Comparison of Cell Arrays and Multi-Well Plates in Microscopy-Based Screening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Materials
2.2. Preparation of on Cell Arrays for Small Interfering RNA Transfection
2.3. Coating of Multi-Well Plates with Small Interfering RNAs
2.4. Cell Cycle Progression Assay
2.5. Epidermal Growth Factor Internalization Assay
2.6. Microscopy
2.7. Image Analysis
2.8. Statistical Data Analysis
3. Results
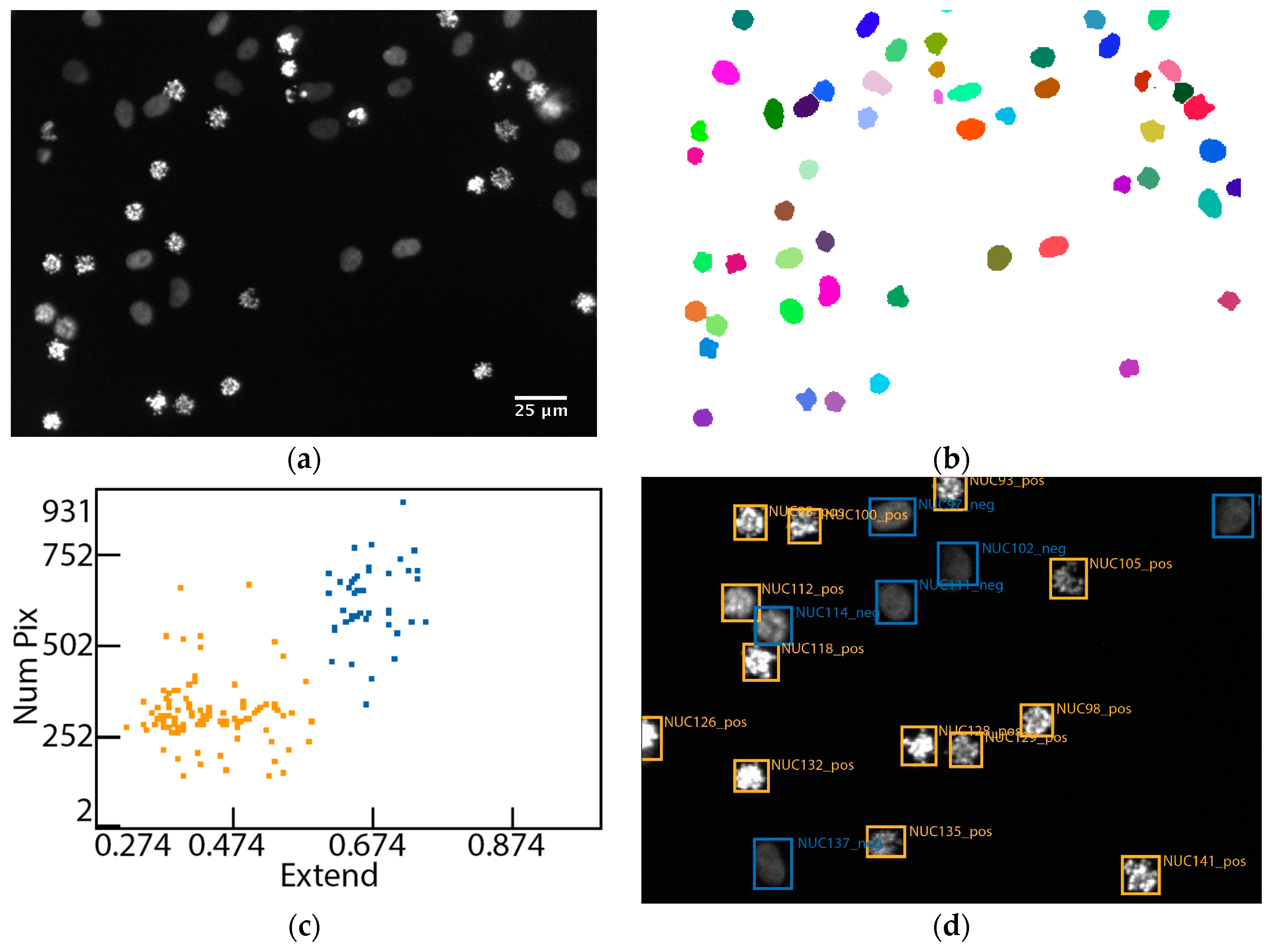
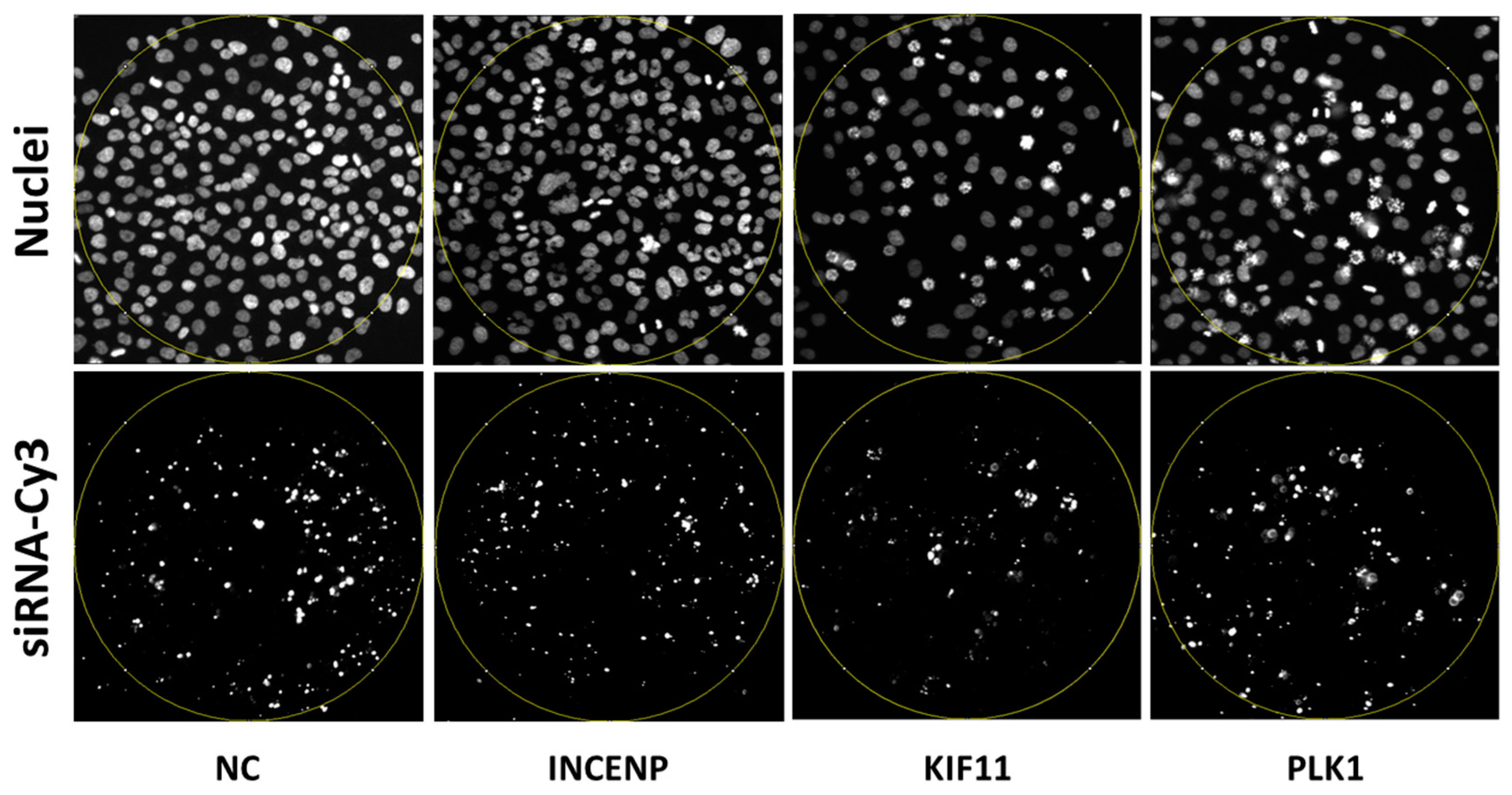
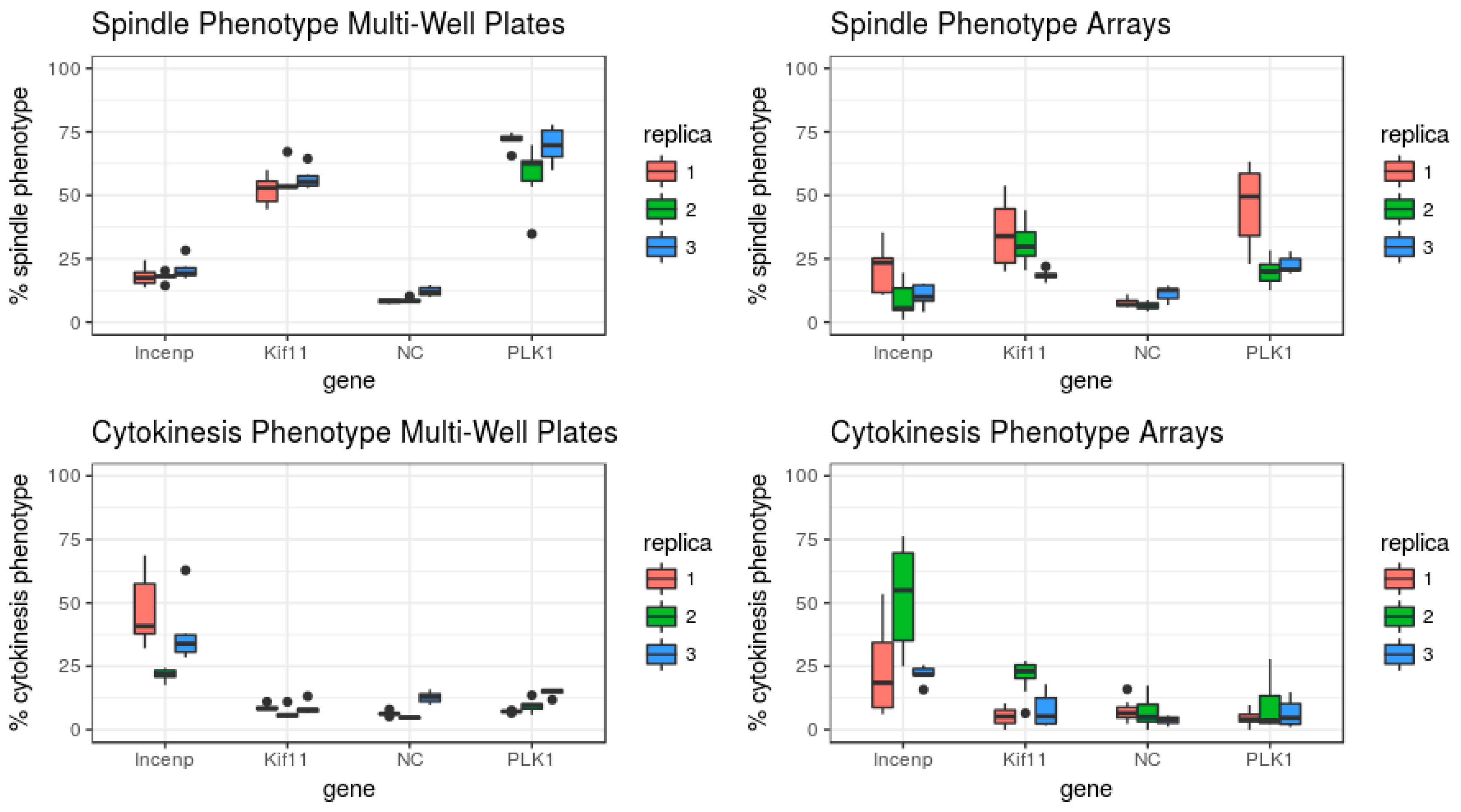
3.1. Set-Up of the Assay to Quantify the Arrest of Cell Cycle Progression
3.2. Evaluation of the Assay for Cell Cycle Progression
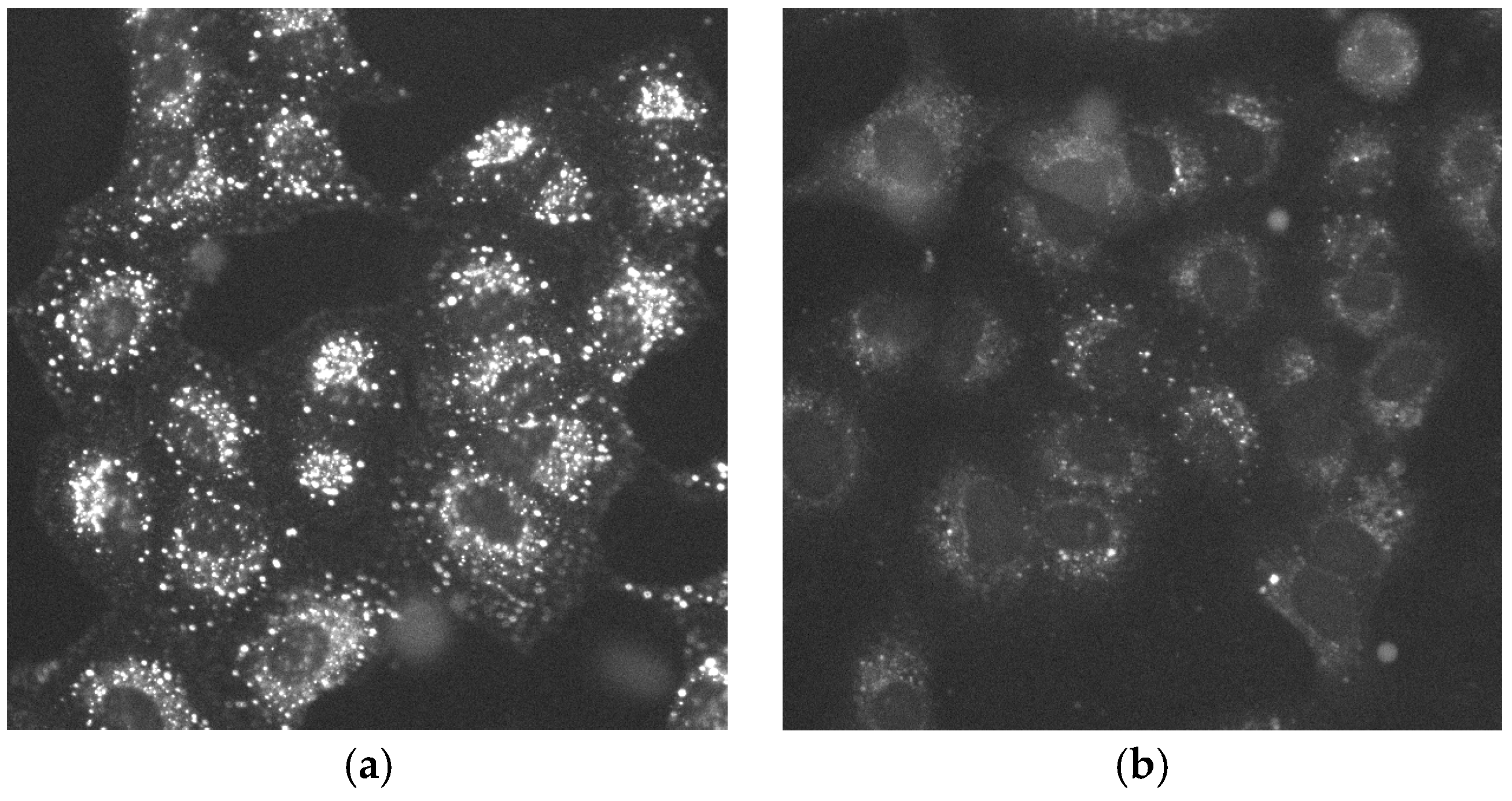
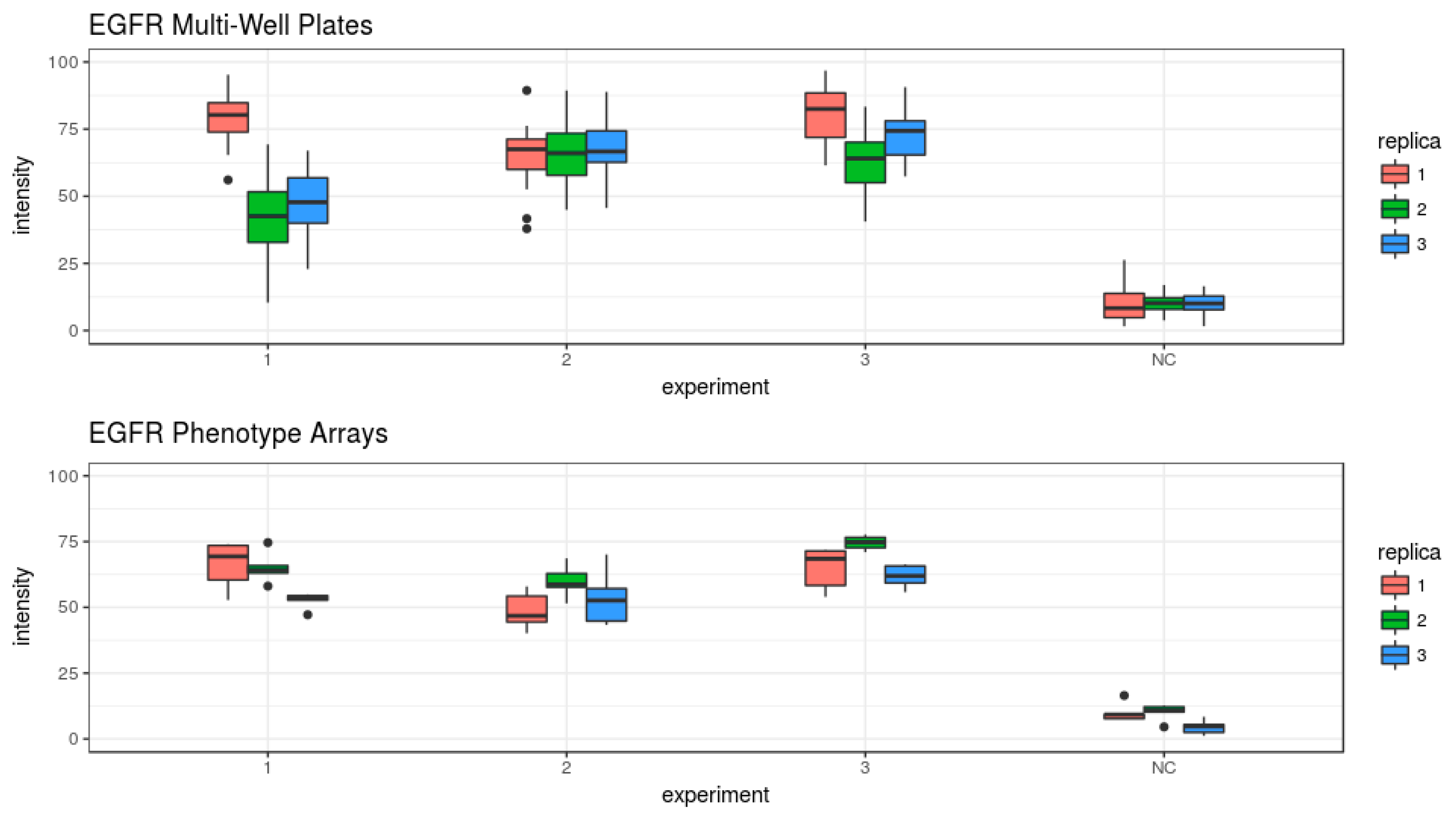
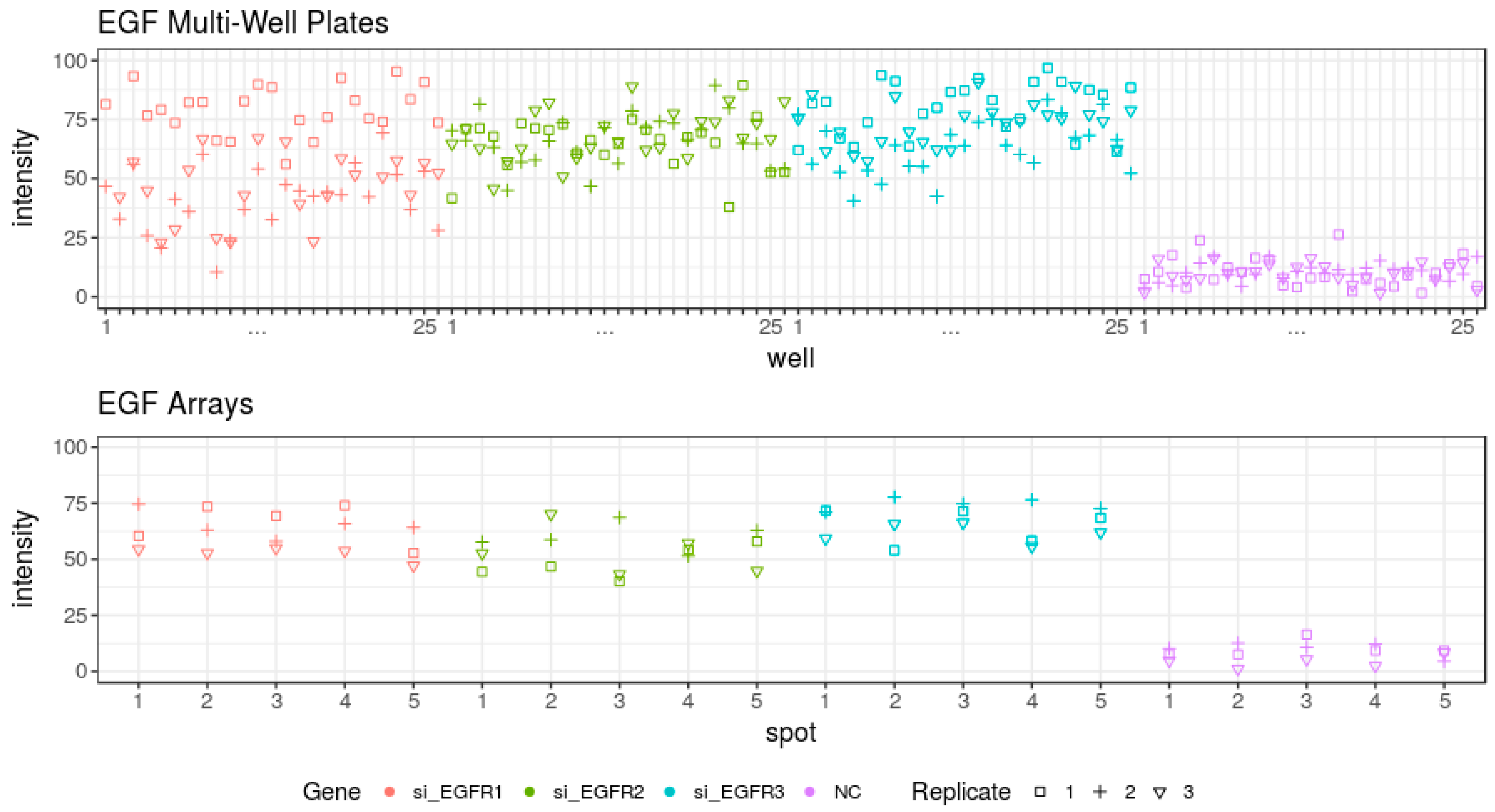
3.3. Evaluation of the Epidermal Growth Factor Endocytosis Assay
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ziauddin, J.; Sabatini, D. Microarrays of cells expressing defined cDNAs. Nature 2001, 411, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Reymann, J.; Beil, N.; Beneke, J.; Kaletta, P.P.; Burkert, K.; Erfle, H. Next-generation 9216-microwell cell arrays for high-content screening microscopy. BioTechniques 2009, 47, 877–878. [Google Scholar] [CrossRef] [PubMed]
- Rantala, J.K.; Mäkelä, R.; Aaltola, A.R.; Laasola, P.; Mpindi, J.P.; Nees, M.; Saviranta, P.; Kallioniemi, O. A cell spot microarray method for production of high density siRNA transfection microarrays. BMC Genom. 2011, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Neumann, B.; Walter, T.; Hériché, J.K.; Bulkescher, J.; Erfle, H.; Conrad, C.; Rogers, P.; Poser, I.; Held, M.; Liebel, U.; et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 2010, 464, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Rantala, J.K.; Pouwels, J.; Pellinen, T.; Veltel, S.; Laasola, P.; Mattila, E.; Potter, C.S.; Duffy, T.; Sundberg, J.P.; Kallioniemi, O.; et al. SHARPIN is an endogenous inhibitor of beta1-integrin activation. Nat. Cell Biol. 2011, 13, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.C.; Joggerst, B.; Laketa, V.; Verissimo, F.; Cetin, C.; Erfle, H.; Bexiga, M.G.; Singan, V.R.; Hériché, J.K.; Neumann, B.; et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat. Cell Biol. 2012, 14, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Eskova, A.; Knapp, B.; Matelska, D.; Reusing, S.; Arjonen, A.; Lisauskas, T.; Pepperkok, R.; Russell, R.; Eils, R.; Ivaska, J.; et al. An RNAi screen identifies KIF15 as a novel regulator of the endocytic trafficking of integrin. J. Cell Sci. 2014, 127, 2433–2447. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.A.; Schillo, S.M.; Demir, K.; Ueda, E.; Nesterov-Mueller, A.; Levkin, P.A. Droplet-Array (DA) Sandwich Chip: A Versatile Platform for High-Throughput Cell Screening Based on Superhydrophobic-Superhydrophilic Micropatterning. Adv. Mater. 2015, 27, 5217–5222. [Google Scholar] [CrossRef] [PubMed]
- Erfle, H.; Simpson, J.C.; Bastiaens, P.I.; Pepperkok, R. siRNA cell arrays for high-content screening microscopy. Biotechniques 2004, 37, 454–462. [Google Scholar] [PubMed]
- Feng, W.; Ueda, E.; Levkin, P.A. Droplet Microarrays: From Surface Patterning to High-Throughput Applications. Adv. Mater. 2018, 23, e1706111. [Google Scholar] [CrossRef] [PubMed]
- Tronser, T.; Popova, A.A.; Levkin, P.A. Miniaturized platform for high-throughput screening of stem cells. Curr. Opin. Biotechnol. 2017, 46, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Erard, N.; Knott, S.R.V.; Hannon, G.J. A CRISPR Resource for Individual, Combinatorial, or Multiplexed Gene Knockout. Mol. Cell 2017, 67, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.; Smith, J.A.; Shamu, C.E.; Neumüller, R.A.; Perrimon, N. RNAi screening comes of age: Improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 2014, 15, 591. [Google Scholar] [CrossRef] [PubMed]
- Deans, R.; Morgens, D.W.; Ökesli, A.; Pillay, S.; Horlbeck, M.A.; Kampmann, M.; Gilbert, L.A.; Li, A.; Mateo, R.; Smith, M.; et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat. Chem. Biol. 2016, 12, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Dietz, C.; Berthold, M.R. KNIME for Open-Source Bioimage Analysis: A Tutorial. Adv. Anat. Embryol. Cell Biol. 2016, 219, 179–197. [Google Scholar] [PubMed]
- Sternberg, S.R. Biomedical Image Processing. Computer 1983, 16, 22–34. [Google Scholar] [CrossRef]
- Nobuyuki, O. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar]
- Mardia, K.V.; Hainsworth, T.J. A spatial thresholding method for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 1988, 10, 919–927. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Samet, H.; Tamminen, M. Efficient Component Labeling of Images of Arbitrary Dimension Represented by Linear Bintrees. IEEE Trans. Pattern Anal. Mach. Intell. 1988, 10, 579–586. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment on Statistical Computing; R foundation for Statistical Computing 2017; R Core Team: Viena, Austria, 2017; Available online: http://www.R-project.org (accessed on 8 May 2018).
- Glotzer, M. The 3Ms of central spindle assembly: Microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009, 10, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Rath, O.; Kozielski, F. Kinesins and cancer. Nat. Rev. Cancer 2012, 12, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Ertych, N.; Bastians, H. A phenotypic screen identifies microtubule plus end assembly regulators that can function in mitotic spindle orientation. Cell Cycle 2015, 14, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Breir, S. Identification of the protein binding region of S-trityl-l-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry 2004, 43, 13072–13082. [Google Scholar] [CrossRef] [PubMed]
- Van De Weerdt, B.C.; Medema, R.H. Polo-like kinases: A team in control of the division. Cell Cycle 2006, 5, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Combes, G.; Alharbi, I.; Braga, L.G.; Elowe, S. Playing polo during mitosis: PLK1 takes the lead. Oncogene 2017, 36, 4819–4827. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.; Turnbull, K.; Desai, A.; Campbell, C.S. An engineered minimal chromosomal passenger complex reveals a role for INCENP/Sli15 spindle association in chromosome biorientation. J. Cell Biol. 2017, 216, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Unsal, E.; Degirmenci, B.; Harmanda, B.; Erman, B.; Ozlu, N. A small molecule identified through an in silico screen inhibits Aurora B-INCENP interaction. Chem. Biol. Drug Des. 2016, 88, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Yabuta, N.; Yoshida, K.; Mukai, S.; Kato, Y.; Torigata, K.; Nojima, H. Large tumor suppressors 1 and 2 regulate Aurora-B through phosphorylation of INCENP to ensure completion of cytokinesis. Heliyon 2016, 2, e00131. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Pinson, X.; Platani, M.; Salloum, Z.; Xu, Z.; Clark, A.; Macisaac, F.; Ogawa, H.; Eggert, U.; Glover, D.M.; et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 2012, 10, e1001250. [Google Scholar] [CrossRef]
- Carpenter, G.; Cohen, S. Human epidermal growth factor and the proliferation of human fibroblasts. J. Cell. Physiol. 1976, 88, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Glenewinkel, H.; Vacheva, I.; Hoeller, D.; Dikic, I.; Eils, R. An ultrasensitive sorting mechanism for EGF receptor endocytosis. BMC Syst. Biol. 2008, 2, 32. [Google Scholar] [CrossRef] [PubMed]
- Serva, A.; Knapp, B.; Tsai, Y.T.; Claas, C.; Lisauskas, T.; Matula, P.; Harder, N.; Kaderali, L.; Rohr, K.; Erfle, H.; et al. miR-17-5p regulates endocytic trafficking through targeting TBC1D2/Armus. PLoS ONE 2012, 7, e52555. [Google Scholar] [CrossRef] [PubMed]
- Collinet, C.; Stöter, M.; Bradshaw, C.R.; Samusik, N.; Rink, J.C.; Kenski, D.; Habermann, B.; Buchholz, F.; Henschel, R.; Mueller, M.S.; et al. Systems survey of endocytosis by multiparametric image analysis. Nature 2010, 464, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Starkuviene, V.; Liebel, U.; Simpson, J.C.; Erfle, H.; Poustka, A.; Wiemann, S.; Pepperkok, R. High-content screening microscopy identifies novel proteins with a putative role in secretory membrane traffic. Genome Res. 2004, 14, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.; Goh, G.; Racine, V.; Ng, S.; Kumar, P.; Bard, F. RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol. Syst. Biol. 2012, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Anitei, M.; Chenna, R.; Czupalla, C.; Esner, M.; Christ, S.; Lenhard, S.; Korn, K.; Meyenhofer, F.; Bickle, M.; Zerial, M.; et al. A high-throughput siRNA screen identifies genes that regulate mannose 6-phosphate receptor trafficking. J. Cell Sci. 2014, 127, 5079–5092. [Google Scholar] [CrossRef] [PubMed]
- Lisauskas, T.; Matula, P.; Claas, C.; Reusing, S.; Wiemann, S.; Erfle, H.; Lehmann, L.; Fischer, P.; Eils, R.; Rohr, K.; et al. Live-cell assays to identify regulators of ER-to-Golgi trafficking. Traffic 2012, 13, 416–432. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Z.; Sadok, A.; Sailem, H.; McCarthy, A.; Xia, X.; Li, L.; Garcia, M.A.; Evans, L.; Barr, A.; Perrimon, N.; et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat. Cell Biol. 2013, 15, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Neumann, B.; Held, M.; Liebel, U.; Erfle, H.; Rogers, P.; Pepperkok, R.; Ellenberg, J. High-throughput RNAi screening by time lapse imaging of live human cells. Nat. Methods 2006, 3, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Paul, G.; Walter, T.; Neumann, B.; Hériché, J.K.; Ellenberg, J.; Huber, W. Dynamic modeling of phenotypes in a genome-wide RNAi live-cell imaging assay. BMC Bioinform. 2013, 14, 308. [Google Scholar]

| Feature | Cell Arrays | Multi-Well Plates |
|---|---|---|
| High-throughput | yes | yes/no |
| Genetic screens | yes | yes |
| Chemical screens | yes/no | yes |
| Reverse transfection | yes | yes |
| Direct transfection | yes/no | yes |
| Assay performance needs robotics | no | yes |
| Edge effects | no | yes |
| Complex assays can be performed | yes | yes/no |
| Assay can be performed in cells which are available in small numbers | yes | no |
| Small amounts of the reagents are required for screening | yes | no |
| Cell migration can be analysed | no | yes |
| Cells with the extended shape (e.g., neurons) can be analysed | no | yes |
| Many cells are available for statistics from one replicate | no | yes |
| Possibility to cross-contaminate the samples | yes/no | no |
| Comparison | Well-Plates | Cell Arrays | ||
|---|---|---|---|---|
| Mean Phenotype | p-Value | Mean Phenotype | p-Value | |
| Spindle phenotype | ||||
| NC | 8.6 ± 1.2% | 9.7 ± 1.9% | ||
| KIF11 | 54.8 ± 5.3% | 6.9 × 10−15 | 28.3 ± 7.4% | 2.6 × 10−9 |
| PLK1 | 66.4 ± 7.6% | 1.6 × 10−15 | 29.2 ± 7.9% | 5.1 × 10−7 |
| INCENP | 18.9 ± 3.3% | 1.5 × 10−9 | 13.4 ± 6.6% | 3.9 × 10−2 |
| Cytokinesis phenotype | ||||
| NC | 5.9 ± 1.2% | 8 ± 3.9% | ||
| KIF11 | 7.8 ± 2% | 1 | 11.5 ± 5.6% | 3 × 10−2 |
| PLK1 | 10.5 ± 1.5% | 2 × 10−1 | 6.6 ± 5.4% | 9.7 × 10−1 |
| INCENP | 35.5 ± 10.1% | 2.6 × 10−6 | 32.7 ± 13.1% | 1.8 × 10−6 |
| Comparison | Well-Plates | Cell Arrays | ||
|---|---|---|---|---|
| Mean Cell Numbers | p-Value | Mean Cell Numbers | p-Value | |
| NC | 100% | 100% | ||
| PLK1 | 49.6% | 9.9 × 10−9 | 68.2% | 1.2 × 10−2 |
| KIF11 | 58.9% | 8.2 × 10−7 | 73.7% | 2.2 × 10−2 |
| INCENP | 59.2% | 4.6 × 10−4 | 93.1% | 9.1 × 10−1 |
| Comparison | Well-Plates | Cell Arrays | ||
|---|---|---|---|---|
| Mean Intracellular EGF Signal | p-Value | Mean Intracellular EGF Signal | p-Value | |
| NC | 89.8 ± 5% | 91.8 ± 3.3% | ||
| EGFR_1 | 44.4 ± 12.8% | 8.8 × 10−14 | 38.7 ± 6.6% | 4.1 × 10−14 |
| EGFR_2 | 33.5 ± 10.8% | 1.5 × 10−14 | 45.9 ± 8.4% | 8.8 × 10−14 |
| EGFR_3 | 28 ± 10.8% | 4.4 × 10−10 | 33 ± 5.5% | 4.7 × 10−14 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becker, A.-K.; Erfle, H.; Gunkel, M.; Beil, N.; Kaderali, L.; Starkuviene, V. Comparison of Cell Arrays and Multi-Well Plates in Microscopy-Based Screening. High-Throughput 2018, 7, 13. https://doi.org/10.3390/ht7020013
Becker A-K, Erfle H, Gunkel M, Beil N, Kaderali L, Starkuviene V. Comparison of Cell Arrays and Multi-Well Plates in Microscopy-Based Screening. High-Throughput. 2018; 7(2):13. https://doi.org/10.3390/ht7020013
Chicago/Turabian StyleBecker, Ann-Kristin, Holger Erfle, Manuel Gunkel, Nina Beil, Lars Kaderali, and Vytaute Starkuviene. 2018. "Comparison of Cell Arrays and Multi-Well Plates in Microscopy-Based Screening" High-Throughput 7, no. 2: 13. https://doi.org/10.3390/ht7020013
APA StyleBecker, A.-K., Erfle, H., Gunkel, M., Beil, N., Kaderali, L., & Starkuviene, V. (2018). Comparison of Cell Arrays and Multi-Well Plates in Microscopy-Based Screening. High-Throughput, 7(2), 13. https://doi.org/10.3390/ht7020013




