Abstract
Humus forms are the morphological results of organic matter decay and distribution in the topsoil, and thus important indicators for decomposer activities in forest ecosystems. The first aim was to examine if humus forms are suitable indicators of microbiological properties of the topsoil in a high mountain forest (Val di Rabbi, Trentino, Italian Alps). The second aim was to predict microbiological parameters based on the topsoil pH value on two slopes of the study area (ca. 1200–2200 m a.s.l.). We investigated humus forms and determined pH values and microbiological parameters (enzymatic activities, carbon/nitrogen (C/N) ratio and the ratio of bacterial/archaeal abundance) of the uppermost mineral horizon. The results reveal significant correlations between pH value and microbiological parameters (except for bacterial/archaeal abundance), which enable upscaling to the landscape scale using linear models. Based on a random forest with kriging of model residuals, predictive maps of humus form, pH value and microbiological parameters show that decomposition processes in our study area correspond with the topography. As compared to locations on south-facing slopes or close to the valley bottom, locations on north-facing slopes or close to the upper treeline exhibit Moder (scarcely Mull or Amphimull), more acidic topsoil (around pH 4), a lower activity of leucine-aminopeptidase, a lower ratio of alkaline/acid phosphomonoesterase activity and a higher soil C/N ratio (above 20). Our results suggest a high potential of humus forms to indicate soil microbiological properties in a high mountain forest. Together with the pH values of the topsoil, humus forms proved to be a useful tool as a basis for predictive maps of leucine-aminopeptidase activity, ratio of alkaline/acid phosphomonoesterase activity and C/N ratio of the mineral topsoil.
1. Introduction
The humus form is an important indicator for decomposition in terrestrial ecosystems [1,2]. Humus forms are the morphological results of different biological activities in the topsoil. They thereby reflect the composition of the decomposer community [3]. Additionally, humus forms are well accepted as integrating indicators for changes in forest ecosystems [4]. Previous studies illustrated a strong correlation between the spatial distribution of enchytraeid species, humus forms and pH values of the topsoil (e.g., [5,6,7]), which has also been shown for high mountain environments [8]. Furthermore, extensive information on forest humus forms in a high mountain area can be used to upscale the spatial distribution of enchytraeid species to the landscape scale [9]. A high soil biological activity as derived from a well-structured mineral topsoil was connected with mull-indicating enchytraeid species at southern slope exposure (in case of sufficient soil moisture), whereas a low soil biological activity indicated by a poorly structured mineral topsoil was connected with moder-indicating enchytraeid species at northern slope exposure [9].
Due to the fact that changes in microbial communities can occur more quickly than remarkable changes in basic soil functions (e.g., filtering pollutants, storing nutrients), the study of microbial parameters is deemed a sensitive indicator when evaluating effects of soil disturbance [10]. In this sense, extracellular enzymes, which are biological catalysts of specific reactions, are considered as sensitive indicators of soil biological processes and soil fertility [11]. Abiotic factors like soil temperature, water potential, pH, substrate availability and complexity, along with biotic processes including enzyme synthesis and secretion, largely influence the activities of enzymes in natural environments [12].
Leucine-aminopeptidase activity has an important role in the nitrogen (N) cycle, as it catalyzes the hydrolysis of leucine and other amino acids from protein or peptide substrates [13]. Bacteria play a relevant role in the production of leucine-aminopeptidases [14] and, accordingly, Bardelli et al. [15] observed a greater activity in south-facing subalpine forest soils where a greater bacterial abundance was recorded in comparison with north-facing slopes.
Phosphorus (P) is taken up by microorganisms and plants largely as orthophosphate in soil solution. Since in many terrestrial ecosystems soil orthophosphate is limiting for plant productivity, the production of extracellular enzymes facilitating the mineralization of organic P compounds is therefore determined by the need for orthophosphate [16]. The phosphomonoesterases include acid and alkaline phosphatases, phytases and nucleotidases [17], and mineralize orthophosphate monoesters such as sugar phosphates, phytate and nucleotides. In particular, acid and alkaline phosphatases are responsible for the mineralization of organic P into phosphate by hydrolyzing phosphoric (mono) ester bonds under acid and alkaline conditions, respectively. They have an important role for P cycling in forest ecosystems, particularly where P availability may limit plant productivity [18]; and an increase in their activity can occur when P is limited, reflecting a demand for this macronutrient [16].
The soil carbon/nitrogen (C/N) ratio is among the most important properties of soil organic matter (SOM) and serves as a reliable proxy of the decomposition rate of SOM. The higher the ratio, the lower the decomposition rate, and as such, the soil C/N ratio can be considered as an estimator of microbial activity and overall as a proxy for soil quality and soil ecological conditions [15,19]. Soil microorganisms use carbon and nitrogen for metabolism, with a C/N ratio of about 20:1 favoring SOM decomposition. The C/N ratio is influenced by a multitude of site-related factors (e.g., [20]).
Soil microorganisms (bacteria, archaea and fungi) and their complex interplay are crucial in biogeochemical cycling in (forest) soils, especially in carbon (C) and nitrogen (N) cycling, as principal drivers of mineral weathering, decomposition/mineralization of organic matter (OM) and pedogenesis. Understanding the ecological role of microbial community dynamics for nutrient cycling is essential for understanding the functional stability of ecosystems and for predicting future scenarios due to changing environmental conditions [21]. Although bacteria and fungi are considered the primary decomposers of SOM, archaea have gained increasing interest as ecological indicators due to their adaptability to harsh environmental conditions (e.g., low temperatures, low pH, nutrient deficiency) and their role within the N cycle, owing to their ammonia oxidizing potential. In fact, ammonia oxidation, the first and rate-limiting step of nitrification, was only recently attributed also to archaea, thanks to the discovery of homologue ammonia monooxygenase (amo) genes [22,23,24]. The comparative assessment of bacteria and archaea provides important descriptive information about soil quality [25], as they can compete for the same nutrients [26] and due to different pH dependencies of fungi, bacteria and archaea [27].
In order to understand spatial decomposition patterns in a high mountain environment, it would be useful if humus forms could be applied as indicators not only of the distribution of enchytraeid species [9,28], but also of microbiological parameters [29]. Hence, the aims of this study were (i) to map humus forms and topsoil pH values in a mountain forest area in the Italian Alps as a basis for upscaling, (ii) to examine correlations between humus forms, pH values and microbiological parameters of the topsoil, (iii) to rank the examined parameters in terms of their usability for upscaling by proxy of humus forms and topsoil pH values, (iv) to upscale microbiological parameters utilizing humus forms and topsoil pH values if feasible according to (iii). In this study, upscaling refers to the extrapolation of microbiological data from the level of a study site to the slope scale using information on the spatial distribution of humus forms and pH values. The following microbiological parameters of the topsoil were addressed in this study: (1) the leucine-aminopeptidase activity as indicator of N cycle processes, (2) the ratio of alkaline/acid phosphomonoesterase activity as indicator of P cycle processes, (3) the C/N ratio as indicator of both C cycle and N cycle processes, (4) the ratio of bacterial/archaeal abundance as indicator of C cycle and N cycle processes, especially ammonia oxidation.
2. Materials and Methods
2.1. Study Area
The study area is located in the northeastern Italian Alpine valley Val di Rabbi in the Autonomous Province of Trento (Figure 1). With reference to geology, the study area is part of the Central Alps, characterized by siliceous bedrock. The dominant parent materials are acidic paragneiss, mica schists, phyllites and orthogneiss [30,31]. The climate of the study area is governed by the high relief intensity of the Alpine environment. Climatic variations are generally due to differences in elevation and slope exposure, with the local climatic situation being more complex [32]. The mean annual air temperature in the study area ranges from about 2 °C close to the tree line to 7 °C at the lowest sites near the valley bottom. The mean annual precipitation is about 800–850 mm [33]. The main soil classes in the study area are Haplic Cambisols (Dystric) and Umbric Podzols below ca. 1900 m a.s.l., whereas Entic Podzols, Albic Podzols and Umbric Podzols are dominant in the forest above ([34], according to [35]).
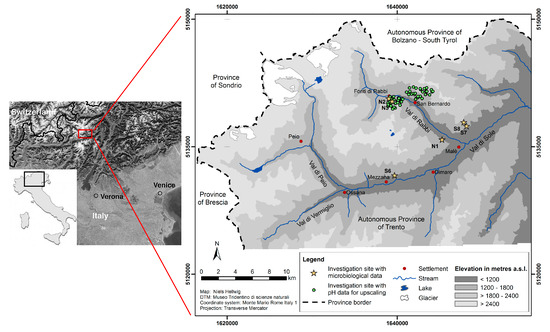
Figure 1.
Study area and investigation sites in the Italian Alps (Autonomous Province of Trento) (left map: modified from Egli et al. [36]).
Upscaling of local data refers to the forested parts of one north-facing slope (~1200–2100 m a.s.l., approx. 2.5 km2) and one south-facing slope (~1200–2200 m a.s.l., approx. 3.8 km2). The main tree species on these slopes are European larch (Larix decidua) and Norway spruce (Picea abies), both of them similarly prevalent at both slopes. Regarding the lower tree layer, Norway spruce is by far the dominant species, whereas young European larch trees only occur above 1800 m a.s.l. [37].
2.2. Sampling
For this study, we considered two sampling sets: the first set comprised 60 sampling sites for humus form determination and topsoil acidity analysis (RN1–RN30 and RS1–RS30); the second set comprised six additional sampling sites for extensive microbiological analyses (N1–N3 and S6–S8) (Table 1).

Table 1.
Soil properties, related investigation sites and sampled objects.
Sampling of humus forms was carried out at a total of 60 sampling sites in Val di Rabbi. Among these sites, 30 of them were located on one north-facing (RN1–RN30) and 30 on one neighboring south-facing (RS1–RS30) slope, respectively (Figure 1). These sites were determined based on conditioned Latin Hypercube Sampling (cLHS) [38]. The application of cLHS allowed to obtain a set of sampling sites most closely representing the investigated slopes in terms of the covariates elevation, slope gradient, slope exposure, slope curvature (planform, profile and general curvature), SAGA wetness index, LS factor (slope length and steepness) and forest type (details are given in [37]). Each of these sites comprised an area of 25 m × 25 m. The number of sampling plots corresponded to the number of ground cover types (one sample per available ground cover type: the ground cover types were litter, consisting of tree litter, mostly needles; grass; moss; fern; shrubs). For all of the ground cover types, percentages of their spatial distributions within the area of the site were estimated. At each sampling plot, a humus form profile was described and a sample for topsoil acidity analysis (two replicates) was collected from the uppermost mineral horizon directly beneath the organic layers. All analyses were carried out at these sites in September 2015.
For microbiological analyses, six additional sampling sites were chosen on the basis of local knowledge from experts involved in previous soil ecological studies (Figure 1). They are located at three different elevations (~1200, 1400 and 1630 m a.s.l.) inside the closed forest, each half of them on north-facing (N1–N3) and south-facing (S6–S8) slopes, respectively. Further environmental characteristics of these sites have been specified elsewhere [15,39]. Soil sampling at these sites was comprised of three replicates with five subsamples each (for quantitative real-time PCR, the subsamples were pooled). A corer (diameter 5 cm) was used to sample three soil depths separately, including organic layers at the surface: 0–5 cm, 5–10 cm, 10–15 cm. Sampling was carried out at these sites in August 2012. All samples were bulked and sieved (<2 mm), aliquoted into 50-mL sterile conical centrifuge tubes, and stored at −20 °C. Apart from topsoil acidity, the analyses of these samples covered total C and N contents, soil enzymatic activities, abundance of bacteria and archaea.
2.3. Soil Analysis
Soil properties determined for the first sampling set (RN1–RN30 and RS1–RS30) comprised humus forms and topsoil acidity; the second sampling set (N1–N3 and S6–S8) was analyzed with respect to topsoil acidity, soil enzymatic activities, total C and N, bacterial and archaeal abundance (Table 1).
2.3.1. Humus Forms
Humus form profiles (width 50–100 cm) were described and classified according to [40]. In addition to the set of humus forms specified therein, the humus form Amphimull (AMU) was used whenever organic layers (OF and OH) existed above a well-structured soil in the uppermost mineral horizon (Figure 2). Amphimull is a humus form usually not present under Central European climatic conditions, except for some mountain areas [41].
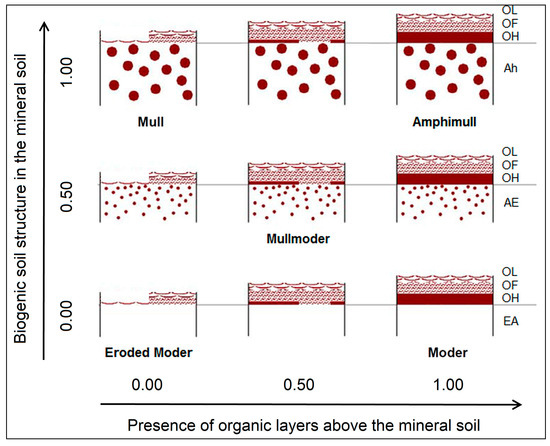
Figure 2.
Humus form classes and parameter values for modeling (only names of described humus forms are given) (modified from Graefe [41]).
2.3.2. Topsoil Acidity
The pH values were determined in soil water extracts (1:10 m/v) from air-dried samples.
2.3.3. Soil Enzymatic Activities
A heteromolecular exchange principle [42] was used to desorb enzymes from soil in order to determine the leucine-aminopeptidase, acid and alkaline phosphomonoesterase potential activities, as described by Bardelli et al. [15]. For the disruption of soil aggregates and microbial cells the procedure involved the use of a 3% solution of lysozyme as desorbant and a bead-beating agent followed by centrifugation at 20,000 g for 5 min. Afterwards, the supernatant containing desorbed enzymes was dispensed into 384-well microplates together with the appropriate buffer and the fluorometric quantification of the enzymes activities was done using 4-methyl-umbelliferyl (MUF) substrate. All the measurements were done in duplicate and the activities were expressed as nanomoles of MUF g−1 dry soil h−1.
2.3.4. Total C and N
Soil samples were homogenized with a mortar prior to analysis. A CN analyzer (TruSpec CHN, LECO, Sao Jose dos Campos, MI, USA) was used to determine total C and N contents in oven-dried soil samples. The temperature used for CN analysis was 950 °C. Due to the siliceous bedrock throughout our study area, total C only includes organic C.
2.3.5. Quantitative Real-Time PCR
The DNA extraction and purification from soil samples (0.5 g, fresh weight) was done using a commercial kit (FastDNA Kit for Soil, MP-Biomedicals) as described in [43]. The Rotorgene 6000 Real Time Thermal Cycler (Corbett Research, Sydney, Australia) was used in combination with the Rotor-Gene Series Software 1.7 in order to quantify the 16S rRNA gene copy number of bacteria and archaea with the primer pairs 1055f/1392r (bacteria, [44]) and Parch519f/Arc915r (archaea, [45]). Standard curves for quantification of both microbial domains were constructed as described by Bardelli et al. [15]. The reaction mix for each qPCR run was performed by using the 1X Sensimix™ SYBR® Hi-rox (Bioline, Taunton, MA, USA) based on the DNA-intercalating dye SYBR Green I as shown in [15]. Each run was accompanied by a melting analysis starting from 60 °C to 95 °C with temperature increments of 0.25 °C and a transition rate of 5 s to check for product specificity and potential primer dimer formation. The purity of the amplified products was also checked by the presence of a single band of the expected length on a 1% agarose gel stained with the DNA stain Midori Green (Nippon Genetics, Duere, Germany) and visualized by UV-transillumination (Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany). The efficiency for the bacterial and archaeal qPCR runs was in a range between 80% and 85%, indicating a good reproducibility.
2.4. Spatial Modeling
Humus forms were modeled in terms of two dimensions: (1) biogenic soil structure in the mineral soil and (2) presence of organic layers above the mineral soil. Relative units (values from 0 to 1) were used for both dimensions according to the determined humus forms (Figure 2, Table 2). At each site, the related samples were aggregated using weights according to the estimated relative distribution of different ground cover types. For example, at a site with Amphimull (AMU) under grass (70% occurrence) and Typischer Moder (MOA) under litter (30% occurrence), the dimension biogenic soil structure in the mineral soil amounts to 0.7 × 1.0 + 0.3 × 0.0 = 0.7 and the presence of organic layers above the mineral soil amounts to 0.7 × 1.0 + 0.3 × 1.0 = 1.0.

Table 2.
Two-dimensional characterization of humus forms for modeling. At each site, the relative distribution of different ground cover types provided weights, which were used to spatially aggregate the related samples.
Spatial modeling consisted of a random forest [46] and ordinary kriging of the model residuals [47]. An approach combining a random forest and residuals kriging has been successfully applied in earlier digital soil mapping studies [48]. During the last years, random forest has been established as one of the most powerful approaches for spatial modeling in the context of predictive mapping in ecology and soil science [49,50,51,52]. In comparison with single-tree-based models, the use of a random forest avoids overfitting tendencies, yet includes the concurrent influences of a large set of environmental variables. In this study, environmental variables included topographic and vegetation parameters. Terrain attributes were derived from a digital terrain model with a grid width of 1 m (Provincia Autonoma di Trento, Ufficio Sistemi Informativi-Servizio autorizzazioni e valutazioni ambientali, LiDAR data from 2006 to 2008, available at http://dati.trentino.it/dataset/lidar-rilievo-2006-2007-2008-link-al-servizio-di-download). These attributes included elevation, slope, slope exposure, general curvature, profile and planform curvature (all slope parameters derived according to [53]) and LS factor (following [54]). Vegetation characteristics included forest type and forest density (obtained from Provincia Autonoma di Trento, Servizio Foreste e Fauna). The random forest models of the humus form parameters and the pH value thereby accounted for the highly heterogeneous conditions of relief and vegetation in the study area. In all random forest models, the number of trees amounted to 10,000 and the terminal nodes had a minimum size of 3 elements. Spatial modeling was performed with the statistical software R [55] and the R package randomForest [56].
Linear models were used to quantify the relationships between the pH value and microbiological parameters. These models were based on the data from the sites N1–N3 and S6–S8. For each sample from these sites, data was taken from that soil depth at which the uppermost mineral horizon was found (Tables S1 and S2). Those linear models with a highly significant correlation (p < 0.01) were used to derive spatial models of the microbiological parameters from the model of topsoil acidity.
2.5. Model Assessment
The random forest models (used for humus form parameters and topsoil acidity) were evaluated by the mean value of the squared model residuals and the explained variance of the model [56]. The predicted values of the model refer to the out-of-bag samples, respectively, i.e., the set of trees where a sample does not belong to the data used for model training.
The linear models underlying the upscaling procedure of microbiological parameters were evaluated with the standard errors of the predictions. Maps of the standard errors were generated to reveal the spatially variable precision of the predicted values.
In order to assess the transferability of the upscaling results from pH values to humus forms, the relationships between the predictions of humus form parameter values on the one hand and pH values, enzyme activities and the soil C/N ratio on the other hand were examined using linear regression analyses.
3. Results
The results from sampling of humus forms and topsoil acidity at the sites RN1–RN30 and RS1–RS30 show a distinct dominance of moder humus forms (particularly in the higher parts of the study area). Mullmoder and mull humus forms occur especially at south-facing sites and Amphimull can be found only below 1600 m a.s.l. The pH values in the uppermost mineral soil horizon range from 4 (at site RN27) to 6 (at site RN1). The data basis for the spatial modeling of humus forms (as characterized by the biogenic soil structure in the mineral soil and the presence of organic layers above the mineral soil) and of topsoil acidity is shown in Table 3 (raw data is presented in Table S3).

Table 3.
Input data for modeling from sampling of humus forms and topsoil acidity.
Chemical and microbiological analyses at the sites N1–N3 and S6–S8 show distinct variations of the investigated parameters with soil depth (especially for the ratio of bacterial/archaeal abundance and the enzymatic activities) (Table 4). At these sites, the range of pH values is comparable to the other 60 sites (from 4.2 to 6.0). In addition, the ratio of alkaline/acid phosphomonoesterase activity shows an increasing trend from northern exposure and high elevation to southern exposure and low elevation. The C/N ratio increases from south-facing to north-facing sites (Table 4).

Table 4.
Chemical and microbiological properties of the soils collected at the six study sites at north-and south-facing areas (N1–N3 and S6–S8, respectively). The results are shown pairwise, i.e., the couples of north- and south-facing sites at the same elevation (N1–S6; N2–S7; N3–S8). Values are means (n = 3) ± standard deviations. Data are expressed on a dry weight basis. At all sites, soil C only includes organic C (Corg). Nitrogen values used for calculation of the soil C/N ratio refer to total nitrogen (Nt).
Maps of humus form parameters (presence of organic layers above the mineral soil and biogenic soil structure in the mineral soil) were obtained from spatial modeling. According to the predictions, organic layers are present almost throughout the whole north-facing slope (values close to 1), whereas there is a relatively heterogeneous pattern at the south-facing slope (Figure 3a). The predictions of the biogenic soil structure in the uppermost mineral horizon embrace a distinct decreasing trend with elevation at the north-facing slope. This holds partially true for the south-facing slope, where the predicted percentage of biogenic soil structure is generally higher as compared to the north-facing slope (Figure 3b).
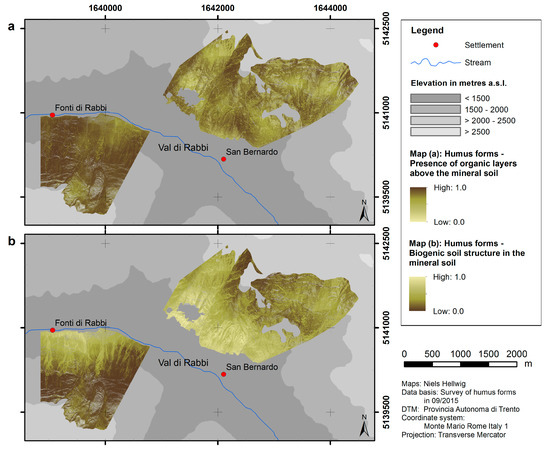
Figure 3.
Predicted distribution of the two modeled humus form dimensions: (a) presence of organic layers above the mineral soil; (b) biogenic soil structure in the mineral soil.
The predicted distribution of pH values of the A horizon is presented in Figure 4. Apart from considerable local patterns of variability, these predictions show a general trend of decreasing topsoil acidity from high to low elevation and from northern to southern slope exposure.
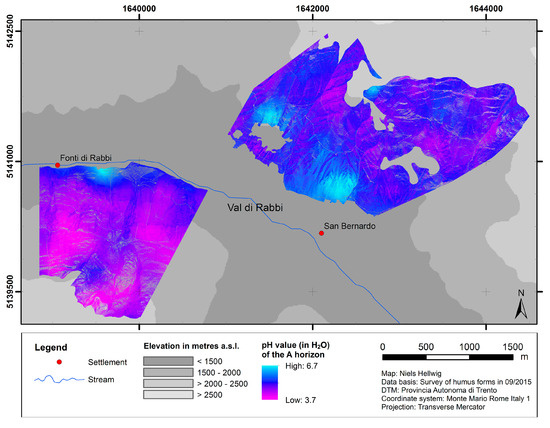
Figure 4.
Predicted distribution of topsoil acidity.
The results of the submodels (random forests and kriging procedures) are shown in Figures S1–S6. Mean values of the squared residuals and explained variances of the underlying random forest models are shown in Table 5.

Table 5.
Quality measures of the random forest models.
The relationships between the pH value and microbiological parameters as quantified by linear models are summarized in Table 6. On the basis of the P values of the linear models, the leucine-aminopeptidase activity, the ratio of alkaline/acid phosphomonoesterase activity and the C/N ratio are rated as usable parameters for upscaling, whereas the ratio of bacterial/archaeal abundance is rated as not usable.

Table 6.
Results of linear modeling of microbiological parameters and the soil C/N ratio as a function of the pH value (n = number of observations).
The results of upscaling of the enzyme activities as well as the C/N ratio are presented as predictive maps (Figure 5). Because of the linear relationship to the topsoil pH value, the patterns concur with the predicted distribution of pH values (Figure 4). The predicted activity of leucine-aminopeptidase in the uppermost mineral horizon ranges from 22.0 to 310.4 nmol MUF g−1 dry soil h−1; the ratio of alkaline/acid phosphomonoesterase has values up to 1.0 (where the activity of alkaline phosphomonoesterase is predicted to equal the activity of acid phosphomonoesterase). Within our study area, both parameters are generally predicted to be lower at northern slope exposure and high elevation as compared to southern slope exposure and low elevation. The predicted values of the C/N ratio range from 16 at south-facing sites with low elevation to 23 at north-facing sites.
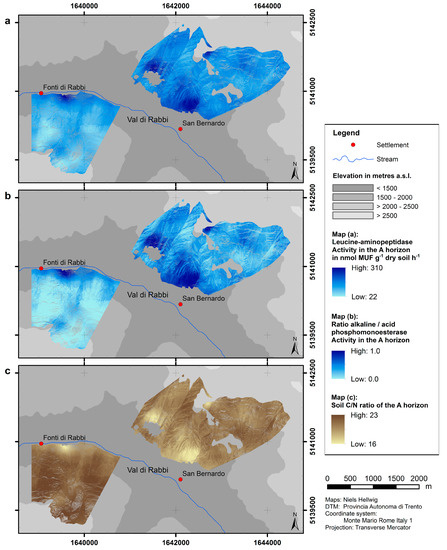
Figure 5.
Predicted distributions of microbiological parameters.
Figure 6 shows the standard error of the predictions from upscaling using the example of leucine-aminopeptidase (see Figures S7 and S8 for alkaline/acid phosphomonoesterase and C/N ratio). The predictions from linear modeling tend to be more imprecise at sites with predicted values below ca. pH 4.2 and above ca. pH 6.0 than at sites with intermediate pH values.
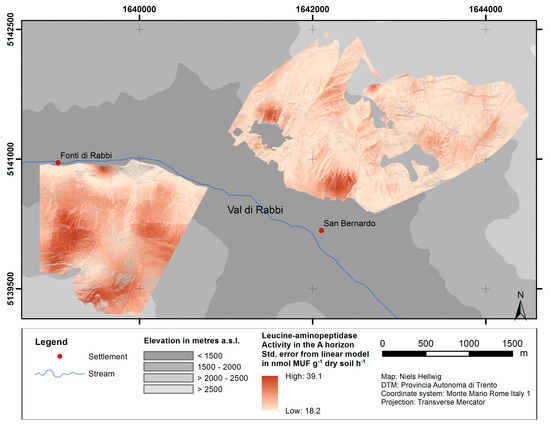
Figure 6.
Standard error of predictions from linear model of leucine-aminopeptidase activity as a function of the pH value.
The linear regression models used for the analysis of the transferability of upscaling results from the pH value to humus forms revealed moderate, but highly significant relationships between the predictions of humus form dimensions (biogenic soil structure in the mineral soil and presence of organic layers above the mineral soil) on the one hand and the predicted pH values, enzyme activities and soil C/N ratio on the other hand (Table 7).

Table 7.
Results of linear regression analysis between predictions for humus forms (x1 = presence of organic layers above the mineral soil, x2 = biogenic soil structure in the mineral soil) (Figure 3) and predictions of topsoil acidity and microbiological parameters (Figure 4 and Figure 5) (n = 5,891,366, number of predicted pixels).
4. Discussion
4.1. Spatial Modeling of Humus Forms and Topsoil Acidity
The spatial models illustrate that humus forms, as well as pH values of the A horizon, in our study area are arranged in patterns corresponding to topographical features. The distributions of both humus form dimensions (presence of organic layers above the mineral soil and biogenic soil structure in the mineral soil) are related to slope exposure. Nevertheless, spatial patterns of both humus form dimensions are relatively dissimilar (Figure 3). This implies that effects of topographic characteristics on the biological activity in the mineral soil are different from those on the formation of organic layers. For example, the percentage of biogenic soil structure in the mineral soil distinctly decreases within the forested area from low to high elevation (which indicates a decline of burrowing and mixing activities of soil organisms), whereas the presence of organic layers does not show discernible variations in terms of elevation. It would be interesting to analyze in more detail if the climatic conditions in our study area have a higher effect on the biogenic soil structure in the mineral soil as compared to the presence of organic layers. The results generally emphasize the dominance of moder-like humus forms on the north-exposed slope, especially at high elevation. Mull-like humus forms, on the contrary, mainly occur at the south-exposed slope, especially at low elevation. However, the correlations of the topography with the biogenic soil structure and with the presence of organic layers are different. Therefore, the humus forms Amphimull and Eroded Moder are also common in our study area.
The distribution of pH values of the A horizon likewise relates to the topographical situation, which influences the microclimatic conditions and the kind of litter: low pH values (around 4.0–4.5) are clearly dominant on the north-exposed slope (except for the very lowest part). In contrast, on the south-exposed slope there is a trend of relatively high pH values (around 6.5) decreasing with elevational gain to lower average values (around 4.5–5.0). Consequently, the analogous distributions of humus forms and pH values generally turn out as expected [6]: the presence of mull-like humus forms characterized by a high biological activity in the mineral soil coincides with higher pH values as compared to moder-like humus forms. Altogether, these findings at the slope scale confirm and amplify the trends from modeling at broader scales [9,57].
Regarding the quality of the presented models, the mean values of the squared residuals from the random forest models are low in relation to the total ranges of values. Nevertheless, the random forest models show relatively low explained variances (about 18–37%). Therefore, we addressed the local variability in the data (which the random forest models were not able to cover) by means of a kriging procedure of the model residuals (Figures S2, S4 and S6). This way, the deviations of the observed values from those values predicted by the random forest model (based on out-of-bag samples) could be included in the final predictions. Therefore, the quality of the presented models is altogether rated as high. However, the model results only account for the effects of topographic variation and spatial forest patterns (random forest models) together with deviations given by the kriging of the model residuals. This implies that the models do not reflect possible effects of variation of siliceous bedrock (as geological conditions are similar throughout the study area). Furthermore, the models are not representative of local peculiarities of un-sampled locations within the study area (e.g., totally different percentages of ground cover types or local disturbances).
Until now, only a few studies have considered spatial modeling of humus forms or organic horizons in high mountain areas. Aberegg et al. [30] used a classification tree approach to model the distribution of mull, moder and mor humus forms. In a more recent study, Hellwig et al. [57] introduced an approach based on decision tree analysis and fuzzy logic for modeling humus forms. This approach was specially designed for highly heterogeneous areas in the high mountains, where only a low amount of sample data is available due to poor accessibility. Recent implementations of this model addressed the presence of OH horizons [9,57]. The humus form model presented in this study also involves the biological activity in the mineral soil, thus accounting for the properties of five different humus form classes (Figure 2).
The lack of studies on spatial modeling of humus forms and other soil parameters related to organic matter decomposition contradicts the benefits of spatial models as compared to conventional mapping approaches. Soil analysis in the field is often time-consuming and costly. This is especially true for high mountain areas, where the spatial variation of soil properties is complex due to highly heterogeneous environmental conditions. In this context, digital soil mapping techniques are valuable tools to utilize and combine extensive information on soil-forming factors, for example from remote sensing [58].
4.2. Upscaling of Microbiological Parameters
Topsoil pH values appear to be useful for upscaling three out of the four parameters investigated in this study: leucine-aminopeptidase activity, ratio of alkaline/acid phosphomonoesterase activity and C/N ratio. The significant correlations that we found between pH values, C/N ratio and enzyme activities correspond to the study of Leifeld and von Lützow [59], who found that microbial soil organic matter decomposition depended on pH and substrate C/N ratio rather than on inherent chemical substrate properties. As linear models depending on pH value were used for upscaling (Table 6), the predicted spatial patterns of enzyme activities and C/N ratio are arranged in the same spatial patterns as the pattern of pH values (Figure 5). The significant relationships that we found between predicted humus form dimensions on the one hand and predicted pH values, enzyme activities and soil C/N ratio on the other hand suggest that also humus forms are a suitable indicator of microbiological processes related to organic matter decomposition.
Regarding leucine-aminopeptidase activity, the positive relationship with topsoil pH values aligns with Sinsabaugh et al. [60], who found an increase of this activity with increasing soil pH levels (pH 4 to 8.5). Furthermore, the slope exposure had a significant impact on leucine-aminopeptidase activity, even though such exposure-effect was elevation-dependent and a higher activity was registered at south than at north exposure only at 1400 m a.s.l. This suggests that at this specific elevation this enzymatic activity might have been more sensitive to the differences in soil temperature between both slopes, bearing in mind that also in alpine soils temperature is an important factor in the regulation of soil N mineralization [61]. In addition, the type of vegetation could also have influenced the potential activity of the leucine-aminopeptidase, since the proportion of easily available monomers and polymers that enter the soil varies greatly depending on plant community composition as pointed out by Sanaullah et al. [62].
Similar to the activity of leucine-aminopeptidase, the ratio of alkaline/acid phosphomonoesterase activity shows a positive relationship with topsoil pH values. This implies that the activity of alkaline phosphomonoesterase increases as compared to the activity of acid phosphomonoesterase under less acidic conditions, which is consistent with earlier studies [63,64]. The ratio of alkaline/acid phosphomonoesterase activity has also previously been reported as being closely related to the soil pH value [65]. Our results confirm this relationship for high mountain forests of the Central Alps.
The soil C/N ratio of the A horizon is significantly, positively correlated with the pH value. Similarly to the results of this study, other studies from mountain areas have shown an increasing C/N ratio of the topsoil with elevation depending on the kind of litter [66,67,68]. Moreover, Cools et al. [20] found a distinct relationship between the topsoil C/N ratio and the humus form.
The linear models used to quantify the relationships between the pH value and the microbiological parameters are all highly significant, but differ in the explained variation as indicated by the coefficient of determination (R²) (Table 6). Accordingly, a ranking of the parameters can be deduced with regard to the quality of the predictions and to the usability for upscaling by proxy of humus forms and topsoil pH values. The ratio of alkaline/acid phosphomonoesterase activity is ranked first; the C/N ratio follows in the second position; the leucine-aminopeptidase activity takes the third position.
Additionally, standard errors of the predictions from the linear models were determined to assess the accuracy of the predicted values in respect of its spatial variability. Although the pattern of the standard errors corresponds with the independent variable (pH value), the maps indicate those areas, where predictions are less accurate (mostly at the north-facing slope). Furthermore, they provide quantitative accuracy values, which allow for the consideration of the magnitude of deviations when using predicted values.
Contrary to the aforementioned parameters, upscaling based on pH values is infeasible for the ratio of bacterial/archaeal abundance (P value ca. 0.22). Prosser and Nicol [23] suggested ammonia limitation, mixotrophy, and pH as the main factors providing niche specialization and differentiation between soil ammonia oxidizing archaea (AOA) and ammonia oxidizing bacteria (AOB) in a wide range of soil types. In a study of forest soils along an elevation gradient in South Tyrol (Italian Alps), Siles and Margesin [69] found that bacterial and fungal diversity properties and community structures were highly correlated to topsoil pH values. However, archaeal, bacterial and fungal abundances were not significantly related to pH values [69]. Corresponding to these findings, the results of our study do not identify a significant relationship between the pH values and the ratio of bacterial/archaeal abundance in the topsoil of a high mountain forest. This might be due to the complex interactions and ecological functions of soil microorganisms or caused by the range of low pH values, in particular at the north-facing slope. Especially for undisturbed forest soils, our knowledge of its autochthonous microorganisms—even in the era of high throughput molecular ecology—remains limited [18,70].
Upscaling of microbiological parameters is based on two sampling sets and two steps of modeling (random forest model together with kriging of the residuals and linear regression analysis). Therefore, the model results are subject to potential uncertainty due to limitations both of the sample data and of the models. The sampling set used for microbiological analyses comprises considerably less investigation sites than the sampling set used for the determination of humus forms. Hence, that sampling set is less representative in terms of factors such as elevation. In addition, those samples for microbiological analyses partly were collected at sites within a few kilometers distance from the slopes where humus forms were described (Figure 1). With respect to upscaling, the spatial patterns of the pH value are used to derive predictions of the microbiological parameters with a linear model. The model evaluation revealed that this is feasible for all parameters except for the ratio of bacterial/archaeal abundance. Nevertheless, the actual relationship remains uncertain (whether linear or not) and other factors potentially relevant for microbiological properties of the mineral soil (e.g., soil C content, depth of the A horizon) are not part of these models.
4.3. Soil Ecological Implications
In mountain ecosystems, topography causes a high spatial variability of microclimatic conditions and slope dynamics. The results of this study emphasize the considerable effects of topography on decomposition processes as expressed by the distribution of humus forms, topsoil pH values and microbiological parameters. Bojko and Kabala [71] established a significant relationship between soil organic carbon pools and humus forms of mountain soils. In that regard, our results confirm the findings from earlier studies that demonstrated the pivotal role of topography (alongside with factors such as vegetation and climate) for the formation of soil organic carbon patterns in high mountain areas [72,73,74]. Spatial variation of organic matter decomposition has been reported to be engendered by differences of soil temperature [75,76,77,78], soil moisture [79,80,81,82], litter quality and quantity [83,84,85], slope processes [86] and seasonality. These factors are likely to be strongly affected by the topographic diversity in our study area, thus they potentially govern also spatial differences that we found in the models of this study.
Furthermore, the results of this study are in line with the patterns of humus forms and soil pH values described in previous studies of mountain forests. Egli et al. [87] investigated mountain soils also within the area of this study and found a higher percentage of weakly degraded organic matter as well as higher soil organic carbon concentrations at northern slope exposure as compared to southern slope exposure. Other studies described patterns similar to this study regarding humus forms, pH values and soil C/N ratio along an elevation gradient (e.g., [66,88]).
However, random forest models alone are not sufficient to predict the spatial distribution of humus forms and pH values. Most of the variance in humus forms and pH values is not explained by those models depending on climatic, topographic and vegetation influences. This insight points to the importance of additional, currently undiscovered mechanisms determining decomposition processes. These may include rather local effects of decomposer organisms [89,90], caused by e.g., litter affinity of decomposer organisms [91,92], temporal shifts of decomposer communities during decomposition [93] and responses to different levels of litter species diversity [81,94].
The results of this study show a strong relationship between topography on the one hand and enzyme activities and the C/N ratio of the topsoil on the other hand (Figure 5). This relationship might be relevant to consider also for projections of future landscape development, as C and N cycling are predicted to be affected by climate change, especially at high-elevation sites [95,96].
When integrated with those of previous studies [8,9,28], our findings show that the humus form proves to be a comprehensive indicator for soil ecological processes in a high mountain environment of the Central Alps, including soil macro- and mesofaunal as well as microbiological properties.
5. Conclusions
Variations of humus forms, pH values and microbiological parameters investigated in this study are arranged in patterns that are related to topography. Although the underlying random forest models explain only between 18% and 37% of the variances of humus form parameters and pH values, the predictive maps reveal distinct patterns especially corresponding to elevation and slope exposure. These patterns are also reflected by the spatial models of microbiological parameters. Unlike the ratio of bacterial/archaeal abundance, all parameters are highly significantly correlated with the pH value. Regarding their usability for upscaling by proxy of humus forms and topsoil pH values, they can be ranked as follows: 1) the ratio of alkaline/acid phosphomonoesterase activity, 2) the C/N ratio, 3) the leucine-aminopeptidase activity.
With this study, we applied concepts from digital soil mapping to the field of soil microbiology. This study illustrates both the capability and the high value of modeling techniques to cope with soil ecological research questions in a spatial context. Thus, we encourage further usage of soil-landscape modeling in the context of soil ecological studies.
Supplementary Materials
The following are available online at http://www.mdpi.com/2571-8789/2/1/12/s1, Figure S1: Results of the random forest model for the biogenic soil structure in the mineral soil, Figure S2: Results of the kriging procedure of the model residuals for the biogenic soil structure in the mineral soil, Figure S3: Results of the random forest model for the presence of organic layers above the mineral soil, Figure S4: Results of the kriging procedure of the model residuals for the presence of organic layers above the mineral soil, Figure S5: Results of the random forest model for the topsoil acidity, Figure S6: Results of the kriging procedure of the model residuals for the topsoil acidity, Figure S7: Standard error of predictions from linear model of the ratio alkaline/acid phosphomonoesterase activity as a function of the pH value, Figure S8: Standard error of predictions from linear model of the soil C/N ratio as a function of the pH value, Table S1: Input data for linear models of soil C/N ratio and enzyme activities depending on pH values, Table S2: Input data for linear model of the ratio bacterial/archaeal abundance depending on pH values, Table S3: Humus forms and pH values sampled at 60 sites in Val di Rabbi.
Acknowledgments
This work was realized in the context of the D.A.CH. project DecAlp and funded by the German Research Foundation (DFG, grant number BR 1106/23-1), the Austrian Science Fund (FWF, I989-B16) and the Swiss National Science Foundation (SNF). The authors thank all colleagues in the project for excellent cooperation. We also thank Dott. Fabio Angeli (Ufficio Distrettuale Forestale di Malè) and the Stelvio National Park for supporting the field work. We are grateful to Alison Simerly for proofreading. María Gómez-Brandón acknowledges support by the Programa Ramón y Cajal (RYC-2016-21231; Ministerio de Economía y Competitividad).
Author Contributions
N.H. performed spatial modeling, model assessment and wrote the paper; N.H. and K.A. performed sampling and soil analysis of the sites RN1–RN30 and RS1–RS30; M.G.-B., J.A.-J. and T.B. performed sampling and soil analysis of the sites N1–N3 and S6–S8 and co-wrote the paper; F.F. and G.P. contributed to soil microbiological analysis; G.B. and H.I. conceived and designed the experiments.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Ponge, J.F. Plant-soil feedbacks mediated by humus forms: A review. Soil Biol. Biochem. 2013, 57, 1048–1060. [Google Scholar] [CrossRef]
- Graefe, U.; Beylich, A. Humus forms as tool for upscaling soil biodiversity data to landscape level? Mitt. Dtsch. Bodenkd. Ges. 2006, 108, 6–7. [Google Scholar]
- Ponge, J.F. Humus forms in terrestrial ecosystems: A framework to biodiversity. Soil Biol. Biochem. 2003, 35, 935–945. [Google Scholar] [CrossRef]
- Moscatelli, M.C.; Bonifacio, E.; Chiti, T.; Cudlin, P.; Dinca, L.; Gömöryova, E.; Grego, S.; La Porta, N.; Karlinski, L.; Pellis, G.; et al. Soil properties as indicators of treeline dynamics in view of anthropogenic pressure and climate change. Clim. Res. 2017, 73, 73–84. [Google Scholar] [CrossRef]
- Graefe, U.; Schmelz, R.M. Indicator values, strategy types and life forms of terrestrial Enchytraeidae and other microannelids. Newsl. Enchytraeidae 1999, 6, 59–67. [Google Scholar]
- Labaz, B.; Galka, B.; Bogacz, A.; Waroszewski, J.; Kabala, C. Factors influencing humus forms and forest litter properties in the mid-mountains under temperate climate of southwestern Poland. Geoderma 2014, 230, 265–273. [Google Scholar] [CrossRef]
- Andreetta, A.; Cecchini, G.; Bonifacio, E.; Cornolli, R.; Vingiani, S.; Carnicelli, S. Tree or soil? Factors influencing humus form differentiation in Italian forests. Geoderma 2016, 264, 195–204. [Google Scholar] [CrossRef]
- Ascher, J.; Sartori, G.; Graefe, U.; Thornton, B.; Ceccherini, M.T.; Pietramellara, G.; Egli, M. Are humus forms, mesofauna and microflora in subalpine forest soils sensitive to thermal conditions? Biol. Fertil. Soils 2012, 48, 709–725. [Google Scholar] [CrossRef]
- Hellwig, N.; Graefe, U.; Tatti, D.; Sartori, G.; Anschlag, K.; Beylich, A.; Gobat, J.M.; Broll, G. Upscaling the spatial distribution of enchytraeids and humus forms in a high mountain environment on the basis of GIS and fuzzy logic. Eur. J. Soil Biol. 2017, 79, 1–13. [Google Scholar] [CrossRef]
- Baldrian, P. Microbial activity and the dynamics of ecosystem processes in forest soils. Curr. Opin. Microbiol. 2017, 37, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Gianfreda, L.; Rao, M.A. Enzymes in Agricultural Sciences; OMICS Group International: California, CA, USA, 2014. [Google Scholar]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Matsui, M.; Fowler, J.H.; Walling, L.L. Leucine aminopeptidases: Diversity in structure and function. Biol. Chem. 2006, 387, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Burke, D.J.; Weintraub, M.N.; Hewins, C.R.; Kalisz, S. Relationship between soil enzyme activities, nutrient cycling and soil fungal communities in a northern hardwood forest. Soil Biol. Biochem. 2011, 43, 795–803. [Google Scholar] [CrossRef]
- Bardelli, T.; Gómez-Brandón, M.; Ascher-Jenull, J.; Fornasier, F.; Arfaioli, P.; Francioli, D.; Egli, M.; Sartori, G.; Insam, H.; Pietramellara, G. Effects of slope exposure on soil physico-chemical and microbiological properties along an altitudinal climosequence in the Italian Alps. Sci. Total Environ. 2017, 575, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Salazar, S.; Sánchez, L.E.; Alvarez, J.; Valverde, A.; Galindo, P.; Igual, J.M.; Peix, A.; Santa-Regina, I. Correlation among soil enzyme activities under different forest ecosystem management practices. Ecol. Eng. 2011, 37, 1123–1131. [Google Scholar] [CrossRef]
- Nannipieri, P.; Giagnoni, L.; Renella, G.; Puglisi, E.; Ceccanti, B.; Masciandaro, G.; Fornasier, F.; Moscatelli, M.C.; Marinari, S. Soil enzymology: Classical and molecular approaches. Biol. Fertil. Soils 2012, 48, 743–762. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Yamakura, T.; Sahunalu, P. Soil Carbon/Nitrogen Ratio as a Site Quality Index for Some South-East Asian Forests. J. Trop. Ecol. 1990, 6, 371–377. [Google Scholar] [CrossRef]
- Cools, N.; Vesterdal, L.; De Vos, B.; Vanguelova, E.; Hansen, K. Tree species is the major factor explaining C:N ratios in European forest soils. For. Ecol. Manag. 2014, 311, 3–16. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E. Environmental Genome Shotgun Sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I.; Nicol, G.W. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Isobe, K.; Ohte, N. Ecological Perspectives on Microbes Involved in N-Cycling. Microbes Environ. 2014, 29, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Sterngren, A.E.; Hallin, S.; Bengtson, P. Archaeal Ammonia Oxidizers Dominate in Numbers, but Bacteria Drive Gross Nitrification in N-amended Grassland Soil. Front. Microbiol. 2015, 6, 1350. [Google Scholar] [CrossRef] [PubMed]
- Bengtson, P.; Barker, J.; Grayston, S.J. Evidence of a strong coupling between root exudation, C and N availability, and stimulated SOM decomposition caused by rhizosphere priming effects. Ecol. Evol. 2012, 2, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Brandón, M.; Ascher-Jenull, J.; Bardelli, T.; Fornasier, F.; Sartori, G.; Pietramellara, G.; Arfaioli, P.; Egli, M.; Beylich, A.; Insam, H.; Graefe, U. Ground cover and slope exposure effects on micro- and mesobiota in forest soils. Ecol. Indic. 2017, 80, 174–185. [Google Scholar] [CrossRef]
- Andreetta, A.; Macci, C.; Giansoldati, V.; Masciandaro, G.; Carnicelli, S. Microbial activity and organic matter composition in Mediterranean humus forms. Geoderma 2013, 209–210, 198–208. [Google Scholar] [CrossRef]
- Aberegg, I.; Egli, M.; Sartori, G.; Purves, R. Modelling spatial distribution of soil types and characteristics in a high Alpine valley (Val di Sole, Trentino, Italy). Stud. Trent. Sci. Nat. 2009, 85, 39–50. [Google Scholar]
- Von Seidlein, C. Petrographie und Struktur des ostalpinen Altkristallins südlich des Ultentales (Trentino, Nord-Italien). Ph.D. Thesis, Ludwig-Maximilians-Universität München, Munich, Germany, March 2000. [Google Scholar]
- Dobrowski, S.Z. A climatic basis for microrefugia: The influence of terrain on climate. Glob. Chang. Biol. 2011, 17, 1022–1035. [Google Scholar] [CrossRef]
- Sboarina, C.; Cescatti, A. Il Clima del Trentino—Distribuzione Spaziale delle Principali Variabili Climatiche, Report Centro Ecologia Alpina 33; Centro di Ecologia Alpina: Sardagna (Trento), Italy, 2004; pp. 1–20. [Google Scholar]
- Sartori, G.; Mancabelli, A. Carta dei suoli del Trentino: Scala 1:250.000. Museo Tridentino di Scienze Naturali di Trento, Centro di Ricerca per l’Agrobiologia e la Pedologia di Firenze: Trento, Italy, 2009. [Google Scholar]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2006; World Soil Resources Report 103; FAO: Rome, Italy, 2006. [Google Scholar]
- Egli, M.; Mirabella, A.; Sartori, G.; Zanelli, R.; Bischof, S. Effect of north and south exposure on weathering rates and clay mineral formation in Alpine soils. Catena 2006, 67, 155–174. [Google Scholar] [CrossRef]
- Anschlag, K.; Tatti, D.; Hellwig, N.; Sartori, G.; Gobat, J.M.; Broll, G. Vegetation-based bioindication of humus forms in coniferous mountain forests. J. Mt. Sci. 2017, 14, 662–673. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B. A conditioned Latin hypercube method for sampling in the presence of ancillary information. Comput. Geosci. 2006, 32, 1378–1388. [Google Scholar] [CrossRef]
- Egli, M.; Hafner, S.; Derungs, C.; Ascher-Jenull, J.; Camin, F.; Sartori, G.; Raab, G.; Bontempo, L. Decomposition and stabilisation of Norway spruce needle-derived material in Alpine soils using a 13C-labelling approach in the field. Biogeochemistry 2016, 131, 321–338. [Google Scholar] [CrossRef]
- Ad-hoc-AG Boden. Bodenkundliche Kartieranleitung, 5th ed.; E. Schweizerbart’sche Verlagsbuchhandlung: Hannover, Germany, 2005; ISBN 978-3-510-95920-4. [Google Scholar]
- Graefe, U. Gibt es in Deutschland die Humusform Amphi? Mitt. Dtsch. Bodenkd. Ges. 2007, 110, 459–460. [Google Scholar]
- Fornasier, F.; Margon, A. Bovine serum albumin and Triton X-100 greatly increase phosphomonoesterases and arylsulphatase extraction yield from soil. Soil Biol. Biochem. 2007, 39, 2682–2684. [Google Scholar] [CrossRef]
- Ascher, J.; Ceccherini, M.T.; Pantani, O.L.; Agnelli, A.; Borgogni, F.; Guerri, G.; Nannipieri, P.; Pietramellara, G. Sequential extraction and genetic fingerprinting of a forest soil metagenome. Appl. Soil Ecol. 2009, 42, 176–181. [Google Scholar] [CrossRef]
- Ferris, M.J.; Muyzer, G.; Ward, D.M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 1996, 62, 340–346. [Google Scholar] [PubMed]
- Coolen, M.J.L.; Hopmans, E.C.; Rijpstra, W.I.C.; Muyzer, G.; Schouten, S.; Volkman, J.K.; Damste, J.S.S. Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: Response of methanogens and methanotrophs to environmental change. Org. Geochem. 2004, 35, 1151–1167. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Heuvelink, G.B.M.; Webster, R. Modelling soil variation: Past, present, and future. Geoderma 2001, 100, 269–301. [Google Scholar] [CrossRef]
- Guo, P.T.; Li, M.F.; Luo, W.; Tang, Q.F.; Liu, Z.W.; Lin, Z.M. Digital mapping of soil organic matter for rubber plantation at regional scale: An application of random forest plus residuals kriging approach. Geoderma 2015, 237, 49–59. [Google Scholar] [CrossRef]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer Classification and Regression Tree Techniques: Bagging and Random Forests for Ecological Prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, J.T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.C.; Lawler, J.J. Random Forests for Classification in Ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Grimm, R.; Behrens, T.; Märker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island – Digital soil mapping using Random Forest analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Heung, B.; Bulmer, C.E.; Schmidt, M.G. Predictive soil parent material mapping at a regional-scale: A Random Forest approach. Geoderma 2014, 214–215, 141–154. [Google Scholar] [CrossRef]
- Zevenbergen, L.W.; Thorne, C.R. Quantitative analysis of land surface topography. Earth Surf. Process. Landf. 1987, 12, 47–56. [Google Scholar] [CrossRef]
- Moore, I.D.; Grayson, R.B.; Ladson, A.R. Digital terrain modelling: A review of hydrological, geomorphological, and biological applications. Hydrol. Process. 1991, 5, 3–30. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.R-project.org/ (accessed on 20 February 2018).
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R. News 2002, 2, 18–22. [Google Scholar]
- Hellwig, N.; Anschlag, K.; Broll, G. A fuzzy logic based method for modeling the spatial distribution of indicators of decomposition in a high mountain environment. Arct. Antarct. Alp. Res. 2016, 48, 623–635. [Google Scholar] [CrossRef]
- Baruck, J.; Nestroy, O.; Sartori, G.; Baize, D.; Traidl, R.; Vrščaj, B.; Bräm, E.; Gruber, F.E.; Heinrich, K.; Geitner, C. Soil classification and mapping in the Alps: The current state and future challenges. Geoderma 2016, 264, 312–331. [Google Scholar] [CrossRef]
- Leifeld, J.; von Lützow, M. Chemical and microbial activation energies of soil organic matter decomposition. Biol. Fertil. Soils 2014, 50, 147–153. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Lauber, C.L.; Weintraub, M.N.; Ahmed, B. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Koch, O.; Tscherko, D.; Kandeler, E. Temperature sensitivity of microbial respiration, nitrogen mineralization, and potential soil enzyme activities in organic alpine soils. Glob. Biogeochem. Cycles 2007, 21. [Google Scholar] [CrossRef]
- Sanaullah, M.; Blagodatskaya, E.; Chabbi, A.; Rumpel, C.; Kuzyakov, Y. Drought effects on microbial biomass and enzyme activities in the rhizosphere of grasses depend on plant composition. Appl. Soil Ecol. 2011, 48, 38–44. [Google Scholar] [CrossRef]
- Eivazi, F.; Tabatabai, M.A. Phosphatases in Soils. Soil Biol. Biochem. 1977, 9, 167–172. [Google Scholar] [CrossRef]
- Juma, N.G.; Tabatabai, M.A. Distribution of Phosphomonoesterases in Soils. Soil Sci. 1978, 126, 101–108. [Google Scholar] [CrossRef]
- Dick, W.A.; Cheng, L.; Wang, P. Soil acid and alkaline phosphatase activity as pH adjustment indicators. Soil Biol. Biochem. 2000, 32, 1915–1919. [Google Scholar] [CrossRef]
- Badía, D.; Ruiz, A.; Girona, A.; Martí, C.; Casanova, J.; Ibarra, P.; Zufiaurre, R. The influence of elevation on soil properties and forest litter in the Siliceous Moncayo Massif, SW Europe. J. Mt. Sci. 2016, 13, 2155–2169. [Google Scholar] [CrossRef]
- Tashi, S.; Singh, B.; Keitel, C.; Adams, M. Soil carbon and nitrogen stocks in forests along an altitudinal gradient in the eastern Himalayas and a meta-analysis of global data. Glob. Chang. Biol. 2016, 22, 2255–2268. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Oelmann, Y.; Schickhoff, U.; Böhner, J.; Scholten, T. Himalayan treeline soil and foliar C:N:P stoichiometry indicate nutrient shortage with elevation. Geoderma 2017, 291, 21–32. [Google Scholar] [CrossRef]
- Siles, J.A.; Margesin, R. Abundance and Diversity of Bacterial, Archaeal, and Fungal Communities along an Altitudinal Gradient in Alpine Forest Soils: What Are the Driving Factors? Microb. Ecol. 2016, 72, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Giagnoni, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2017, 68, 12–26. [Google Scholar] [CrossRef]
- Bojko, O.; Kabala, C. Organic carbon pools in mountain soils—Sources of variability and predicted changes in relation to climate and land use changes. Catena 2017, 149, 209–220. [Google Scholar] [CrossRef]
- Oueslati, I.; Allamano, P.; Bonifacio, E.; Claps, P. Vegetation and Topographic Control on Spatial Variability of Soil Organic Carbon. Pedosphere 2013, 23, 48–58. [Google Scholar] [CrossRef]
- Prietzel, J.; Christophel, D. Organic carbon stocks in forest soils of the German Alps. Geoderma 2014, 221–222, 28–39. [Google Scholar] [CrossRef]
- Chen, L.-F.; He, Z.-B.; Du, J.; Yang, J.-J.; Zhu, X. Patterns and environmental controls of soil organic carbon and total nitrogen in alpine ecosystems of northwestern China. Catena 2016, 137, 37–43. [Google Scholar] [CrossRef]
- Davidson, E.A.; Janssens, I.A. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 2006, 440, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Conant, R.T.; Ryan, M.G.; Ågren, G.I.; Birge, H.E.; Davidson, E.A. Temperature and soil organic matter decomposition rates – synthesis of current knowledge and a way forward. Glob. Chang. Biol. 2011, 17, 3392–3404. [Google Scholar] [CrossRef]
- Von Lützow, M.; Kögel-Knabner, I. Temperature sensitivity of soil organic matter decomposition—what do we know? Biol. Fertil. Soils 2009, 46, 1–15. [Google Scholar] [CrossRef]
- Klimek, B.; Jelonkiewicz, Ł.; Niklińska, M. Drivers of temperature sensitivity of decomposition of soil organic matter along a mountain altitudinal gradient in the Western Carpathians. Ecol. Res. 2016, 31, 609–615. [Google Scholar] [CrossRef]
- A’Bear, A.D.; Jones, T.H.; Kandeler, E.; Boddy, L. Interactive effects of temperature and soil moisture on fungal-mediated wood decomposition and extracellular enzyme activity. Soil Biol. Biochem. 2014, 70, 151–158. [Google Scholar] [CrossRef]
- Brockett, B.F.T.; Prescott, C.E.; Grayston, S.J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 2012, 44, 9–20. [Google Scholar] [CrossRef]
- Santonja, M.; Fernandez, C.; Proffit, M.; Gers, C.; Gauquelin, T.; Reiter, I.M.; Cramer, W.; Baldy, V. Plant litter mixture partly mitigates the negative effects of extended drought on soil biota and litter decomposition in a Mediterranean oak forest. J. Ecol. 2017, 105, 801–815. [Google Scholar] [CrossRef]
- García-Palacios, P.; Prieto, I.; Ourcival, J.-M.; Hättenschwiler, S. Disentangling the Litter Quality and Soil Microbial Contribution to Leaf and Fine Root Litter Decomposition Responses to Reduced Rainfall. Ecosyst. 2016, 19, 490–503. [Google Scholar] [CrossRef]
- De Long, J.R.; Dorrepaal, E.; Kardol, P.; Nilsson, M.C.; Teuber, L.M.; Wardle, D.A. Understory plant functional groups and litter species identity are stronger drivers of litter decomposition than warming along a boreal forest post-fire successional gradient. Soil Biol. Biochem. 2016, 98, 159–170. [Google Scholar] [CrossRef]
- Makkonen, M.; Berg, M.P.; Handa, I.T.; Hättenschwiler, S.; van Ruijven, J.; van Bodegom, P.M.; Aerts, R. Highly consistent effects of plant litter identity and functional traits on decomposition across a latitudinal gradient. Ecol. Lett. 2012, 15, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Hättenschwiler, S.; Gasser, P. Soil animals alter plant litter diversity effects on decomposition. Proc. Natl. Acad. Sci. USA 2005, 102, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Berhe, A.A.; Harden, J.W.; Torn, M.S.; Harte, J. Linking soil organic matter dynamics and erosion-induced terrestrial carbon sequestration at different landform positions. J. Geophys. Res. 2008, 113. [Google Scholar] [CrossRef]
- Egli, M.; Sartori, G.; Mirabella, A.; Favilli, F.; Giaccai, D.; Delbos, E. Effect of north and south exposure on organic matter in high Alpine soils. Geoderma 2009, 149, 124–136. [Google Scholar] [CrossRef]
- Bayranvand, M.; Kooch, Y.; Hosseini, S.M.; Alberti, G. Humus forms in relation to altitude and forest type in the Northern mountainous regions of Iran. For. Ecol. Manag. 2017, 385, 78–86. [Google Scholar] [CrossRef]
- Bradford, M.A.; Berg, B.; Maynard, D.S.; Wieder, W.R.; Wood, S.A. Understanding the dominant controls on litter decomposition. J. Ecol. 2016, 104, 229–238. [Google Scholar] [CrossRef]
- Prescott, C.E.; Maynard, D.G.; Laiho, R. Humus in northern forests: Friend or foe? For. Ecol. Manag. 2000, 133, 23–36. [Google Scholar] [CrossRef]
- Austin, A.T.; Vivanco, L.; González-Arzac, A.; Pérez, L.I. There’s no place like home? An exploration of the mechanisms behind plant litter-decomposer affinity in terrestrial ecosystems. New Phytol. 2014, 204, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Milcu, A.; Manning, P. All size classes of soil fauna and litter quality control the acceleration of litter decay in its home environment. Oikos 2011, 120, 1366–1370. [Google Scholar] [CrossRef]
- García-Palacios, P.; Shaw, E.A.; Wall, D.H.; Hättenschwiler, S. Temporal dynamics of biotic and abiotic drivers of litter decomposition. Ecol. Lett. 2016, 19, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, N.N.; Vanhellemont, M.; De Schrijver, A.; Schelfhout, S.; Baeten, L.; Verheyen, K. Mixing effects on litter decomposition rates in a young tree diversity experiment. Acta Oecol. 2016, 70, 79–86. [Google Scholar] [CrossRef]
- Gutiérrez-Girón, A.; Díaz-Pinés, E.; Rubio, A.; Gavilán, R.G. Both altitude and vegetation affect temperature sensitivity of soil organic matter decomposition in Mediterranean high mountain soils. Geoderma 2015, 237–238, 1–8. [Google Scholar] [CrossRef]
- Dawes, M.A.; Schleppi, P.; Hättenschwiler, S.; Rixen, C.; Hagedorn, F. Soil warming opens the nitrogen cycle at the alpine treeline. Glob. Chang. Biol. 2017, 23, 421–434. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).