the Yaqui Valley, Mexico. Plant Dis. 2016, 100, 2329. [Google Scholar] [CrossRef]
- Ayala Zepeda, M.; Valenzuela Ruiz, V.; Parra Cota, F.I.; Chinchilla-Soto, C.; de la Cruz Torres, E.; Ibba, M.I.; Estrada Alvarado, M.I.; de los Santos Villalobos, S. Genomic insights of a native bacterial consortium for wheat production sustainability. Curr. Res. Microb. Sci. 2024, 6, 100230. [Google Scholar] [CrossRef] [PubMed]
- Rojas Padilla, J.; Chaparro Encinas, L.A.; Robles Montoya, R.I.; de los Santos Villalobos, S. Promoción de crecimiento en trigo (Triticum turgidum L. subsp. durum) por la co-inoculación de cepas nativas de Bacillus aisladas del Valle del Yaqui, México. Nova Sci. 2020, 12. [Google Scholar] [CrossRef]
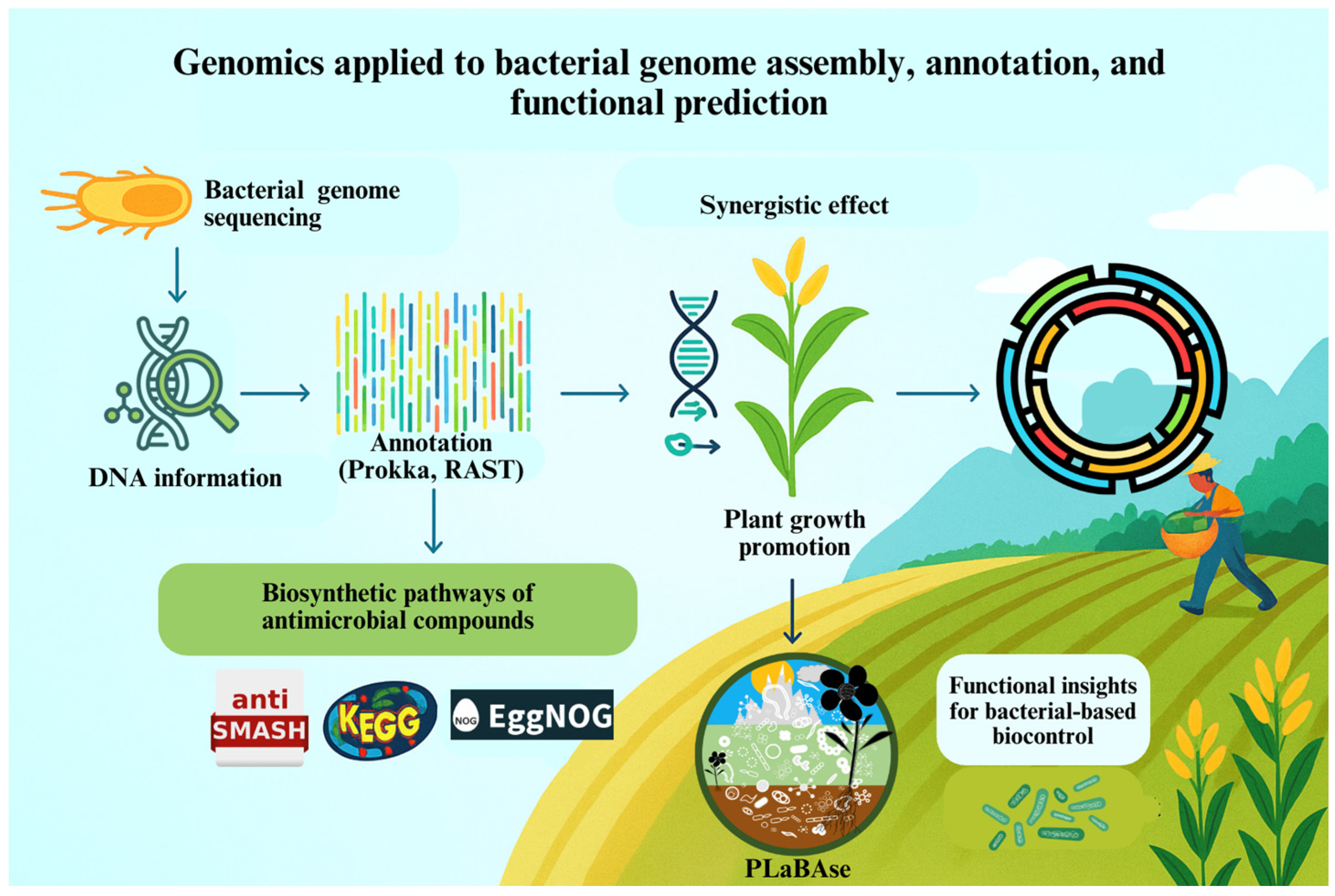
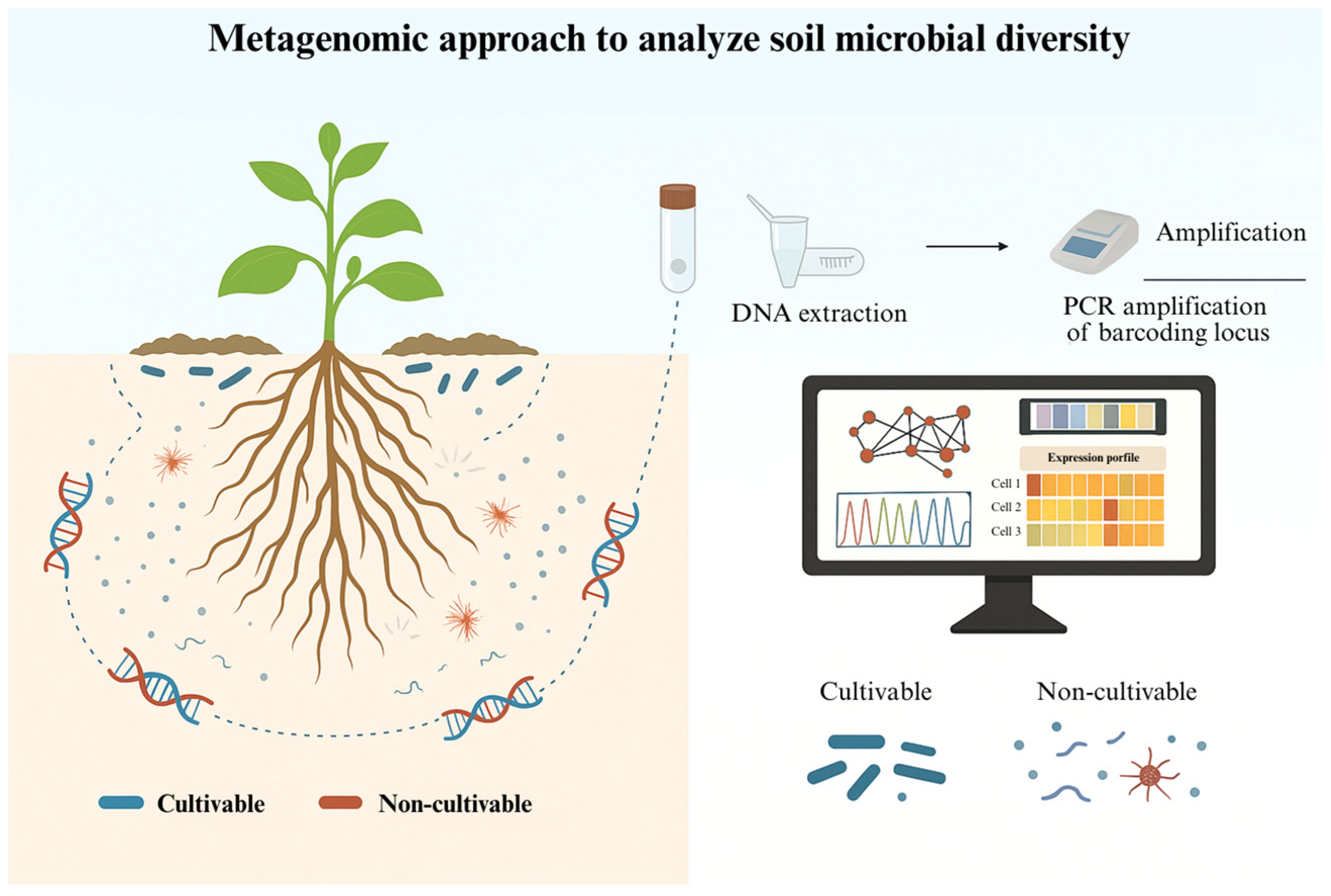
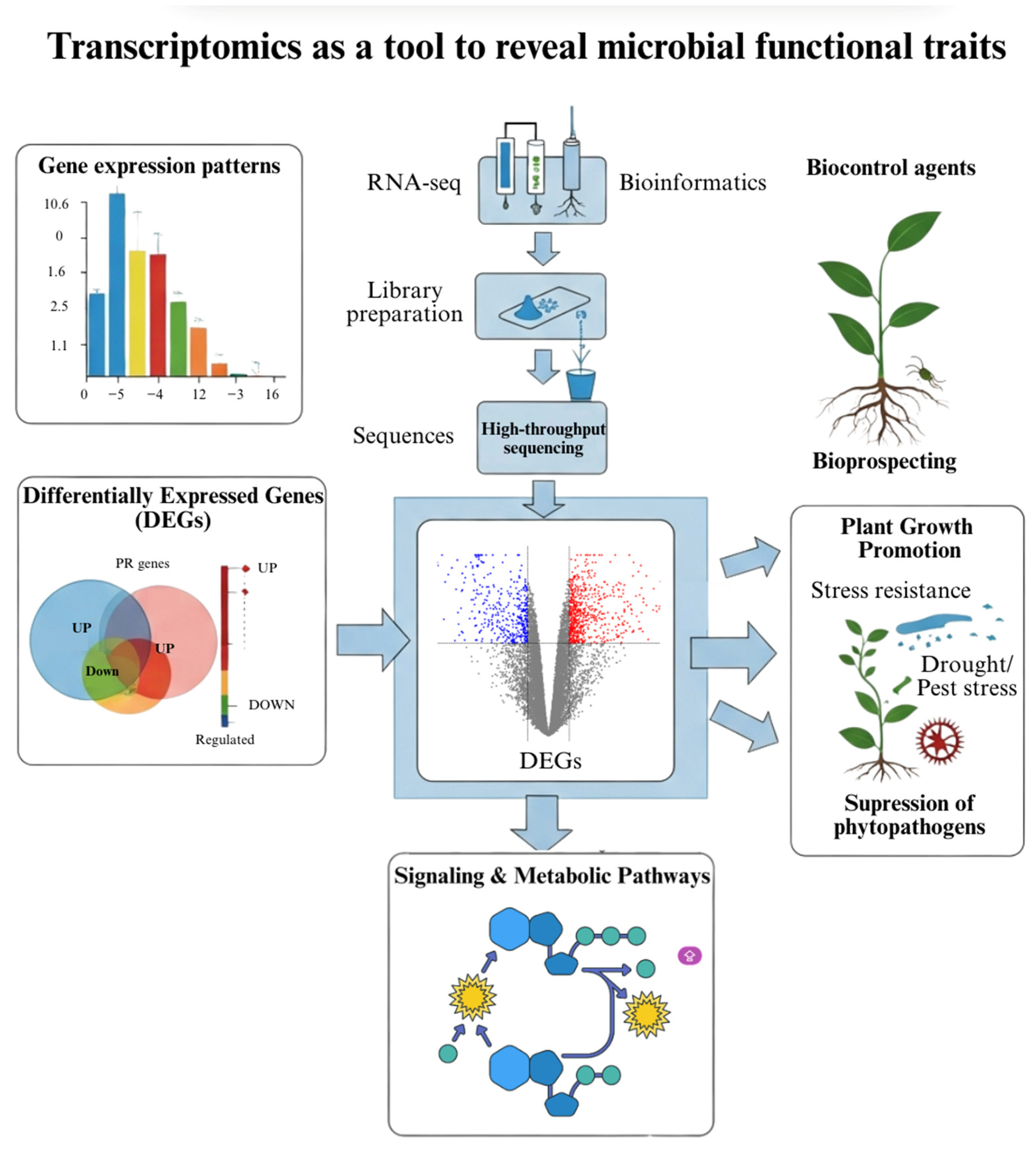
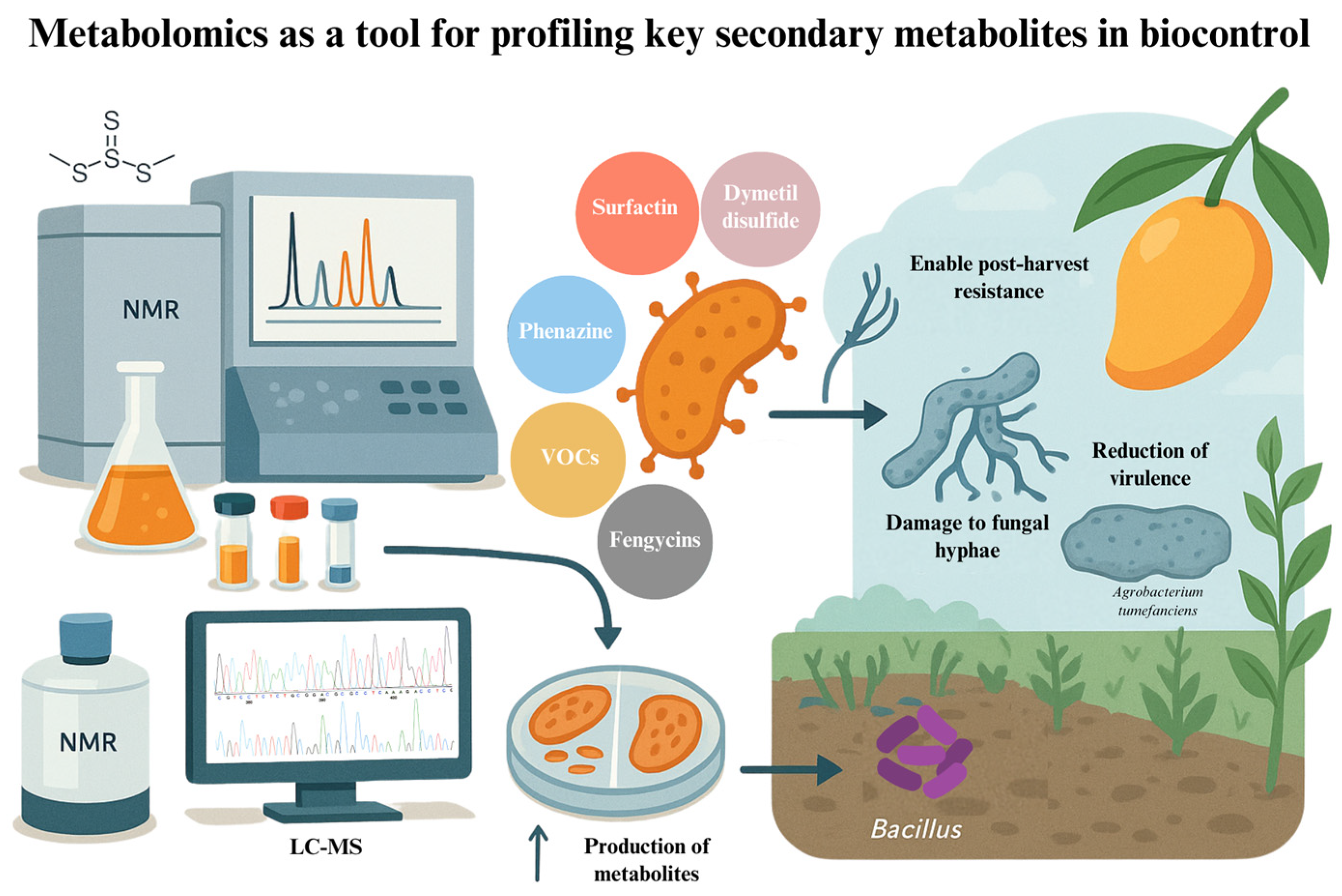
| Strain | Species | Key Traits | Genomic Tools Used | Agricultural Application | Reference |
|---|---|---|---|---|---|
| NBAIR-BtAr | Bacillus thuringiensis | Lipopeptides active against Sclerotium rolfsii | Prokka, RAST, antiSMASH | Antifungal biocontrol | [35] |
| TS022 | Bacillus inaquosorum | Surfactin, Bacillaene, Fengycin, Pipastain, Bacillibactin, among others | ANI, GGDC, Prokka, RAST, antiSMASH | Plant growth promotion, biocontrol against B. sorokiniana | [28] |
| [29] | Burkholderia ambaria | Broad-spectrum antifungal activity | ANI, dDDH, OGRIs, antiSMASH, Realphy | Biocontrol | [29] |
| BN | Bacillus velezensis | Fengycin and surfactin | antiSMASH, NR, Swiss-Prot, Pfam, EggNOG, GO, KEGG | Broad-spectrum antimicrobial | [32] |
| NEB573 | Brevibacillus brevis | Unexplored secondary metabolites | Phylogenetic software, pan-genome, secondary metabolite mining tools | Plant disease management, growth promotion | [34,36] |
| AF23 | Bacillus halotolerans | Genes for salt stress tolerance, biocontrol, and plant growth promotion | Comparative genomics | Plant growth promotion in tomato; synergistic with AF12 | [36] |
| TE3T | Bacillus cabrialesii subsp. cabrialesii | Lipopeptides, siderophores, antimicrobial compounds | RAST, PROKKA, PGAP, antiSMASH, | Biocontrol against B. sorokiniana | [37,38,39] |
| TRQ65 | Bacillus paralicheniformis | Lipopeptides, siderophores, antimicrobial compounds | RAST, antiSMASH, BAGEL | Biocontrol against Botrytis, Fusarium, Bipolaris, and other phytopathogens | [37,40] |
| Technology | Description | Advantages | Limitations | Typical Applications in Biocontrol |
|---|---|---|---|---|
| LC-MS (liquid chromatography–mass spectrometry) | Combines chromatographic separation with mass spectrometry for sensitive detection of metabolites | High sensitivity and broad metabolite coverage; suitable for complex mixtures; can detect low-abundance compounds | Requires sample preparation, potential matrix effects, and instrument cost | Discovery and quantification of antimicrobial secondary metabolites (e.g., lipopeptides, phenazines) from bacterial and fungal BCAs |
| GC-MS (Gas Chromatography–Mass Spectrometry) | Separation of volatile and semi-volatile metabolites followed by mass spectrometry detection | Excellent for volatile compounds; high resolution and reproducibility | Limited to volatile/thermally stable metabolites; derivatization often needed | Profiling volatile organic compounds involved in pathogen inhibition and signaling |
| NMR (Nuclear Magnetic Resonance) Spectroscopy | Spectroscopic technique providing structural information of metabolites based on nuclear spin properties | Non-destructive, highly reproducible, minimal sample preparation; structural elucidation | Lower sensitivity compared to MS; requires larger sample amounts | Structural characterization of novel antimicrobial compounds; in situ metabolic profiling |
| Ion Mobility Spectrometry (IMS) coupled with MS | Adds an ion mobility separation step before MS to separate isomers and conformers | Improved separation of complex mixtures; faster analysis | Requires specialized instrumentation; data complexity | Enhanced resolution of structurally similar bioactive metabolites |
| Direct Injection MS (DIMS) | Direct introduction of the sample into the MS without prior chromatographic separation | Very high throughput; minimal sample prep | Reduced metabolite coverage due to ion suppression/matrix effects | Rapid screening of metabolite profiles in large BCA libraries |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela Ruiz, V.; Cervantes Enriquez, E.P.; Vázquez Ramírez, M.F.; Bivian Hernández, M.d.l.Á.; Cárdenas-Manríquez, M.; Parra Cota, F.I.; de los Santos Villalobos, S. A New Era in the Discovery of Biological Control Bacteria: Omics-Driven Bioprospecting. Soil Syst. 2025, 9, 108. https://doi.org/10.3390/soilsystems9040108
Valenzuela Ruiz V, Cervantes Enriquez EP, Vázquez Ramírez MF, Bivian Hernández MdlÁ, Cárdenas-Manríquez M, Parra Cota FI, de los Santos Villalobos S. A New Era in the Discovery of Biological Control Bacteria: Omics-Driven Bioprospecting. Soil Systems. 2025; 9(4):108. https://doi.org/10.3390/soilsystems9040108
Chicago/Turabian StyleValenzuela Ruiz, Valeria, Errikka Patricia Cervantes Enriquez, María Fernanda Vázquez Ramírez, María de los Ángeles Bivian Hernández, Marcela Cárdenas-Manríquez, Fannie Isela Parra Cota, and Sergio de los Santos Villalobos. 2025. "A New Era in the Discovery of Biological Control Bacteria: Omics-Driven Bioprospecting" Soil Systems 9, no. 4: 108. https://doi.org/10.3390/soilsystems9040108
APA StyleValenzuela Ruiz, V., Cervantes Enriquez, E. P., Vázquez Ramírez, M. F., Bivian Hernández, M. d. l. Á., Cárdenas-Manríquez, M., Parra Cota, F. I., & de los Santos Villalobos, S. (2025). A New Era in the Discovery of Biological Control Bacteria: Omics-Driven Bioprospecting. Soil Systems, 9(4), 108. https://doi.org/10.3390/soilsystems9040108






