The Prenatal Microbiome: A New Player for Human Health
Abstract
:1. Introduction
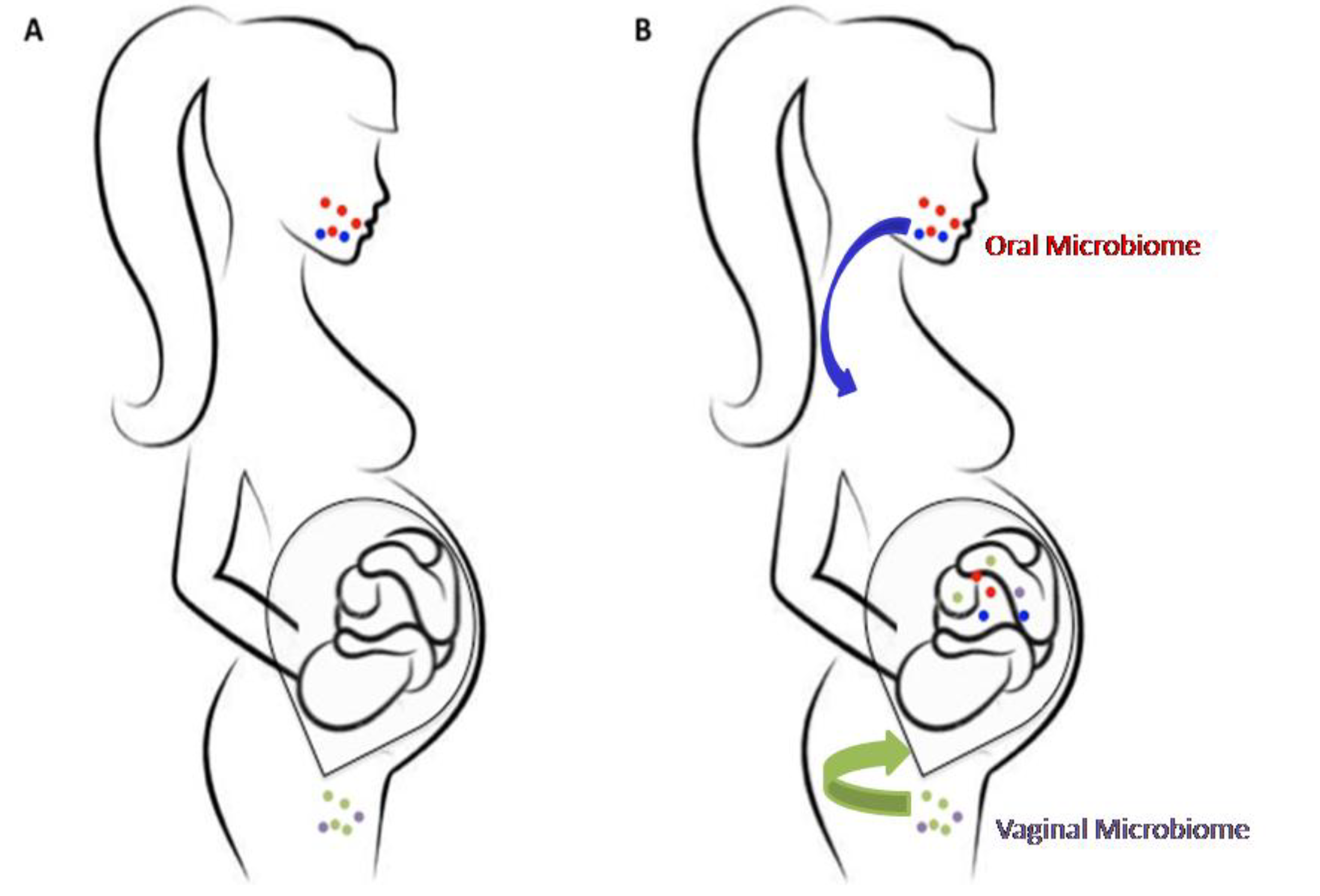
2. The Prenatal Microbiome
3. Correlation between Fetal Microbiome and Newborn’s Health
4. Manipulation of Prenatal Microbiome for Healthy Status Maintenance
5. Conclusions
Author Contributions
Conflicts of Interest
References
- D’Argenio, V.; Salvatore, F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 2015, 451, 97–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Argenio, V. Human Microbiome Acquisition and Bioinformatic Challenges in Metagenomic Studies. Int. J. Mol. Sci. 2018, 19, 383. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Selber-Hnatiw, S.; Rukundo, B.; Ahmadi, M.; Akoubi, H.; Al-Bizri, H.; Aliu, A.F.; Ambeaghen, T.U.; Avetisyan, L.; Bahar, I.; Baird, A.; et al. Human Gut Microbiota: Toward an Ecology of Disease. Front. Microbiol. 2017, 8, 1265. [Google Scholar] [CrossRef] [PubMed]
- Kantarci, A.; Hasturk, H. Microbes and Host Response: A relationship in health and disease. Oral Dis. 2017, 24, 1385–1387. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 8, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Brusaferro, A.; Cavalli, E.; Farinelli, E.; Cozzali, R.; Principi, N.; Esposito, S. Gut dysbiosis and paediatric Crohn’s disease. J. Infect. 2018. [Google Scholar] [CrossRef] [PubMed]
- Imhann, F.; Vich Vila, A.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Del Vecchio Blanco, G.; et al. Metagenomics Reveals Dysbiosis and a Potentially Pathogenic N. flavescens Strain in Duodenum of Adult Celiac Patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Del Vecchio Blanco, G.; et al. No Change in the Mucosal Gut Microbiome is Associated with Celiac Disease-Specific Microbiome Alteration in Adult Patients. Am. J. Gastroenterol. 2016, 111, 1659–1661. [Google Scholar] [CrossRef]
- Wen, C.; Zheng, Z.; Shao, T.; Liu, L.; Xie, Z.; Le Chatelier, E.; He, Z.; Zhong, W.; Fan, Y.; Zhang, L.; et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017, 18, 142. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Torino, M.; Precone, V.; Casaburi, G.; Esposito, M.V.; Iaffaldano, L.; Malapelle, U.; Troncone, G.; Coto, I.; Cavalcanti, P.; et al. The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing. Int. J. Mol. Sci. 2017, 18, 109. [Google Scholar] [CrossRef] [PubMed]
- Iaffaldano, L.; Granata, I.; Pagliuca, C.; Esposito, M.V.; Casaburi, G.; Salerno, G.; Colicchio, R.; Piccirillo, M.; Ciacci, C.; Del Vecchio Blanco, G.; et al. Oropharyngealmicrobiome evaluation highlights Neisseria abundance in active celiac patients. Sci. Rep. 2018, 8, 11047. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.M.; Jin, H.Z. Skin Microbiome: An Actor in the Pathogenesis of Psoriasis. Chin. Med. J. 2018, 131, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Nycz, B.T.; Dominguez, S.R.; Friedman, D.; Hilden, J.M.; Ir, D.; Robertson, C.E.; Frank, D.N. Evaluation of bloodstream infections, Clostridium difficile infections, and gut microbiota in pediatric oncology patients. PLoS ONE 2018, 13, e0191232. [Google Scholar] [CrossRef]
- Rinninella, E.; Mele, M.C.; Merendino, N.; Cintoni, M.; Anselmi, G.; Caporossi, A.; Gasbarrini, A.; Minnella, A.M. The Role of Diet, Micronutrients and the Gut Microbiota in Age-Related Macular Degeneration: New Perspectives from the Gut⁻Retina Axis. Nutrients 2018, 10, 1677. [Google Scholar] [CrossRef]
- D’Ippolito, S.; Di Nicuolo, F.; Pontecorvi, A.; Gratta, M.; Scambia, G.; Di Simone, N. Endometrial microbes and microbiome: Recent insights on the inflammatory and immune “players” of the human endometrium. Am. J. ReprodImmunol. 2018, 80, e13065. [Google Scholar] [CrossRef]
- Precone, V.; Del Monaco, V.; Esposito, M.V.; De Palma, F.D.; Ruocco, A.; Salvatore, F.; D’Argenio, V. Cracking the Code of Human Diseases Using Next-Generation Sequencing: Applications, Challenges, and Perspectives. Biomed. Res. Int. 2015, 2015, 161648. [Google Scholar] [CrossRef]
- D’Argenio, V. The High-Throughput Analyses Era: Are We Ready for the Data Struggle? High-Throughput 2018, 7, 8. [Google Scholar] [CrossRef]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial Changes during Pregnancy, Birth, and Infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef]
- Neu, J. The microbiome during pregnancy and early postnatal life. Semin. Fetal Neonatal Med. 2016, 21, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Aldo, P.; Alvero, A.B. The unique immunological and microbial aspects of pregnancy. Nat. Rev. Immunol. 2017, 17, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, J.M.; Dattilo, A.M. Early development of intestinal microbiota: Implications for future health. Gastroenterol. Clin. N. Am. 2012, 41, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Stinson, L.F.; Payne, M.S.; Keelan, J.A. Planting the seed: Origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit. Rev. Microbiol. 2017, 43, 352–369. [Google Scholar] [CrossRef] [PubMed]
- Koleva, P.T.; Kim, J.S.; Scott, J.A.; Kozyrskyj, A.L. Microbial programming of health and disease starts during fetal life. Birth Defects Res. C Embryo Today 2015, 105, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.; Sakwinska, O.; Soh, S.E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Lee, Y.S.; Yap, F.; Chong, Y.S.; et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 2015, 6. [Google Scholar] [CrossRef]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Kawashima, K.; Nagata, S.; Yamashiro, Y. Gut dysbiosis following C-section instigates higher colonisation of toxigenic Clostridium perfringens in infants. Benef. Microbes 2017, 8, 353–365. [Google Scholar] [CrossRef]
- Escherich, T. Die Darmbakterien des Säuglings; ArbA D Path InstzuMünchen: Stuttgart, Germany, 1886; pp. 1–180. [Google Scholar]
- Perez-Muñoz, M.E.; Arrieta, M.C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Jiménez, E.; Marìn, M.L.; Martìn, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef]
- Mshvildadze, M.; Neu, J.; Shuster, J.; Theriaque, D.; Li, N.; Mai, V. Intestinal microbialecology in premature infants assessed with non-culture-based techniques. J. Pediatr. 2010, 156, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Madan, J.C.; Salari, R.C.; Saxena, D.; Davidson, L.; O’Toole, G.A.; Moore, J.H.; Sogin, M.L.; Foster, J.A.; Edwards, W.H.; Palumbo, P.; et al. Gut microbial colonization in premature neonates predicts neonatal sepsis. Arch. Dis. Child. Fetal Neonatal Ed. 2012, 97, F456–F462. [Google Scholar] [CrossRef] [PubMed]
- Ardissone, A.N.; de la Cruz, D.M.; Davis-Richardson, A.G.; Rechcigl, K.T.; Li, N.; Drew, J.C.; Murgas-Torrazza, R.; Sharma, R.; Hudak, M.L.; Triplett, E.W.; et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE 2014, 9, e90784. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonization may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Nomura, Y.; Bashir, A.; Fernandez-Hernandez, H.; Itzkowitz, S.; Pei, Z.; Stone, J.; Loudon, H.; Peter, I. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE 2013, 8, e78257. [Google Scholar] [CrossRef] [PubMed]
- Moles, L.; Gòmez, M.; Heilig, H.; Bustos, G.; Fuentes, S.; de Vos, W.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 2013, 8, e66986. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Tapiainen, T.; Paalanne, N.; Tejesvi, M.V.; Koivusaari, P.; Korpela, K.; Pokka, T.; Salo, J.; Kaukola, T.; Pirttilä, A.M.; Uhari, M.; et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr. Res. 2018. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Gosalbes, M.J.; Llop, S.; Vallès, Y.; Moya, A.; Ballester, F.; Francino, M.P. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 2013, 43, 198–211. [Google Scholar] [CrossRef]
- Hansen, R.; Scott, K.P.; Khan, S.; Martin, J.C.; Berry, S.H.; Stevenson, M.; Okpapi, A.; Munro, M.J.; Hold, G.L. First-pass meconium samples from healthy term vaginally-delivered neonates: An analysis of the microbiota. PLoS ONE 2015, 10, e0133320. [Google Scholar] [CrossRef] [PubMed]
- Pettker, C.M.; Buhimschi, I.A.; Magloire, L.K.; Sfakianaki, A.K.; Hamar, B.D.; Buhimschi, C.S. Value of placental microbial evaluation in diagnosing intra-amniotic infection. Obstet. Gynecol. 2007, 109, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed]
- Sanschagrin, S.; Yergeau, E. Next-generation sequencing of 6S ribosomal RNA gene amplicons. J. Vis. Exp. 2014, 90, e51709. [Google Scholar] [CrossRef]
- Romero, R.; Espinoza, J.; Gonçalves, L.F.; Kusanovic, J.P.; Friel, L.; Hassan, S. The role of inflammation and infection in preterm birth. Semin. Reprod. Med. 2007, 25, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, R.L.; Andrews, W.W.; Goepfert, A.R.; Faye-Petersen, O.; Cliver, S.P.; Carlo, W.A.; Hauth, J.C. The Alabama preterm birth study: Umbilical cord blood Ureaplasmaurealyticum and Mycoplasma hominis cultures in very preterm newborns. Am. J. Obstet. Gynecol. 2008, 198, 43. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef]
- Steel, J.H.; Malatos, S.; Kennea, N.; Edwards, A.D.; Miles, L.; Duggan, P.; Reynolds, P.R.; Feldman, R.G.; Sullivan, M.H. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr. Res. 2005, 57, 404–411. [Google Scholar] [CrossRef]
- Jones, H.E.; Harris, K.A.; Azizia, M.; Bank, L.; Carpenter, B.; Hartley, J.C.; Klein, N.; Peebles, D. Differing prevalence and diversity of bacterial species in fetal membranes from very preterm and term labor. PLoS ONE 2009, 4, e8205. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Torres, A.; Jones-Carson, J.; Bäumler, A.J.; Falkow, S.; Valdivia, R.; Brown, W.; Le, M.; Berggren, R.; Parks, W.T.; Fang, F.C. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 1999, 401, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, E.; Fernández, L.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodríguez, J.M. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W.; Ikegami, A.; Bissada, N.F.; Herbst, M.; Redline, R.W.; Ashmead, G.G. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J. Clin. Microbiol. 2006, 44, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Fardini, Y.; Chung, P.; Dumm, R.; Joshi, N.; Han, Y.W. Transmission of diverse oral bacteria to murine placenta: Evidence for the oral microbiome as a potential source of intrauterine infection. Infect. Immun. 2010, 78, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Gut microbiota—At the intersection of everything? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.; Nanan, R. Foetal immune programming: Hormones, cytokines, microbes and regulatory T cells. J. Reprod. Immunol. 2014, 104–105, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Romano-Keeler, J.; Weitkamp, J.H. Maternal influences on fetal microbial colonization and immune development. Pediatr. Res. 2015, 77, 189–195. [Google Scholar] [CrossRef]
- Kollmann, T.R.; Levy, O.; Montgomery, R.R.; Goriely, S. Innate immune function by Toll-like receptors: Distinct responses in newborns and the elderly. Immunity 2012, 37, 771–783. [Google Scholar] [CrossRef]
- Brugman, S.; Perdijk, O.; van Neerven, R.J.; Savelkoul, H.F. Mucosal immune development in early life: Setting the stage. Arch. Immunol. Ther. Exp. 2015, 63, 251–268. [Google Scholar] [CrossRef]
- Strunk, T.; Currie, A.; Richmond, P.; Simmer, K.; Burgner, D. Innate immunity in human newborn infants: Prematurity means more than immaturity. J. Matern. Fetal Neonatal Med. 2011, 24, 25–31. [Google Scholar] [CrossRef]
- Smolen, K.K.; Ruck, C.E.; Fortuno, E.S.; Ho, K.; Dimitriu, P.; Mohn, W.W.; Speert, D.P.; Cooper, P.J.; Esser, M.; Goetghebuer, T.; et al. Pattern recognition receptor-mediated cytokine response in infants across 4 continents. J. Allergy Clin. Immunol. 2014, 133, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Corbett, A.J.; Eckle, S.B.; Birkinshaw, R.W.; Liu, L.; Patel, O.; Mahony, J.; Chen, Z.; Reantragoon, R.; Meehan, B.; Cao, H.; et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014, 509, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Leeansyah, E.; Loh, L.; Nixon, D.F.; Sandberg, J.K. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat. Commun. 2014, 5, 3143. [Google Scholar] [CrossRef] [PubMed]
- Kjer-Nielsen, L.; Patel, O.; Corbett, A.J.; Le Nours, J.; Meehan, B.; Liu, L.; Bhati, M.; Chen, Z.; Kostenko, L.; Reantragoon, R.; et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012, 491, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.M.; Antony, K.M.; Ma, J.; Prince, A.L.; Showalter, L.; Moller, M.; Aagaard, K.M. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, S.N.; Madan, J.C.; Emond, J.A.; Morrison, H.G.; Christensen, B.C.; Karagas, M.R.; Hoen, A.G. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome 2018, 6, 109. [Google Scholar] [CrossRef]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: A prospective follow-up study initiated in early pregnancy. Am. J. Clin. Nutr. 2010, 92, 1023–1030. [Google Scholar] [CrossRef]
- Lammert, C.R.; Frost, E.L.; Bolte, A.C.; Paysour, M.J.; Shaw, M.E.; Bellinger, C.E.; Weigel, T.K.; Zunder, E.R.; Lukens, J.R. Cutting Edge: Critical Roles for Microbiota-Mediated Regulation of the Immune System in a Prenatal Immune Activation Model of Autism. J. Immunol. 2018, 201, 845–850. [Google Scholar] [CrossRef]
- Zmora, N.; Zeevi, D.; Korem, T.; Segal, E.; Elinav, E. Taking it Personally: Personalized Utilization of the Human Microbiome in Health and Disease. Cell Host Microbe 2016, 19, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Brusaferro, A.; Cozzali, R.; Orabona, C.; Biscarini, A.; Farinelli, E.; Cavalli, E.; Grohmann, U.; Principi, N.; Esposito, S. Is It Time to Use Probiotics to Prevent or Treat Obesity? Nutrients 2018, 10, 1613. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.S.; Kim, S.Y.; Suk, K.T.; Kim, B.Y. Modulation of gut microbiome in nonalcoholic fatty liver disease: Pro-, pre-, syn-, and antibiotics. J. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, M.; Yu, M.; Shen, M.; Wang, Q.; Yu, Y.; Xie, J. Natural polysaccharides exhibit anti-tumor activity by targeting gut microbiota. Int. J. Biol. Macromol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Strzępa, A.; Lobo, F.M.; Majewska-Szczepanik, M.; Szczepanik, M. Antibiotics and autoimmune and allergy diseases: Causative factor or treatment? Int. Immunopharmacol. 2018, 65, 328–341. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Precone, V.; Casaburi, G.; Miele, E.; Martinelli, M.; Staiano, A.; Salvatore, F.; Sacchetti, L. An altered gut microbiome profile in a child affected by Crohn’s disease normalized after nutritional therapy. Am. J. Gastroenterol. 2013, 108, 851–852. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Li, J.; Panagiotou, G. A Molecular-Level Landscape of Diet-Gut Microbiome Interactions: Toward Dietary Interventions Targeting Bacterial Genes. mBio 2015, 6, e01263-15. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Yacyshyn, B.R.; Yacyshyn, M.B. Gut microbiota and obesity: An opportunity to alter obesity through Fecal Microbiota Transplant (FMT). Diabetes Obes. Metab. 2018. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Argenio, V. The Prenatal Microbiome: A New Player for Human Health. High-Throughput 2018, 7, 38. https://doi.org/10.3390/ht7040038
D’Argenio V. The Prenatal Microbiome: A New Player for Human Health. High-Throughput. 2018; 7(4):38. https://doi.org/10.3390/ht7040038
Chicago/Turabian StyleD’Argenio, Valeria. 2018. "The Prenatal Microbiome: A New Player for Human Health" High-Throughput 7, no. 4: 38. https://doi.org/10.3390/ht7040038
APA StyleD’Argenio, V. (2018). The Prenatal Microbiome: A New Player for Human Health. High-Throughput, 7(4), 38. https://doi.org/10.3390/ht7040038




