Abstract
This study examines culturable diazotrophs and non-culturable bacteria found in the rhizospheres and root pseudonodules of wild blackberry plants (Rubus ulmifolius) that dwell on an unmanaged calcareous nitrogen-deficient soil. The DNA was extracted from the nodules and rhizospheres, and 16S rRNA gene metabarcoding was carried out. The metagenome functions were predicted with bioinformatic approaches. The soil samples were analyzed for the physico-chemical properties. The culturable diazotrophs were isolated and evaluated for the biochemical and plant growth-promoting properties. The soil was classified as nutrient-depleted calcareous soil. The microbial communities of the nodules and rhizospheres showed marked differences. The Pseudomonadota was the nodules’ dominant phyla (90%), while the Actinobacteriota was the most abundant (63%) in the rhizospheres. Stenotrophomonas was the dominant genus (55%) in the nodules, while the Streptomyces genus was widely present (39%) in the rhizospheres. The differences among the nodule and rhizosphere microbial communities were also highlighted by the metagenome function predictions. The gene copies (KOs) revealed the most interesting findings. Similar KOs involved in the nitrogen fixation were found to be similar in terms of the nodules and rhizospheres. However, the nitrate reduction was higher in the rhizosphere, while the denitrification was more prominent in the nodules. Nine diazotrophs were isolated from the nodules and rhizospheres. The plant growth promoting traits’ characterization has shown the interesting potential of the isolates in improving the acquisition of nutrients in plants, promoting their growth, and tolerating stress. Based on interesting biochemical and plant growth-promoting traits, the isolate N2A was further characterized and identified as Pantoea agglomerans.
1. Introduction
Often overlooked, the soil is one of the most important components of our environments, playing a vital role in sustaining life. Soil comprises the second largest carbon reservoir of the planet, the keystone of many biogeochemical cycles, and is inhabited by countless living beings closely associated with soil particles [1]. Among the soil microorganisms, bacteria are the most basic and widespread dwellers [2,3]. It is estimated that we could sample up to 10,000,000,000 bacterial cells in a single gram of soil [4], consisting of about 4000 to 50,000 different species [5,6]. Despite this richness, we only know a little about this immense vastness, only a small percentage of bacterial phyla can be cultured, and their metabolism is not known [7].
Bacteria shape our world; they are the primary converters of carbon dioxide into molecular oxygen; they refill soil with nutrients and make plants grow [8]. Thanks to the latest molecular analysis techniques, we have begun to understand the complexity of the soil microbial communities and their metabolisms [9]. Rather than an inert surface, the soil is alive and constantly changing due to the activity of bacteria and many other organisms. Microorganisms weave refined webs of a nutrient interexchange between themselves and other living beings, often forming symbiotic relationships [10].
Microbial communities are shaped by the physico-chemical properties of the soil [11]. Among the latter, the nutrient content is a major factor in shaping microflora [12]. Changes in nitrogen, phosphorus, and potassium modify soil microbial communities [13]. The nitrogen fertilization of natural ecosystems, for example, decreased the microbial communities’ richness and diversity [14]. At the same time, rhizobacteria play beneficial roles for plants, improving their nutrient uptake, growth rate, and disease resistance [15]. Plants cope better with abiotic stress and biological threats thanks to these relationships, and they sometimes cannot thrive or even grow without interconnections with bacteria [16]. Thus, plants have evolved in exceptional ways to communicate chemically with microorganisms [17]. The application of plant growth-promoting rhizobacteria (PGPR) in an agriculture has grown in recent decades due to their stimulating effects on plant growth [18,19] and the production of different metabolites of interest [20].
One of the most studied classes of PGPR is that of diazotrophs, or nitrogen-fixing bacteria, as they can help reduce our dependence on nitrogen fertilizers. These compounds helped improve the yields and feed a growing population during the 20th century [21]. However, while nitrogen fertilizers are useful, there is growing concern about their overuse. They are a source of environmental pollution (e.g., soil acidification, eutrophication, and even the emission of greenhouse gases) [22]. Therefore, scientific research is attempting to find alternative and more sustainable solutions to mitigate the effects of their dispersal or reduce their use. PGPR are a promising tool, but there are currently some limitations when applied, mainly because bacterial formulations do not always survive in open-field inoculations, and root colonization is challenged by the presence of in situ microorganisms [23].
To increase the success of a microbial inoculant, the strain or strains of interest should be isolated from the target species in which the product is to be applied. Plants, in fact, establish cultivar-specific associations with microbiota [24]. Within these associations, the Frankia (phylum Actinomycetota) genus forms a specific symbiotic relationship with actinorhizal plants [25]. Actinorhizal symbiosis is a polyphyletic trait. Therefore, the association with Frankia in Rosales probably arose independently multiple times in the co-evolutionary history of these plants (and vice versa) [26]. Frankia and other actinobacteria can colonize roots forming vesicles that stimulate the formation of a vascular core surrounded by cortex cells known as pseudo-nodules [27]. Within these structures, these bacteria fix nitrogen in an oxygen-free environment [28]. These structures are different from nodules, which are the cortical cell-derived root outgrowth structures formed by the symbiotic association with rhizobia [29]. This article presents a case study of microbiota of wild blackberry (Rubus ulmifolius) root pseudo-nodules (hereafter, nodules) and rhizospheres. This species belongs to the family Rosaceae, order Rosales. A few cases dealing with the endophytic associations between the members of Rosales and nitrogen-fixing rhizobacteria are described. Bond reported the first case of nodular formations on a specimen of Rubus ellipticus [30]. Later, Becking confirmed Frankia as the causative agent of these structures [31]. Rubus ferdinandi-muelleri was also described as an actinorhizal plant [32]. However, to the best of our knowledge, there are no cases of R. ulmifolius hosting pseudo-nodules.
Culturable diazotrophs, unculturable bacteria, and the soil physico-chemical characteristics were studied. Based on the presence of nodules and given the nutrient deficiency in the soil, we hypothesized a microbial community composed of numerous N2-fixing bacteria. Within the microbiota we also hypothesized the presence of culturable diazotroph strains with plant growth-promoting (PGP) traits. The plant samples and their rhizospheres were collected from an unmanaged olive grove. The nodules and rhizospheres were subjected to a DNA extraction and diazotrophs isolation. Rhizosphere soil was also subjected to a physico-chemical characterization. The DNA samples were subjected to 16S rRNA gene metabarcoding to study the nodules and rhizosphere microbiota. The metabarcoding data were used to predict the metagenome functions. The isolates were screened for their bacteriological, biochemical, and plant growth-promoting traits to evaluate the biostimulant abilities.
2. Materials and Methods
2.1. Plant and Soil Sampling and Analysis
Fifteen plants were sampled in an unmanaged olive grove in the municipality of Scheggino (province of Perugia, Italy—12.8272, 42.6864–471 m a.s.l.) by digging them with the soil surrounding the plants (20 cm depth) and transferring them to the laboratory in sterile bags. The plant specimens were analyzed by our botanist (Prof. Loretta Pace), classified as Rubus ulmifolius Schott., and deposited in the Herbarium Aquilanum of the University of L’Aquila (Environmental Sciences Section, MeSVA Department, L’Aquila, Italy). After the plant’s identification, the nodules were excised and bulked. The rhizosphere samples were also bulked.
Three aliquots of the bulked samples were immediately processed for the isolation of culturable microflora. Further, three aliquots of the bulked samples were placed in the freezer (−20 °C) for DNA isolation. An aliquot of a bulked soil sample was dried at room temperature for 2 days and analyzed for the physico-chemical properties by the Laboratori ISPA—Istituto Sperimentale Problematiche Ambientali Panetta S.r.l. (Atina, Italy), following the Italian standards methods [33] as previously described [34].
2.2. DNA Extraction, 16S rRNA Gene Metabarcoding, and Metagenome Functions Prediction
The nodules were bulked across all the samples and homogenized with a mortar–pestle with the help of liquid nitrogen. The soil samples were also bulked and processed by sieving (<2 mm). The genomic DNA was collected using 500 mg bead beating techniques from the homogenous samples utilizing a NucleoSpin®Soil kit, according to the company protocol (Macherey Nagel, Düren, Germany). Using a Qubit fluorometer (Thermo ScientificTM), the extracted samples were examined spectrophotometrically and fluorometrically to determine the DNA content and purity. Three replicates from each sample were mixed to form an equimolar solution. According to the previously reported analytical method [35] and paired-end 16S rRNA gene community sequencing on the MiSeq Illumina platform (Bio-Fab Research, Rome, Italy), a customized 16S rRNA technique was performed to amplify the V3 and V4 (16S Amplicon PCR Forward Primer 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and 16S Amplicon PCR Reverse Primer 5′GTCTCGTGGGCTCG GAGATGTGTATAAGAGACAGGA CTACHVGGGTATCTAATCC) regions of the 16S rRNA gene. The reads were initially checked for their quality and counted after filtering. QIIME2 (qiime2-2020.2 version) was used for the ASV (Amplicon Sequence Variant) assembly with the DADA2 plugin [36]. From the 16S file obtained from the SILVA 132 database (https://www.arb-silva.de/, accessed on 2 October 2021), the V3–V4 specific region was extracted and utilizedfor classifier training by the fit-classifier-naive-bayes plugin 1 or the assignment of the taxa.
We used the PICRUSt2 program to estimate the functional abundances of the microbial communities using information from the 16S rRNA gene sequencing. Based on the ASV sequence profiles and abundances, the PICRUSt2 predictions were calculated [37]. We considered three gene family databases: the Kyoto Encyclopedia of Genes and Genomes (KEGG), orthologs (KOs), Enzyme Commission numbers (ECs), and MetaCyc pathways abundances (PWYs). The PICRUSt2 outputs (KOs, ECs, and PWYs) were analyzed by QIIME2 software. The KOs involved in the nitrogen cycle were also studied following Das and colleagues [38].
We calculated the alpha-diversity metrics (i.e., Simpson, Shannon, and Chao1 indices) by PAST 4.03 software. We studied the differences among the ECs, KOs, and PWYs predicted for the nodules and rhizospheres by GraphpadPrism 9 using a Paired t-test (p < 0.001).
2.3. Isolation and Biochemical Characterization of Diazotrophs
The diazotrophic strains were isolated from the root nodules and rhizospheres of blackberries. A total of 1g of root segment was added to 9 mL of 1% of sterile Tween 20 saline solution and agitated for 30 min at room temperature. Then, the serial dilutions (from 10−2 to 10−9) were prepared and 0.1 mL were plated on a nitrogen-free combined carbon (NFCC) selective medium. The plates were incubated at 30 °C for 48 h.
The strains obtained were further investigated for the bacteriological and biochemical traits. The Gram nature was identified by a Gram staining kit (Sigma-Aldrich, St. Louis, MO, USA). The biochemical characteristics were investigated using VITEK 2 equipment and procedures (Biomerieux, Bagno a Ripoli, Italy). Based on the Gram nature, the isolates were processed with either Gram-positive or Gram-negative biochemical traits cards following the manufacturer’s standard procedures.
2.4. Plant Growth-Promoting Traits Investigation
Several PGP tests were evaluated to test the isolates’ ability to promote the plant’s growth. The production of hydrocyanic acid (HCN), ammonia (NH3) and indole-3-acetic acid (IAA), phosphate solubilization, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity were estimated.
2.4.1. Hydrocyanic Acid and Ammonia Production
To assess the HCN production, a trypticase soy agar (TSA) medium supplemented with glycine (4.4 g L−1) was plated with 100 µL of each liquid culture in a nutrient broth (NB) medium. Each inoculated Petri dish had a Whatman paper identical to the dish’s diameter that had been immersed in picric acid (0.5%) and sodium carbonate (2%), closed with parafilm, and incubated at 30 °C for 48 h. The change in the color of the paper (from yellowish to an orange or brown color) was used to measure the HCN production [39].
The NH3 production was analyzed on peptone water (PW) liquid medium. A total of 100 µL of liquid culture in the NB medium of each isolate were inoculated in 10 mL of peptone water and incubated for 48 h at 30 °C. After the incubation, 0.5 mL of Nessler’s reagent was added to each tube. The occurrence of a yellow color was assumed as a positive result [40].
2.4.2. Production of Indole-3-Acetic Acid
The IAA production was evaluated as explained by Djebaili et al. (2020) [41] and was performed as follows: 100 µL of a liquid culture in the NB medium of each strain were inoculated in 10 mL of the NB medium added with tryptophan (0.2% w/v) and the mixture was incubated at 30 °C for 48 h with moderated shaking at 150 rpm. The cultures were centrifuged after incubation for 20 min at 3000 rpm. After that, 4 mL of Salkowski’s reagent were combined with 1 mL of supernatant [42]. The solution was kept in the dark at 37 °C for 30 min. The optical density was measured at 530 nm and the IAA standard curve was used to quantify the IAA concentration in each sample [43].
2.4.3. Phosphate Solubilization
The isolates were smeared on the National Botanical Research Institute’s Growth medium (NBRIP), which contains Ca3(PO4)2 as the sole source of phosphate, to assess the ability of the strains to solubilize the inorganic phosphate [44]. This ability was observed by a solubilization halo around the colony formation after an incubation of 48 h at 30 °C. The solubilized phosphate was quantified using a liquid NBRIP media. Briefly, 10 mL of the liquid NBRIP medium was inoculated with 100 µL of each suspension culture in a nutrient broth (NB) medium. The mixture was incubated at 30 °C for 48 with a moderate agitation. The cultures were then centrifuged after incubation at 3000 rpm for 20 min [45] and the Olsen and Sommers colorimetric method was used to measure the solubilized phosphorus in the supernatant [46].
2.4.4. 1-Aminocyclopropane-1-Carboxilate Deaminase Estimation
The method outlined by Brigido et al. was used to determine the activity of the ACC deaminase [47]. A total of 200 µL of the liquid culture of each strain were inoculated in 15 mL of an NB medium and incubated for 48 h at 30 °C in a rotatory shaker at 200 rpm. A centrifugation was carried out at 3000 rpm following two washings with 10 mL of Dworkin and Foster (DF) salts minimum medium deprived of a nitrogen source [48]. The pellets of each culture were homogenized in 15 mL of a minimal DF salt medium added with 3 mM of ACC and were then shaken for 72 h at 30 °C. After incubation, the cell suspensions were obtained by centrifuging the cultures and washing them in 5 mL of 0.1 M Tris-HCl (pH 7.6). After centrifugation at 10,000 rpm, the obtained pellet was utilized for the enzymatic activity assay.
It was dissolved in 400 µL of 0.1 M Tris-HCl (pH 8.0) with 20 µL of toluene. Then, 50 µL from each cell lysate was distributed into three microtubes, where 5 µL of ACC (0.5 M) was added in two tubes, while the third tube was used as a negative control. A second negative control was also made, comprising 5 µL of 0.5 M of ACC and 50 µL of 0.1 M Tris-HCl (pH 8.0). The cell suspensions were agitated for 5 s after the addition of the ACC and incubated for 30 min at 30 °C. Each tube received 500 µL of 0.56 M HCl followed by a vortexing for 5 s. A centrifugation was performed and the standard was effectuated with a solution of α-ketobutyrate (Sigma-Aldrich, St. Louis, MO, USA) in 0.1 M of TRIS-HCl (pH 8.0). The optical density was read at 540 nm. The determination of the ACC deaminase activity was conducted using the calibration curve of α-ketobutyrate (5, 10, 15, 20, and 25 µmol mL−1) and expressed as a µmol α-ketobutyrate h−1 mg protein−1.
The Bradford method [49] was used to determine the protein amount using bovine serum albumin (BSA) as a standard and the total protein concentration of the extracts was calculated using a calibration curve with BSA (1.25, 2.5, 5, and 10 µg/mL).
2.5. 16S rRNA Gene Barcoding and Phylogenetic Analysis
The 16S rRNA gene barcoding was carried out on the most promising strain (Microbion, Verona, Italy). The DNA was first extracted from an overnight bacterial culture. The DNA was then amplified using universal bacterial primers (8F/1541R). The genetic database of the NCBI (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov, accessed on 11 July 2022) was used to compare the consensus sequence (~1400 bp) with those present in the database, utilizing the local base alignment search (BLAST). More than 99% of the sequence’s similarity was taken into consideration. MEGA X was used to conduct the phylogenetic analysis [50]. The tree was inferred by using the maximum likelihood method. The initial tree(s) for the heuristic search were obtained automatically by applying neighbor join and BioNJ algorithms to a matrix of pairwise distances estimated using the Jukes–Cantor model [51] (1000 bootstrap), selected based on the lowest Bayesian information criterion scores [52].
3. Results
3.1. Plant and Soil Analyses
Figure 1 shows a nodular formation on the plant samples, developed where the stem joins the root below the soil’s surface. All the nodules which were sampled had woody external and internal structures. The inner tissues of the nodules had a peculiar red-pink pigmentation.

Figure 1.
Plant nodular formations present on Rubus ulmifolius where the stem joins the roots below the soil surface.
The characterization of the soil sample is given in Table 1. Based on the pH, electrical conductivity (EC), and low nutrient contents, the sample was classified as a calcareous nutrient-depleted soil.

Table 1.
Soil physico-chemical properties.
3.2. 16S rRNA Gene Metabarcoding, and Metagenome Functions Prediction
The outcomes of the 16S rRNA gene metabarcoding were examined to evaluate the richness and diversity of the samples and the results are shown in Table 2. The nodule samples showed more taxa, individuals, and species numbers in the community (Chao-1) than in the rhizosphere samples. However, according to the diversity indexes, the rhizosphere samples presented more richness and diversity (0.8 and 2.7) for Simpson_1-D and Shannon_H indices than the nodule samples.

Table 2.
Diversity indices generated from the results of 16S rRNA metabarcoding using PAST 4.03.
The classification of the ASV taxa revealed the presence of 4 common bacterial phyla on both nodule and rhizosphere samples and 22 genera distributed differently between the two samples. At the phylum level (Figure 2 and Table S1 of the Supplementary Materials), Pseudomonadota was the dominant phyla (90%) on the nodule samples, followed by Actinobacteriota, Bacteroidota, and Bacillota with an equivalent dispersion. For the rhizosphere samples, Actinobacteriota was the most abundant (63%), followed by Bacteroidota and Pseudomonadota (24 and 9%, respectively).
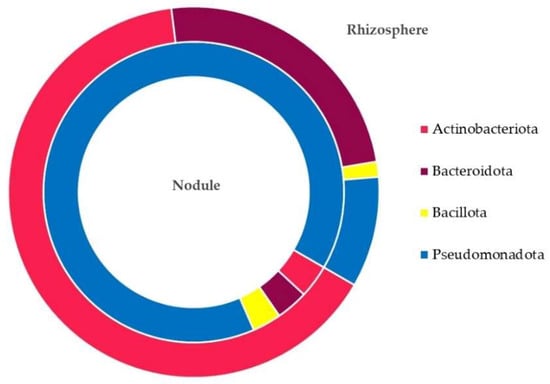
Figure 2.
Amplicon sequence variants (ASVs) distribution at the phylum level in the nodule and rhizosphere samples.
At the genus level (Figure 3 and Table S2 of Supplementary Materials), the ASVs were mainly associated with 22 taxa (cutoff of relative abundances at 0.5%). Stenotrophomonas was the dominant genus (55%) in the nodules, followed by Pseudomonas (21%). Seven lineages were only present in the nodules, i.e., Brevundimonas, Cupriavidus, Flavobacterium, Paenibacillus, Pseudomonas, Sphingomonas, and Stenotrophomonas. The other genera were also found in the rhizosphere samples. The common presence of the diazotroph group ANPR (Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium) was important. The genus Streptomyces was widely present (39%) in the rhizospheres, followed by Chitinophaga (23%), Glycomyces (9%), and Promicromonospora (7%). Glycomyces, Inquilinus, Kribbella, Nocardia, Nonomuracea, Streptomyces, and TM7a were exclusive to rhizospheres.
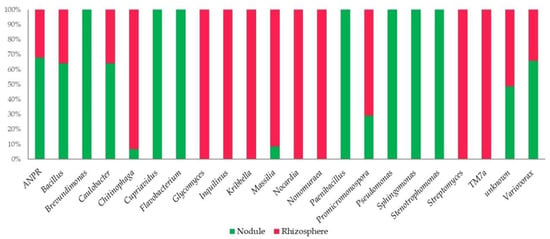
Figure 3.
Amplicon sequence variants (ASVs) distribution at the genus level (0.5% cutoff) in the nodule and rhizosphere samples. In Figure 3, ANPR refers to Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium.
The different composition and structure of the nodule and rhizosphere microbial communities were also made evident by the predicted functions of the metagenome. Figure 4 shows the correlation plot for the predicted ECs. A marked difference in the predicted ECs count was found between the nodules and rhizospheres (Paired t-test: p < 0.001). As depicted, only a few ECs achieved relative counts greater than 100000 in both the samples (labeled black with EC codes). Marked differences were shown by EC:32.2.1.23 (beta-galactosidase), with the highest counts in the nodules, and EC:2.5.1.18 (glutathione transferase), with the highest counts in the rhizospheres.
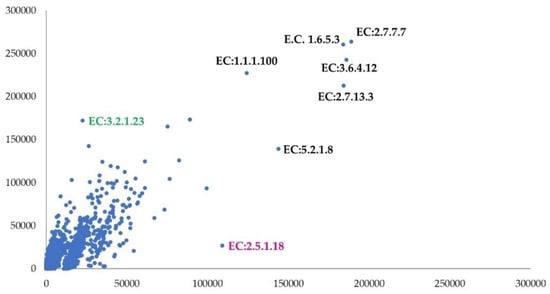
Figure 4.
Correlation plot on enzymes predicted by PICRUSt 2 software [37] (nodule y axis and rhizosphere x axis). In Figure 4, the EC codes refer to the enzyme commission numbers of the KEGG database. Only ECs with relative counts higher than 100,000 were labeled in black. The green label highlights the EC with higher counts in nodule sample while the magenta one shows the EC with higher counts in the rhizosphere. Correlation coefficient: 0.84.
The predicted KEGG orthologs relative counts also presented a marked difference between the samples (Paired t-test: p < 0.001) (Figure 5). The KOs with counts higher than 100,000 in both samples were few (black labels), with prominent counts of K03406 (methyl-accepting chemotaxis protein) in the rhizospheres and K19737 (type II protein arginine methyltransferase) in the nodule. Counts lower than 100,000 were recorded for all the rhizosphere predicted pathways (PWY, Figure S1 of the Supplementary Materials) with a significant difference from those predicted for the nodule (Paired t-test: p < 0.001).
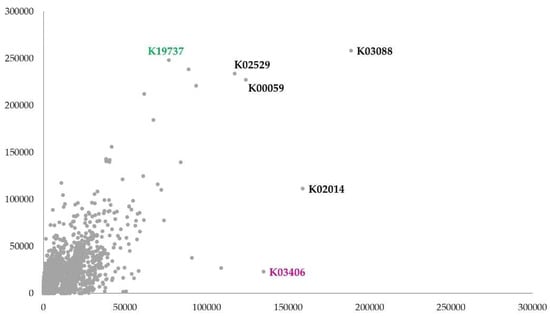
Figure 5.
Correlation plot on functional orthologs predicted by PICRUSt 2 software (nodule y axis and rhizosphere x axis). In Figure the KO codes refer to the KEGG Orthologs of KEGG database. Only KOs with relative counts higher than 100,000 were labeled in black. The label in magenta shows the KO with higher counts in rhizosphere. Correlation coefficient: 0.73.
To study the predicted KOs associated with the nitrogen metabolism, we investigated them separately and considered the process to which they belonged (Table 3). The analysis revealed that the predicted gene copies involved in the nitrogen fixation are similar in the nodule and rhizosphere. No relative counts for the KOs associated with the nitrification process were observed. Some dissimilarities were found for the other KOs involved in the other processes linked to the nitrogen cycle. The relative counts of the KOs associated with assimilatory and dissimilatory nitrate reduction were higher in the rhizosphere than in the nodule. In contrast, denitrification was more prominent in the nodule than in the rhizosphere.

Table 3.
Relative counts of the selected KEGG orthologs associated with nitrogen cycle predicted through PICRUSt 2.
3.3. Isolation and Biochemical Characterization of Diazotrophs
Nine diazotrophic strains (N1, N2A, N2B, N3, N5, N6A, N6B, N6C, and N7) were obtained from the root nodules and rhizosphere samples on the NFCC selective medium. Their N2-fixation ability was detected by evaluating the intensity of the color change in the NFCC medium from green to blue.
According to the Gram staining, six strains were classified as Gram-positive (N1, N2B, N6A, N6B, N6C, and N7) and three as Gram-negative (N2A, N3, and N5). The biochemical characterization shown in Tables S3 and S4 showed metabolic traits for the isolates N7, N2A, and N5. The phenotypic identification of the strains based on these biochemical traits was only possible for N2A and N5, with an identification rate of 98% and 96%, respectively. Both isolates were associated with Pantoea agglomerans species. The other isolates received a phenotypic association lower than 80%.
3.4. Plant Growth-Promoting Traits Investigation
3.4.1. Hydrocyanic Acid and Ammonia Production
The hydrocyanic acid and ammonia production was estimated for the nine isolated strains with a N2-fixing ability (Table 4). HCN was produced by all the isolates, except by the N1 and N7 strains, while all the strains seemed to be able to produce NH3. Two strains (N2A and N5) showed results considering both the production of HCN and NH3.

Table 4.
HCN and NH3 production by the isolated strains. The estimate was indicated as follows: no production (−); low production (+); medium production (++); high production (+++).
3.4.2. Indole-3-Acetic Acid Production
The production of IAA by the different strains is shown in Figure 6. All the tested isolates demonstrated an ability to produce IAA up to 10 µg mL−1 of IAA. The highest amount of IAA was observed for the N2A strain (14.79 µg mL−1), followed by N5 (13.91 µg mL−1) and N3 (13.03 µg mL−1). The lowest amounts were observed for the N2B and N6B strains, with 10.73 µg mL−1 and 10.81 µg mL−1, respectively.
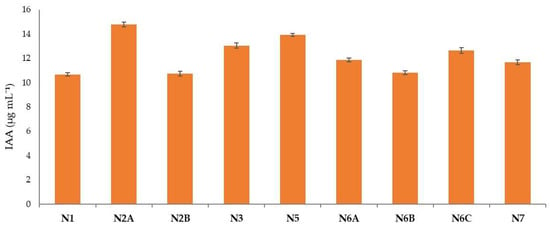
Figure 6.
IAA concentrations produced by the different strains.
3.4.3. Phosphate Solubilization
The phosphate (PO43−) solubilization activity was further investigated as a PGP trait through qualitative and quantitative approaches. The solubilization ability on a solid NBRIP medium was shown by all the isolates with the presence of a solubilization halo around the colony. The estimated PO43− solubilized in a liquid NBRIP medium (Figure 7) was reported in the N2B strain showing the highest amount of solubilized PO43− (82.33 µg mL−1), followed by N3, N6A, and N2A with 77.59 µg mL−1, 75.14 µg mL−1, and 72.14 µg mL−1, respectively. The lowest amounts were obtained for strain N6B (61.85 µg mL−1) and N7 (61.51µg mL−1).
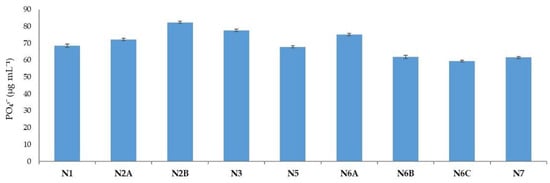
Figure 7.
Amounts of solubilized phosphate (PO43−) by different isolates cultivated on liquid NBRIP medium.
3.4.4. 1-Aminocyclopropane-1-Carboxilate Deaminase Estimation
In Figure 8, the estimation of the ACC deaminase activity was reported. The highest amount was observed in N1 (3.0 µmol α-KB mg proteins −1 h−1), followed by N2A (1.82 µmol α-KB mg proteins −1 h−1). The lowest amount was observed for the strain N5 (0.09 µmol α-KB mg proteins −1 h−1). The other strains showed values ranging from 0.54 µmol α-KB mg proteins −1 h−1 to 0.82 µmol α-KB mg proteins −1 h−1.
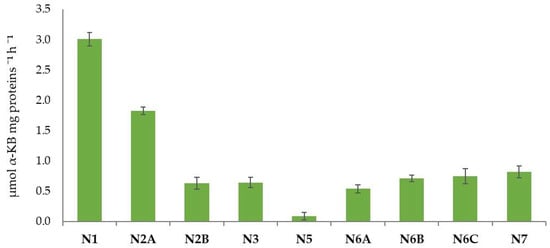
Figure 8.
The 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity (µmol α-KB mg proteins −1 h−1) expressed by the isolated strains.
3.5. 16S rRNA Gene Barcoding and Phylogenetic Analysis
We further characterized the most promising isolate by 16s rRNA barcoding and conducted a phylogenetic analysis. The N2A isolate was identified as Pantoea agglomerans with an identity percentage of 99.9% (Figure 9). These analyzes allowed us to confirm the VITEK 2 identification of the N2A isolate.
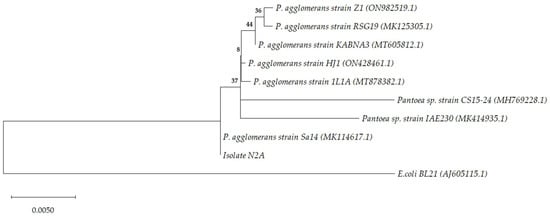
Figure 9.
Phylogenetic tree inferred using the maximum likelihood method and Jukes–Cantor model [38]. The tree with the highest log likelihood (−2671.15) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 10 nucleotide sequences. There were a total of 1488 positions in the final dataset. NCBI accession numbers are provided in parenthesis.
4. Discussion
There is still much to learn about soil microbial communities. Increasing our knowledge of their composition, structure, functionality, processes, and relationships is a fundamental component in the research. It can directly help us understand how to improve agriculture through the modulation, management, and manipulation of the interactions between plants and microbes. In this study, we investigated wild root-nodulating R. ulmifolius plants from calcareous nitrogen-deficient soil, describing for the first time the microbiota of its nodule and rhizosphere and unveiling its potential as an isolation source of diazotrophic PGP strains. The findings showed that the soil was calcareous and nutrient depleted. The 16S rRNA gene analysis showed marked differences among the nodule and rhizosphere microbial populations. The most prevalent phylum in the rhizosphere (63%) was Actinobacteriota, whereas Pseudomonadota dominated the nodule one (90%). The Streptomyces genus was widely distributed in the rhizospheres (39%), although Stenotrophomonas was the dominant genus (55%) in the nodules. Due to the absence of the Frankia genus, the association of R. ulmifolius with actinorhizal plants was not possible. In the nodules and rhizospheres, the predicted gene copies implicated in the nitrogen fixation were similar. Although denitrification was more prevalent in the rhizosphere, the nitrate reduction was higher in the nodules. The nodules and rhizospheres yielded nine diazotrophs with PGP traits. The most interesting isolate was further identified as P. agglomerans N2A.
The occurrence R. ulmifolius as part of the shrubland towards the woodland is linked to the unmanaged cropping history. This occurrence is common in Italian abandoned olive groves and has been already described for the Tyrrhenian district of Central Italy [53]. Even if Rubus is reported as thriving in nitrophilic-type habitats, the association with nitrogen-fixing bacteria and the presence of pseudo-nodules participated in the colonization of this plant in a such a nitrogen-poor habitat. The plants associated microbes have an active role in the plant growth promotion, enhancing the plant’s ecological fitness under harsh environmental conditions [54,55]. Plant-associated microbes are divided into rhizosphere microorganisms (which reside in the soil near the roots), epiphytic bacteria (which colonize the phyllosphere), and endophytic microbes (which live inside plant tissue) [56,57,58]. Endophytic bacteria play a crucial role in plants’ physiological and phytosanitary status [59,60]. In our case, root nodules and microbiota studies suggested a particular plant-microbe interaction and the involvement of diazotrophs in their formation. However, the absence of the Frankia genus in the nodule samples suggests that Rubus ulmifolius should not be described as an actinorhizal plant. Root nodules result from the host plant’s selection of compatible nitrogen-fixing soil bacteria, with the subsequent proliferation and induction of the nodule’s formation [61]. The presence of nodules in non-leguminous plants is described for several species, including Rubus spp. [62]. The pigments found within the inner tissues of the nodule could be linked to something akin to a respiratory pigment that is mutualistically produced by the plant to enhance the plant–diazotrophs interaction, the globins. Based on the literature reports and considering the stressful conditions of the sampling area, this pigment could be non-symbiotic hemoglobin (NsHb). NsHb is present in some non-leguminous plants, with an over-expression in stressful conditions [63].
The 16S rRNA metabarcoding revealed an abundant and diverse microbial community of nodules, mainly represented by Pseudomonadota. This phylum is frequent in the roots and the rhizosphere and includes several diazotrophs with the ability to create root nodule symbiosis [64]. Within Pseudomonadota, the most abundant genus was Stenotrophomonas, followed by Pseudomonas. These genera are present in a wide range of environments and play a key role in the nitrogen cycle [65,66]. Additionally, worthy of attention was the presence of Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, including genera relevant to a nitrogen fixation and nodule formation [67]. Pseudomonadota, Bacillota, and Actinobacteria are the most prevalent phyla discovered in bacterial plant endophytes [68]. Others are less frequently recorded as endophytes, including Acidobacteria, Bacteroidetes, Planctomycetes, and Verrucomicrobia [59]. Within these phyla, the most widespread endosymbionts belong to Pseudomonas, Bacillus, Burkholderia, Stenotrophomonas, Micrococcus, Pantoea, Microbacterium, Enterobacter, Azospirillum, and Serratia [68].
The rhizosphere bacterial community was different from the nodule ones, consisting mainly of Actinobacteriota belonging to the genus Streptomyces. Streptomyces, together with Micromonospora, is the most common genus of soil actinomycetes accounting for up to 20% of culturable bacteria [69]. This genus colonizes the rhizosphere and the rhizoplane and can also be found as an endophyte [70]. Streptomyces is fundamental in the breakdown of the soil organic matter and participates in nutrient cycling [69]. Its complex life cycle is the result of multiple pioneering efforts with multicellular transformations and evolution [71]. These characteristics allow actinomycetes to survive in harsh conditions (e.g., alkaline soils) [72]. Streptomyces and Nocardia are well represented in rhizosphere soils, comprising about 95% of soil actinobacterial microbiomes [57,73]. The other relevant genera were Chitinophaga, Glycomyces, and Promicromonospora, genera usually described in rhizospheres with degradation abilities towards a wide variety of carbohydrates, chitin, and lignocellulose biomasses [74,75,76].
The prediction of the metagenome function showed a huge difference among the ECs, KOs, and PWYs of nodule and rhizosphere. The most interesting results were discovered with the study of the KOs involved in the nitrogen cycle. The absence of the KOs involved in the nitrification process (microbial-induced conversion of NH4+ into NO3−) is due to the ability of the plant and rhizosphere microbiota to inhibit this process, especially in N-limited ecosystems [77]. This finding is also supported by the KOs involved in the denitrification processes (NO3− to N2) and assimilatory and dissimilatory nitrate reduction present in both plant districts [78].
In recent years, there has been a growing interest in developing biological tools for agriculture to reduce our dependence on chemical fertilizers. Microbial inoculants with plant growth-promoting properties are a promising field of study with several ongoing applications [79]. To fully exploit the potential of a microbial stimulation and overcome the limitations, more field research is needed to find species and strains with characteristics deemed useful in agriculture [80]. Bacteria are among the pioneering organisms that colonize challenging environments by adapting their metabolisms [81]. Therefore, the isolating of bacteria from difficult habitats could represent a valid strategy to obtain valid strains useful in agriculture [82]. The PGP traits of the isolates showed a good potential for agricultural applications, particularly the P. agglomerans N2A strain. Several P. agglomerans isolates have already been isolated from the plant rhizosphere and described as diazotrophs and PGPR [83,84,85,86,87].
The results we obtained with the analysis of the PGP traits on the isolated strains lead us to think they can be used as inoculants in agriculture. Nitrogen fixation, IAA, phosphate solubilization, ACC deaminase, NH3, and HCN: these properties are the most studied and reliable PGP traits for an effective indirect and direct plant growth promotion [28]. Hydrocyanic acid induces plant resistance by altering the cytochrome oxidase pathway and exerting deleterious impacts on pathogens [88]. Ammonia appears to be linked to the root and shoot elongation as well as to the increase in the plant biomass. Furthermore, it contributes to the decomposition of organic materials, increasing the phytoparasite tolerance and plant growth [89]. Indole-3-acetic acid is a compound that enhances the plant development by increasing the cell division, root elongation, and root hair development through several metabolic pathways [90]. It is produced by several PGPR and has a variety of benefits, including promoting the germination of seeds, seedling length, and dry matter. Several studies indicate that moderate amounts of IAA are necessary for the development of primary roots and that PGP activity is present in bacteria that can secrete amounts of indole compounds higher than 13.5 g mL−1 [91]. The use of Ca3(PO4)2 is crucial to produce phytohormones and, consequently, indirectly enhances plant growth [43]. Another microbial mechanism of mineral phosphate solubilization in the rhizosphere involved in the formation of low molecular weight organic acids, which chelate cations to bind phosphates through their hydroxyl and carboxyl groups. ACC deaminase as a PGP trait is useful for promoting the regulation of ethylene levels, thus protecting plants from stressful conditions [92]. Several studies have confirmed that PGPR with ACC deaminase activity stimulates plant growth and resilience toward a pathogens attack, with an important role in biocontrol activity [93].
Future studies should further investigate the colonization abilities of the isolates, the interactions with plants, and the suitability of these PGP strains for use as biostimulating agents. The traits described suggest a potential application in agriculture. However, it is mandatory to further study their N2-fixing abilities in association with different plant species and in different pedoclimatic conditions. Moreover, to obtain successful bacteria-based formulations, the transfer of knowledge from the laboratory to the field will be crucial, addressing technical, regulatory, and marketing challenges [80].
5. Conclusions
To the best of our knowledge, we have described for the first time the microbiota of Rubus ulmifolius. As soil-dwelling microbial communities and their interactions are largely still unknown, research in this area is important in increasing our knowledge of the environment. These studies could pave the way for new bacterial traits, genes, and metabolites useful in sustainable biotechnologies. The presence of nodules and these specific PGPR suggests a pedoclimatic-driven pressure, mainly represented by the soil type. A nutrient deficiency and alkaline conditions promoted a beneficial bacterial recruitment from plants, inducing nodule formation and rhizosphere microbiota shaping. Future research may investigate the plant-diazotrophs association, nodule pigment, and nitrogen-fixing gene expression. Future research should also examine the ability of isolates to colonize new environments, their interactions with plants, and their eligibility for use as biostimulating agents. In addition, transferring knowledge from the laboratory to the field will be essential to create viable bacterial-based formulations and address technical, regulatory, and commercial issues.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/soilsystems6040096/s1, Table S1: Amplicon Sequence Variants (ASVs) relative abundances (%) at the phylum level (no cut-off); Table S2: Amplicon Sequence Variants (ASVs) relative abundances (%) at the genus level (cut-off 0.5%); Table S3: Biochemical traits of the Gram-positive strains with N2-fixing ability. The green color indicates positive results, while the grey color negative ones; Table S4: Biochemical traits of the Gram-negative strains with N2-fixing ability. The green color indicates positive results, while the grey color negative ones; Figure S1: Correlation plot on pathways predicted by PICRUSt 2 software (nodule y axis and rhizosphere x axis). Correlation coefficient: 0.73.
Author Contributions
Conceptualization, M.P. and M.D.G.; methodology, L.P.; software, M.P.; validation, M.P., R.D. and L.P.; formal analysis, B.F. and A.M.; investigation, B.F. and A.M.; resources, M.D.G.; data curation, M.P. and R.D.; writing—original draft preparation, B.F., A.M. and R.D.; writing—review and editing, M.D.G. and M.P.; visualization, L.P.; supervision, R.D. and M.P.; project administration, M.P.; funding acquisition, M.P. and M.D.G. Both B.F. and A.M. contributed equally and have the right to list their name first in the CV. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. The nucleotide sequences of the V3-V4 16S rRNA gene region have been deposited in the NCBI database repository, BioProject: PRJNA907239 (http://www.ncbi.nlm.nih.gov/bioproject/907239, accessed on 24 November 2021).
Acknowledgments
The authors wish to thank Alessandro Pietrosanti and Amedeo Mignini for the VITEK2 identifications (Gruppo INI, Italy).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schmidt, M.W.I.; Torn, M.S.; Abiven, S.; Dittmar, T.; Guggenberger, G.; Janssens, I.A.; Kleber, M.; Kögel-Knabner, I.; Lehmann, J.; Manning, D.A.C.; et al. Persistence of Soil Organic Matter as an Ecosystem Property. Nature 2011, 478, 49–56. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a Bacterial World, a New Imperative for the Life Sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef]
- Schmidt, C. Living in a Microbial World. Nat. Biotechnol. 2017, 35, 401–403. [Google Scholar] [CrossRef]
- Raynaud, X.; Nunan, N. Spatial Ecology of Bacteria at the Microscale in Soil. PLoS ONE 2014, 9, e87217. [Google Scholar] [CrossRef]
- Torsvik, V.; Goksøyr, J.; Daae, F.L. High Diversity in DNA of Soil Bacteria. Appl. Environ. Microbiol. 1990, 56, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Roesch, L.F.W.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.O.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing Enumerates and Contrasts Soil Microbial Diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.C.; Leff, J.W.; Lauber, C.L.; Fierer, N. Cell Size Distributions of Soil Bacterial and Archaeal Taxa. Appl. Environ. Microbiol. 2013, 79, 7610–7617. [Google Scholar] [CrossRef] [PubMed]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A New View of the Tree of Life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- van der Heijden, M.G.; de Bruin, S.; Luckerhoff, L.; van Logtestijn, R.S.; Schlaeppi, K. A Widespread Plant-Fungal-Bacterial Symbiosis Promotes Plant Biodiversity, Plant Nutrition and Seedling Recruitment. ISME J. 2016, 10, 389–399. [Google Scholar] [CrossRef]
- Dasgupta, D.; Brahmaprakash, G.P. Soil Microbes Are Shaped by Soil Physico-Chemical Properties: A Brief Review of Existing Literature. Int. J. Plant Soil Sci. 2021, 33, 59–71. [Google Scholar] [CrossRef]
- Marschner, P.; Crowley, D.; Yang, C.H. Development of Specific Rhizosphere Bacterial Communities in Relation to Plant Species, Nutrition and Soil Type. Plant Soil 2004, 261, 199–208. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, Resilience, and Redundancy in Microbial Communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X.; et al. Long-Term Nitrogen Fertilization Decreases Bacterial Diversity and Favors the Growth of Actinobacteria and Proteobacteria in Agro-Ecosystems across the Globe. Glob. Change Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef]
- Figueiredo, M.D.V.B.; Bonifacio, A.; Rodrigues, A.C.; de Araujo, F.F. Plant Growth-Promoting Rhizobacteria: Key Mechanisms of Action. In Microbial-Mediated Induced Systemic Resistance in Plants; Choudhary, D.K., Varma, A., Eds.; Springer: Singapore, 2016; pp. 23–37. [Google Scholar]
- Munir, N.; Hanif, M.; Abideen, Z.; Sohail, M.; El-Keblawy, A.; Radicetti, E.; Mancinelli, R.; Haider, G. Mechanisms and Strategies of Plant Microbiome Interactions to Mitigate Abiotic Stresses. Agronomy 2022, 12, 2069. [Google Scholar] [CrossRef]
- Cheng, Y.T.; Zhang, L.; He, S.Y. Plant-Microbe Interactions Facing Environmental Challenge. Cell Host Microbe 2019, 26, 183–192. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M.P. Plant Growth-Promoting Rhizobacteria (PGPR): Their Potential as Antagonists and Biocontrol Agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Pellegrini, M.; Pagnani, G.; Bernardi, M.; Mattedi, A.; Spera, D.M.; Gallo, M. del Cell-Free Supernatants of Plant Growth-Promoting Bacteria: A Review of Their Use as Biostimulant and Microbial Biocontrol Agents in Sustainable Agriculture. Sustainability 2020, 12, 9917. [Google Scholar] [CrossRef]
- Igiehon, N.; Babalola, O. Rhizosphere Microbiome Modulators: Contributions of Nitrogen Fixing Bacteria towards Sustainable Agriculture. Int. J. Environ. Res. Public Health 2018, 15, 574. [Google Scholar] [CrossRef]
- O’Callaghan, M.; Ballard, R.A.; Wright, D. Soil Microbial Inoculants for Sustainable Agriculture: Limitations and Opportunities. Soil Use Manag. 2022, 38, 1340–1369. [Google Scholar] [CrossRef]
- Sessitsch, A.; Pfaffenbichler, N.; Mitter, B. Microbiome Applications from Lab to Field: Facing Complexity. Trends Plant Sci. 2019, 24, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Swensen, S.M.; Mullin, B.C. Phylogenetic relationships among actinorhizal plants. The impact of molecular systematics and implications for the evolution of actinorhizal symbioses. Physiol. Plant 1997, 99, 565–573. [Google Scholar] [CrossRef]
- Kohlen, W.; Ng, J.L.P.; Deinum, E.E.; Mathesius, U. Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Bot. 2018, 69, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ratet, P.; Magne, K. Nodule diversity, evolution, organogenesis and identity. In Advances in Botanical Research; Frendo, P., Frugier, F., Masson-Boivin, C., Eds.; Academic Press: London, UK; San Diego, CA, USA; Waltham, MA, USA; Oxford, UK, 2020; Volume 94, pp. 119–148. [Google Scholar]
- Wall, L.G. The Actinorhizal Symbiosis. J. Plant Growth Regul. 2000, 19, 167–182. [Google Scholar] [CrossRef]
- Zahran, H.H. Rhizobium-Legume Symbiosis and Nitrogen Fixation under Severe Conditions and in an Arid Climate. Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef]
- Bond, G. The results of the IBP survey of root-nodule formation in non-leguminous angiosperms. In Symbiotic Nitrogen Fixation in Plants; Nutman, P.S., Ed.; Cambridge University Press: Cambridge, UK, 1976; pp. 443–474. [Google Scholar]
- Becking, J.H. Identification of the endophypte of Dryas and Rubus (Rosaceae). In Frankia Symbioses; Akkermans, A.D.L., Baker, D., Huss-Danell, K., Tjepkema, J.D., Eds.; Springer: Dordrecht, The Netherlands, 1984; pp. 105–128. [Google Scholar]
- Bond, G. Taxonomy and distribution of non-legume nitrogen-fixing systems. In Biological Nitrogen Fixation in Forest Ecosystems: Foundations and Applications; Gordon, J.C., Wheeler, C.T., Eds.; Springer: Dordrecht, The Netherlands, 1983; pp. 55–87. [Google Scholar]
- Ministero Delle Politiche Agricole Alimentari e Forestali Approvazione Dei “Metodi Ufficiali Di Analisi Chimica Del Suolo”. Gazz. Uff. Della Repubb. Ital. 1999, 1–222.
- Mantoni, C.; Pellegrini, M.; Dapporto, L.; del Gallo, M.; Pace, L.; Silveri, D.; Fattorini, S. Comparison of Soil Biology Quality in Organically and Conventionally Managed Agro-Ecosystems Using Microarthropods. Agriculture 2021, 11, 1022. [Google Scholar] [CrossRef]
- Farda, B.; Djebaili, R.; del Gallo, M.; Ercole, C.; Bellatreccia, F.; Pellegrini, M. The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria. Biology 2022, 11, 1204. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Das, B.K.; Ishii, S.; Antony, L.; Smart, A.J.; Scaria, J.; Brözel, V.S. The Microbial Nitrogen Cycling, Bacterial Community Composition, and Functional Potential in a Natural Grassland Are Stable from Breaking Dormancy to Being Dormant Again. Microorganisms 2022, 10, 923. [Google Scholar] [CrossRef]
- Donate-Correa, J.; León-Barrios, M.; Pérez-Galdona, R. Screening for Plant Growth-Promoting Rhizobacteria in Chamaecytisus Proliferus (Tagasaste), a Forage Tree-Shrub Legume Endemic to the Canary Islands. Plant Soil. 2005, 266, 261–272. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology a Laboratory Manual; The Benjamin Cummings Publishing Co. Inc.: San Francisco, CA, USA, 1996. [Google Scholar]
- Djebaili, R.; Pellegrini, M.; Smati, M.; del Gallo, M.; Kitouni, M. Actinomycete Strains Isolated from Saline Soils: Plant-Growth-Promoting Traits and Inoculation Effects on Solanum Lycopersicum. Sustainability 2020, 12, 4617. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric Estimation of Indoleacetic Acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Fatmawati, U.; Meryandini, A.; Nawangsih, A.A.; Wahyudi, A.T. Screening and Characterization of Actinomycetes Isolated from Soybean Rhizosphere for Promoting Plant Growth. Biodiversitas 2019, 20. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Hafsa, C.S.; Allaoua, S.; Mostefa, G.; Bilal, Y.; Fouzia, A. Solubilization of Phosphate by the Bacillus under Salt Stress and in the Presence of Osmoprotectant Compounds. Afr. J. Microbiol. Res. 2013, 7, 4562–4571. [Google Scholar] [CrossRef]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis Part 2 Chemical and Microbiological Properties; Page, A.L., Ed.; American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Brígido, C.; Duan, J.; Glick, B.R. Methods to Study 1-Aminocyclopropane-1-Carboxylate (ACC) Deaminase in Plant Growth-Promoting Bacteria. In Handbook for Azospirillum; Springer International Publishing: Cham, Switzerland, 2015; pp. 287–305. [Google Scholar]
- Dworkin, M.; Foster, J.W. Experiments with Some Microorganisms which Utilize Ethane and Hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, R., Allison, J.B., Eds.; Elsevier: Amsterdam, The Netherlands, 1969; pp. 21–132. [Google Scholar]
- Neath, A.A.; Cavanaugh, J.E. The Bayesian Information Criterion: Background, Derivation, and Applications. WIREs Comput. Stat. 2012, 4, 199–203. [Google Scholar] [CrossRef]
- Blasi, C.; Di Pietro, R.; Fortini, P. A phytosociological analysis of abandoned terraced olive grove shrublands in the Tyrrhenian district of Central Italy. Plant Biosyst.-Int. J. Deal All Asp. Plant Biol. 2000, 134, 305–331. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant-Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef]
- Santos, L.F.; Olivares, F.L. Plant microbiome structure and benefits for sustainable agriculture. Curr. Plant Biol. 2021, 26, 100198. [Google Scholar] [CrossRef]
- Suman, A.; Yadav, A.N.; Verma, P. Endophytic Microbes in Crops: Diversity and Beneficial Impact for Sustainable Agriculture. In Microbial Inoculants in Sustainable Agricultural Productivity; Springer: New Delhi, India, 2016; pp. 117–143. [Google Scholar]
- Yadav, A.N.; Verma, P.; Kumar, S.; Kumar, V.; Kumar, M.; Kumari Sugitha, T.C.; Singh, B.P.; Saxena, A.K.; Dhaliwal, H.S. Actinobacteria from Rhizosphere. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 13–41. [Google Scholar]
- Zhou, D.; Huang, X.-F.; Chaparro, J.M.; Badri, D.V.; Manter, D.K.; Vivanco, J.M.; Guo, J. Root and Bacterial Secretions Regulate the Interaction between Plants and PGPR Leading to Distinct Plant Growth Promotion Effects. Plant Soil 2016, 401, 259–272. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Latha, P.; Karthikeyan, M.; Rajeswari, E. Endophytic Bacteria: Prospects and Applications for the Plant Disease Management. In Plant Health Under Biotic Stress; Springer: Singapore, 2019; pp. 1–50. [Google Scholar]
- Agrios, G.N. Plant Diseases Caused by Prokaryotes: Bacteria and Mollicutes. In Plant Pathology; Elsevier: Amsterdam, The Netherlands, 2005; pp. 615–703. [Google Scholar]
- Becking, J.H. N2-FIxing Tropical Non-Iegumes. In Microbiology of Tropical Soils and Plant Productivity; Dommerguesand, Y.R., Diem, H.G., Eds.; Martinus Niihoff/Dr. W. Junk Publishers: The Hague, The Netherlands; Boston, MA, USA; London, UK, 1982; pp. 110–146. [Google Scholar]
- Garrocho-Villegas, V.; Gopalasubramaniam, S.K.; Arredondo-Peter, R. Plant Hemoglobins: What We Know Six Decades after Their Discovery. Gene 2007, 398, 78–85. [Google Scholar] [CrossRef]
- Zgadzaj, R.; Garrido-Oter, R.; Jensen, D.B.; Koprivova, A.; Schulze-Lefert, P.; Radutoiu, S. Root Nodule Symbiosis in Lotus Japonicus Drives the Establishment of Distinctive Rhizosphere, Root, and Nodule Bacterial Communities. Proc. Natl. Acad. Sci. USA 2016, 113, E7996–E8005. [Google Scholar] [CrossRef]
- Bergan, T. Human- and Animal-Pathogenic Members of the Genus Pseudomonas. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 1981; pp. 666–700. [Google Scholar]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; van der Lelie, D.; Dow, J.M. The Versatility and Adaptation of Bacteria from the Genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Kuykendall, L.D.; Dazzo, F.B. Allorhizobium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- Oukala, N.; Aissat, K.; Pastor, V. Bacterial Endophytes: The Hidden Actor in Plant Immune Responses against Biotic Stress. Plants 2021, 10, 1012. [Google Scholar] [CrossRef] [PubMed]
- Olanrewaju, O.S.; Babalola, O.O. Streptomyces: Implications and Interactions in Plant Growth Promotion. Appl. Microbiol. Biotechnol. 2019, 103, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Sousa, J.A.D.J.; Olivares, F.L. Plant Growth Promotion by Streptomycetes: Ecophysiology, Mechanisms and Applications. Chem. Biol. Technol. Agric. 2016, 3, 24. [Google Scholar] [CrossRef]
- Jones, S.E.; Elliot, M.A. Streptomyces Exploration: Competition, Volatile Communication and New Bacterial Behaviours. Trends Microbiol. 2017, 25, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Farda, B.; Djebaili, R.; Vaccarelli, I.; del Gallo, M.; Pellegrini, M. Actinomycetes from Caves: An Overview of Their Diversity, Biotechnological Properties, and Insights for Their Use in Soil Environments. Microorganisms 2022, 10, 453. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Thomas, L.; Ram, H.; Kumar, A.; Singh, V.P. Production, Optimization, and Characterization of Organic Solvent Tolerant Cellulases from a Lignocellulosic Waste-Degrading Actinobacterium, Promicromonospora Sp. VP111. Appl. Biochem. Biotechnol. 2016, 179, 863–879. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Y.; Li, J.; Zhu, P.; Liang, B. Exploring Dynamics and Associations of Dominant Lignocellulose Degraders in Tomato Stalk Composting. J. Environ. Manag. 2021, 294, 113162. [Google Scholar] [CrossRef]
- McKee, L.S.; Martínez-Abad, A.; Ruthes, A.C.; Vilaplana, F.; Brumer, H. Focused Metabolism of β-Glucans by the Soil Bacteroidetes Species Chitinophaga Pinensis. Appl. Environ. Microbiol. 2019, 85, e02231-18. [Google Scholar] [CrossRef]
- Nardi, P.; Laanbroek, H.J.; Nicol, G.W.; Renella, G.; Cardinale, M.; Pietramellara, G.; Weckwerth, W.; Trinchera, A.; Ghatak, A.; Nannipieri, P. Biological Nitrification Inhibition in the Rhizosphere: Determining Interactions and Impact on Microbially Mediated Processes and Potential Applications. FEMS Microbiol. Rev. 2020, 44, 874–908. [Google Scholar] [CrossRef] [PubMed]
- Shapleigh, J.P. Dissimilatory and Assimilatory Nitrate Reduction. In The Purple Phototrophic Bacteria; Hunter, C.N., Daldal, F., Thurnauer, M.C., Beatty, J.T., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 623–642. [Google Scholar]
- Cherif-Silini, H.; Silini, A.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Bouremani, N.; Nowakowska, J.A.; Oszako, T.; Belbahri, L. Tailoring Next Generation Plant Growth Promoting Microorganisms as Versatile Tools beyond Soil Desalinization: A Road Map towards Field Application. Sustainability 2021, 13, 4422. [Google Scholar] [CrossRef]
- Massa, F.; Defez, R.; Bianco, C. Exploitation of Plant Growth Promoting Bacteria for Sustainable Agriculture: Hierarchical Approach to Link Laboratory and Field Experiments. Microorganisms 2022, 10, 865. [Google Scholar] [CrossRef] [PubMed]
- Borin, S.; Ventura, S.; Tambone, F.; Mapelli, F.; Schubotz, F.; Brusetti, L.; Scaglia, B.; D’Acqui, L.P.; Solheim, B.; Turicchia, S.; et al. Rock Weathering Creates Oases of Life in a High Arctic Desert. Environ. Microbiol. 2010, 12, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Gaete, A.; Mandakovic, D.; González, M. Isolation and Identification of Soil Bacteria from Extreme Environments of Chile and Their Plant Beneficial Characteristics. Microorganisms 2020, 8, 1213. [Google Scholar] [CrossRef]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum Wheat Stress Tolerance Induced by Endophyte Pantoea Agglomerans with Genes Contributing to Plant Functions and Secondary Metabolite Arsenal. Int. J. Mol. Sci. 2019, 20, 3989. [Google Scholar] [CrossRef]
- Asis, C.A.; Adachi, K. Isolation of Endophytic Diazotroph Pantoea Agglomerans and Nondiazotroph Enterobacter Asburiae from Sweetpotato Stem in Japan. Lett. Appl. Microbiol. 2004, 38, 19–23. [Google Scholar] [CrossRef]
- Quecine, M.C.; Araújo, W.L.; Rossetto, P.B.; Ferreira, A.; Tsui, S.; Lacava, P.T.; Mondin, M.; Azevedo, J.L.; Pizzirani-Kleiner, A.A. Sugarcane Growth Promotion by the Endophytic Bacterium Pantoea Agglomerans 33.1. Appl. Environ. Microbiol. 2012, 78, 7511–7518. [Google Scholar] [CrossRef]
- Mishra, A.; Chauhan, P.S.; Chaudhry, V.; Tripathi, M.; Nautiyal, C.S. Rhizosphere Competent Pantoea Agglomerans Enhances Maize (Zea Mays) and Chickpea (Cicer Arietinum L.) Growth, without Altering the Rhizosphere Functional Diversity. Antonie Van Leeuwenhoek 2011, 100, 405–413. [Google Scholar] [CrossRef]
- Feng, Y.; Shen, D.; Song, W. Rice Endophyte Pantoea Agglomerans YS19 Promotes Host Plant Growth and Affects Allocations of Host Photosynthates. J. Appl. Microbiol. 2006, 100, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Hammerbacher, A.; Coutinho, T.A.; Gershenzon, J. Roles of Plant Volatiles in Defence against Microbial Pathogens and Microbial Exploitation of Volatiles. Plant Cell Environ. 2019, 42, 2827–2843. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.S.; Gomes, C.B.; Souza Júnior, I.T.; Moura, A.B. Bacterial Selection for Biological Control of Plant Disease: Criterion Determination and Validation. Braz. J. Microbiol. 2017, 48, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Barazani, O.; Friedman, J. Is IAA the Major Root Growth Factor Secreted from Plant-Growth-Mediating Bacteria? J. Chem. Ecol. 1999, 25, 2397–2406. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas Putida Indoleacetic Acid in Development of the Host Plant Root System. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef]
- Glick, B.R.; Penrose, D.M.; Li, J. A Model for the Lowering of Plant Ethylene Concentrations by Plant Growth-Promoting Bacteria. J. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC Deaminase Producing PGPR from Rice Rhizosphere and Evaluating Their Plant Growth Promoting Activity under Salt Stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).