Study of the Humoral Immune Response towards HCV Genotype 4 Using a Bead-Based Multiplex Serological Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Sera
2.2. Hepatitis C Virus Antigens
2.3. Antigen Immobilization
2.4. Bead-Based Multiplex Anti-Hepatitis C Virus Assay Procedure
2.5. Technical Assay Validation
2.6. Cut-off Definition and Methods for Sample Classification
2.7. Statistical Analysis
3. Results
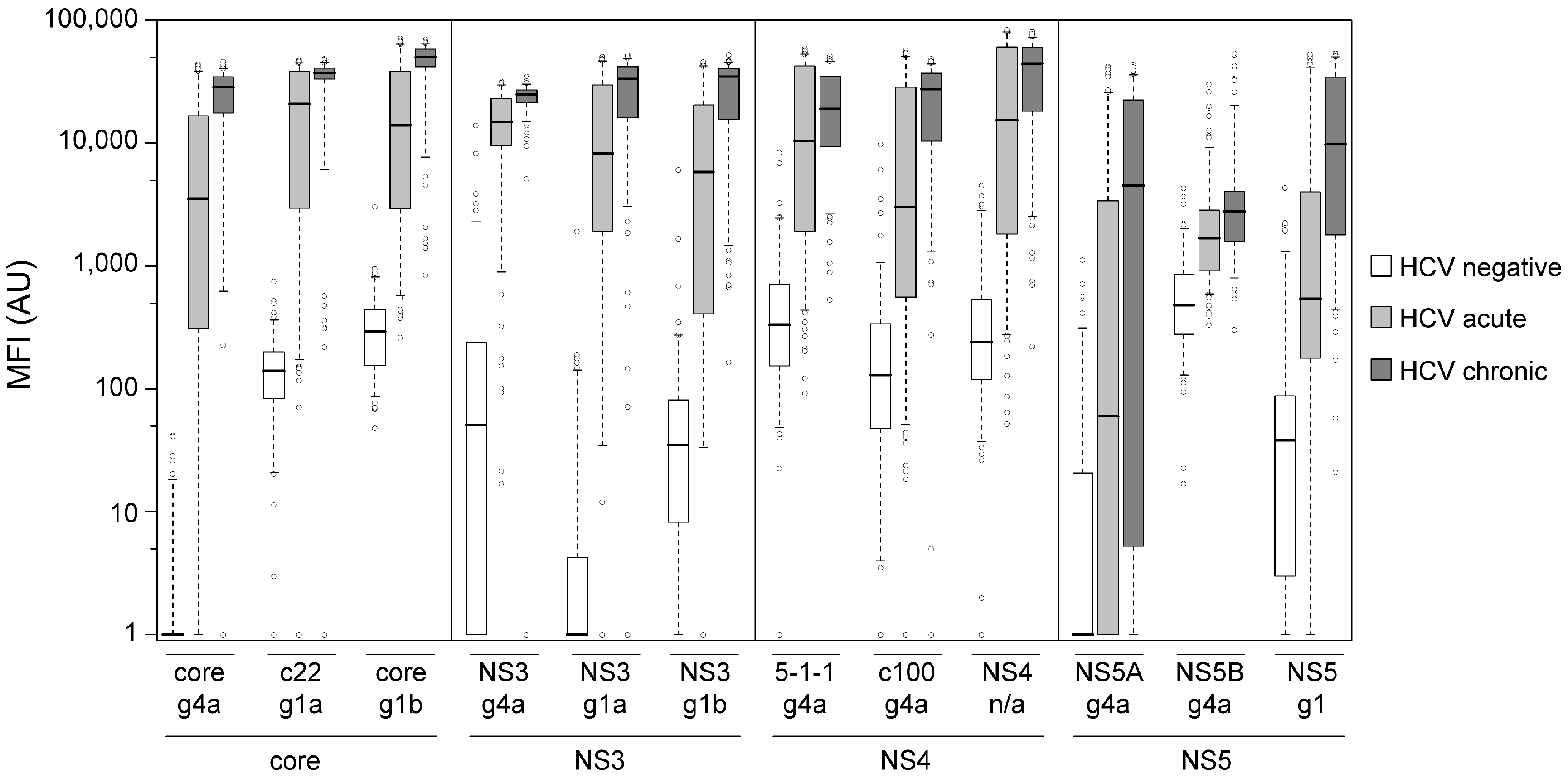
3.1. Technical and Clinical Assay Validation
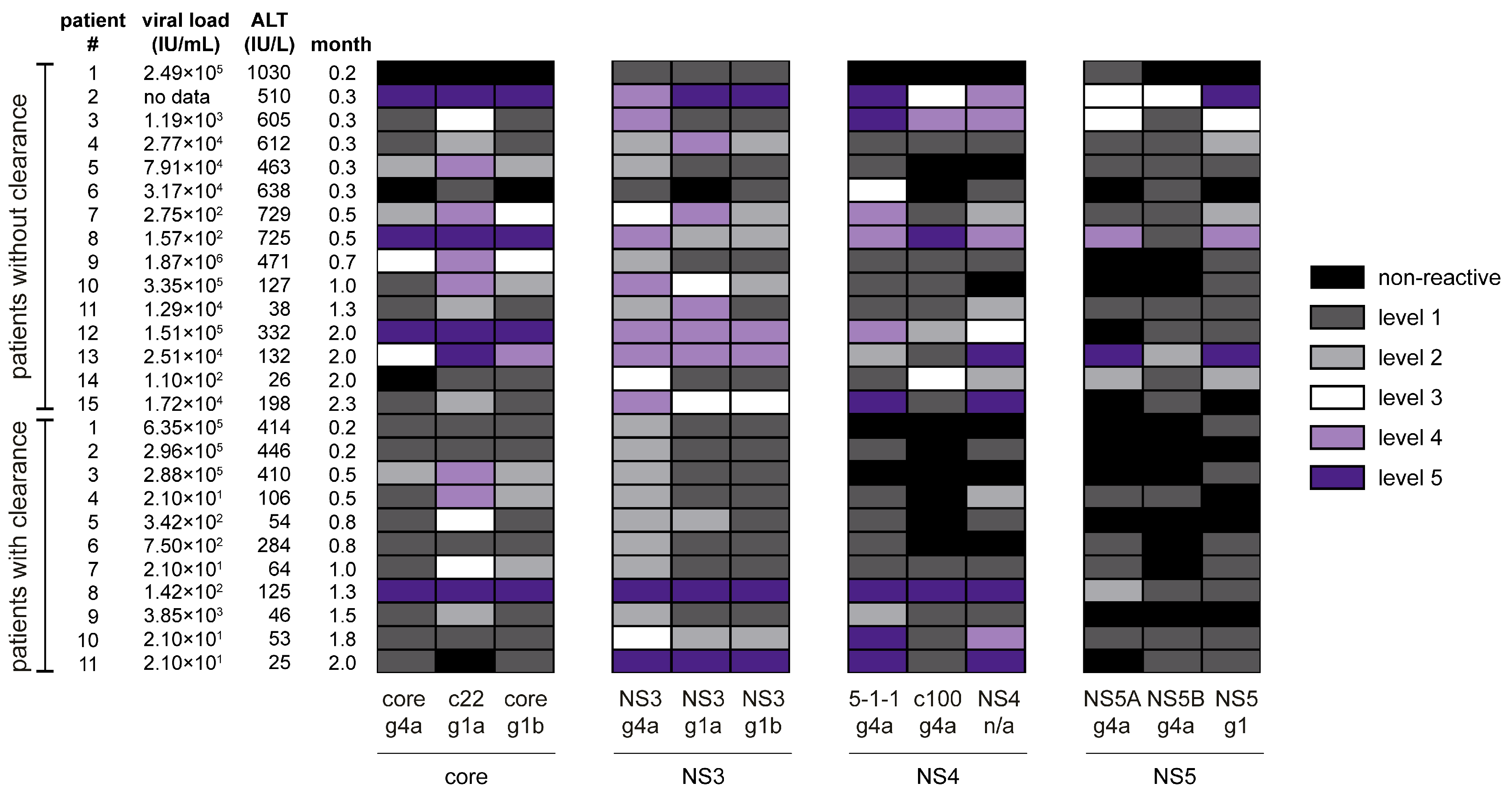
3.2. Classification of Hepatitis C Virus Patients into Acute or Chronic Stage of Infection
3.3. Immune Response towards Hepatitis C Virus Genotype 4
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix
| HCV Protein | Antigen Name Including Genotype | S/CO Value, where Level Starts | Maximal S/CO Value | ||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||
| core | core g4a | 1 | 404 | 808 | 1211 | 1614 | 2018 |
| c22 g1a | 1 | 25 | 50 | 74 | 98 | 123 | |
| core g1b | 1 | 17 | 32 | 48 | 64 | 80 | |
| NS3 | NS3 g4a | 1 | 17 | 32 | 48 | 64 | 79 |
| NS3 g1a | 1 | 188 | 374 | 561 | 747 | 934 | |
| NS3 g1b | 1 | 64 | 128 | 191 | 254 | 317 | |
| NS4 | 5-1-1 g4a | 1 | 10 | 19 | 29 | 38 | 47 |
| c100 g4a | 1 | 16 | 31 | 46 | 61 | 77 | |
| NS4 n/a | 1 | 17 | 34 | 50 | 66 | 82 | |
| NS5 | NS5A g4a | 1 | 170 | 340 | 509 | 678 | 847 |
| NS5B g4a | 1 | 8 | 15 | 22 | 29 | 36 | |
| NS5 g1 | 1 | 56 | 110 | 165 | 220 | 274 | |
References
- Cooke, G.S.; Lemoine, M.; Thursz, M.; Gore, C.; Swan, T.; Kamarulzaman, A.; DuCros, P.; Ford, N. Viral hepatitis and the global burden of disease: A need to regroup. J. Viral Hepat. 2013, 20, 600–601. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Humphreys, I.; Flaxman, A.; Brown, A.; Cooke, G.S.; Pybus, O.G.; Barnes, E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015, 61, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Analysis. World map of HCV prevalence. Available online: http://www.centerforda.com/hcv.htm (accessed on 20 July 2016).
- Seeff, L.B. Natural history of chronic hepatitis C. Hepatology 2002, 36, S35–S46. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.L.; Seeff, L.B. Natural history of hepatitis C. Clin. Liver Dis. 2005, 9, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.M. Use and interpretation of virological tests for hepatitis C. Hepatology 2002, 36, S65–S73. [Google Scholar] [CrossRef] [PubMed]
- Farci, P.; Alter, H.J.; Wong, D.; Miller, R.H.; Shih, J.W.; Jett, B.; Purcell, R.H. A long-term study of hepatitis C virus replication in non-A, non-B hepatitis. N. Engl. J. Med. 1991, 325, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Marcellin, P. Hepatitis C: The clinical spectrum of the disease. J. Hepatol. 1999, 31 (Suppl. 1), 9–16. [Google Scholar] [CrossRef]
- Vogel, W.; Graziadei, I.; Umlauft, F.; Datz, C.; Hackl, F.; Allinger, S.; Grunewald, K.; Patsch, J. High-dose interferon-α2b treatment prevents chronicity in acute hepatitis C: A pilot study. Dig. Dis. Sci. 1996, 41, 81S–85S. [Google Scholar] [CrossRef] [PubMed]
- Jaeckel, E.; Cornberg, M.; Wedemeyer, H.; Santantonio, T.; Mayer, J.; Zankel, M.; Pastore, G.; Dietrich, M.; Trautwein, C.; Manns, M.P. Treatment of acute hepatitis C with interferon α-2b. N. Engl. J. Med. 2001, 345, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- Licata, A.; Di Bona, D.; Schepis, F.; Shahied, L.; Craxi, A.; Camma, C. When and how to treat acute hepatitis C? J. Hepatol. 2003, 39, 1056–1062. [Google Scholar] [CrossRef] [Green Version]
- Gerlach, J.T.; Diepolder, H.M.; Zachoval, R.; Gruener, N.H.; Jung, M.C.; Ulsenheimer, A.; Schraut, W.W.; Schirren, C.A.; Waechtler, M.; Backmund, M.; et al. Acute hepatitis C: High rate of both spontaneous and treatment-induced viral clearance. Gastroenterology 2003, 125, 80–88. [Google Scholar] [CrossRef]
- Kamal, S.M.; Fouly, A.E.; Kamel, R.R.; Hockenjos, B.; Al Tawil, A.; Khalifa, K.E.; He, Q.; Koziel, M.J.; El Naggar, K.M.; Rasenack, J.; et al. Peginterferon α2b therapy in acute hepatitis C: Impact of onset of therapy on sustained virologic response. Gastroenterology 2006, 130, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Deterding, K.; Grüner, N.; Buggisch, P.; Wiegand, J.; Galle, P.R.; Spengler, U.; Hinrichsen, H.; Berg, T.; Potthoff, A.; Malek, N.; et al. Delayed versus immediate treatment for patients with acute hepatitis C: A randomised controlled non-inferiority trial. Lancet Infect. Dis. 2013, 3099, 497–506. [Google Scholar] [CrossRef]
- Planatscher, H.; Rimmele, S.; Michel, G.; Potz, O.; Joos, T.; Schneiderhan-Marra, N. Systematic reference sample generation for multiplexed serological assays. Sci. Rep. 2013, 3, 3259. [Google Scholar] [CrossRef] [PubMed]
- M’Kada, H.; Munteanu, M.; Perazzo, H.; Ngo, Y.; Ramanujam, N.; Imbert-Bismut, F.; Ratziu, V.; Bonnefont-Rousselot, D.; Souberbielle, B.; Schuppe-Koistinen, I.; et al. What are the best reference values for a normal serum alanine transaminase activity (ALT)? Impact on the presumed prevalence of drug induced liver injury (DILI). Regul. Toxicol. Pharmacol. 2011, 60, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.C.; Astrakhantseva, I.V.; Fields, H.A.; Kamili, S. Distinguishing acute from chronic hepatitis C virus (HCV) infection based on antibody reactivities to specific HCV structural and nonstructural proteins. J. Clin. Microbiol. 2011, 49, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Barrera, J.M.; Bruguera, M.; Ercilla, M.G.; Sanchez-Tapias, J.M.; Gil, M.P.; Costa, J.; Gelabert, A.; Rodes, J.; Castillo, R. Incidence of non-A, non-B hepatitis after screening blood donors for antibodies to hepatitis C virus and surrogate markers. Ann. Intern. Med. 1991, 115, 596–600. [Google Scholar] [CrossRef]
- Xu, D.; Lu, H.; Nu, J.; Meng, Z.; Shang, D.; Kang, T. Dynamic study of anti-NS5 and ALT in post-transfusion hepatitis C. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 1997, 11, 344–347. (In Chinese) [Google Scholar] [PubMed]
- Rosa, D.; Campagnoli, S.; Moretto, C.; Guenzi, E.; Cousens, L.; Chin, M.; Dong, C.; Weiner, A.J.; Lau, J.Y.; Choo, Q.L.; et al. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: Cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 1996, 93, 1759–1763. [Google Scholar] [CrossRef] [PubMed]
- Meuleman, P.; Bukh, J.; Verhoye, L.; Farhoudi, A.; Vanwolleghem, T.; Wang, R.Y.; Desombere, I.; Alter, H.; Purcell, R.H.; Leroux-Roels, G. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology 2011, 53, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Keck, Z.Y.; Foung, S.K. Neutralizing antibody response to hepatitis C virus. Viruses 2011, 3, 2127–2145. [Google Scholar] [CrossRef] [PubMed]
- Stamataki, Z.; Coates, S.; Evans, M.J.; Wininger, M.; Crawford, K.; Dong, C.; Fong, Y.L.; Chien, D.; Abrignani, S.; Balfe, P.; et al. Hepatitis C virus envelope glycoprotein immunization of rodents elicits cross-reactive neutralizing antibodies. Vaccine 2007, 25, 7773–7784. [Google Scholar] [CrossRef] [PubMed]
- Maylin, S.; Martinot-Peignoux, M.; Ripault, M.P.; Moucari, R.; Cardoso, A.C.; Boyer, N.; Giuily, N.; Castelnau, C.; Pouteau, M.; Asselah, T.; et al. Sustained virological response is associated with clearance of hepatitis C virus RNA and a decrease in hepatitis C virus antibody. Liver Int. 2009, 29, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Kumada, T.; Kiriyama, S.; Sone, Y.; Tanikawa, M.; Hisanaga, Y.; Kuzuya, T.; Honda, T.; Hayashi, K.; Nakano, I.; et al. Changes in hepatitis C virus (HCV) antibody status in patients with chronic hepatitis C after eradication of HCV infection by interferon therapy. Clin. Infect. Dis. 2005, 40, e49–e54. [Google Scholar] [CrossRef] [PubMed]
- Moran, M.; Chapman, N.; Abela-Oversteegen, L.; Chowdhary, V.; Doubell, A.; Whittall, C.; Howard, R.; Farrell, P.; Halliday, D.; Hirst, C. G-Finder Report 2015; Medicines for Malaria Venture: Geneva, Switzerland, 2015. [Google Scholar]

| HCV Status | Origin | n Samples | n Female | n Male | n Samples w/o Data | Mean Age Female | Mean Age Male |
|---|---|---|---|---|---|---|---|
| Acute | Egypt | 166 | 61 | 103 | 2 | 35.0 ± 10.2 | 35.1 ± 10.6 |
| Chronic | Egypt | 33 | 18 | 15 | - | 47.8 ± 8.6 | 38.5 ± 12.1 |
| Chronic | France | 108 | 46 | 61 | 1 | 59.5 ± 10.1 | 55.1 ± 9.0 |
| Negative | Egypt | 26 | n/a | n/a | 26 | n/a | n/a |
| Negative | France | 61 | 25 | 36 | - | 35.7 ± 11.0 | 38.8 ± 14.2 |
| Negative | USA | 30 | 15 | 15 | - | 37.7 ± 13.7 | 45.5 ± 13.8 |
| Time ourse * (Acute/Chronic) | Egypt | 107 (26 patients) | 10 | 16 | - | 34.4 ± 9.5 | 35.8 ± 11.7 |
| HCV Protein | Antigen Name Including Genotype | Cut-off (MFI) | p-Value HCV Neg. vs. HCV Pos. | S/CO > 1 | Diagnostic Accuracy (%) | ||
|---|---|---|---|---|---|---|---|
| HCV Negative (n = 117) n (%) | HCV Acute (n = 166) n (%) | HCV Chronic (n = 141) n (%) | |||||
| core | core g4a | 23 | 1.12 × 10−54 | 4 (3.4%) | 142 (85.5%) | 135 (95.7%) | 92.0 |
| c22 g1a | 399 | 1.97 × 10−53 | 4 (3.4%) | 145 (87.4%) | 136 (96.5%) | 92.9 | |
| core g1b | 900 | 2.34 × 10−54 | 3 (2.6%) | 146 (88.0%) | 140 (99.3%) | 94.3 | |
| NS3 | NS3 g4a | 442 | 4.14 × 10−54 | 19 (16.3%) | 159 (95.8%) | 140 (99.3%) | 93.6 |
| NS3 g1a | 56 | 2.58 × 10−55 | 13 (11.1%) | 157 (94.6%) | 140 (99.3%) | 94.6 | |
| NS3 g1b | 165 | 9.77 × 10−52 | 12 (10.3%) | 141 (84.9%) | 140 (99.3%) | 91.0 | |
| NS4 | 5-1-1 g4a | 1258 | 5.65 × 10−49 | 12 (10.3%) | 142 (85.5%) | 138 (97.9%) | 90.8 |
| c100 g4a | 751 | 1.13 × 10−44 | 9 (7.7%) | 116 (69.9%) | 135 (95.7%) | 84.7 | |
| NS4 n/a | 1129 | 3.05 × 10−48 | 12 (10.3%) | 135 (81.3%) | 138 (97.9%) | 89.2 | |
| NS5 | NS5A g4a | 52 | 2.87 × 10−45 | 21 (18.0%) | 85 (51.2%) | 103 (73.1%) | 67.0 |
| NS5B g4a | 1631 | 9.24 × 10−42 | 8 (6.8%) | 86 (51.8%) | 102 (72.3%) | 70.0 | |
| NS5 g1 | 199 | 2.75 × 10−46 | 16 (13.7%) | 119 (71.7%) | 138 (97.9%) | 84.4 | |
| Number of NS3 and Core Antigens with a S/CO > 1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | ||
| HCV status | Negative | 56 | 16 | 4 | 2 | 0 | 0 | 0 |
| Acute | 3 | 4 | 3 | 6 | 8 | 9 | 51 | |
| Chronic | 0 | 0 | 0 | 0 | 2 | 3 | 58 | |
| Training Set | Test Set | Complete Sample Set | ||
|---|---|---|---|---|
| n Neg. HCV | 78 | 39 | 117 | |
| n Acute HCV | 84 | 82 | 166 | |
| n Chronic HCV | 63 | 78 | 141 | |
| Classification | True positive | 136 | 160 | 296 |
| False positive | 0 | 0 | 0 | |
| False negative | 11 | 0 | 11 | |
| True negative | 78 | 39 | 117 | |
| Diagnostic accuracy (%) | 95.1 | 100 | 97.4 | |
| Sensitivity (%) | 92.5 | 100 | 96.4 | |
| Specificity (%) | 100 | 100 | 100 | |
| Positive predictive value (%) | 100 | 100 | 100 | |
| Negative predictive value (%) | 87.6 | 100 | 91.4 | |
| Multiplex HCV Assay | INNOTEST HCV Ab IV | ||||||
|---|---|---|---|---|---|---|---|
| n Neg. HCV | 116 | 116 | 116 | 116 | 116 | 116 | |
| n Acute HCV | 118 | 118 | 0 | 118 | 118 | 0 | |
| n Chronic HCV | 134 | 0 | 134 | 134 | 0 | 134 | |
| Classification | True positive | 243 | 109 | 134 | 240 | 106 | 134 |
| False positive | 0 | 0 | 0 | 0 | 0 | 0 | |
| False negative | 9 | 9 | 0 | 12 | 12 | 0 | |
| True negative | 116 | 116 | 116 | 116 | 116 | 116 | |
| Diagnostic accuracy (%) | 97.6 | 96.2 | 100 | 96.7 | 94.9 | 100 | |
| Sensitivity (%) | 96.6 | 92.4 | 100 | 95.5 | 89.8 | 100 | |
| Specificity (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Positive predictive value (%) | 100 | 100 | 100 | 100 | 100 | 100 | |
| Negative predictive value (%) | 92.8 | 92.8 | 100 | 90.6 | 90.6 | 100 | |
| Classified as | |||
|---|---|---|---|
| Acute | Chronic | ||
| Clinical HCV status | Acute (n = 155) | 130 (83.9%) | 25 (16.1%) |
| Chronic (n = 141) | 19 (13.5%) | 122 (86.5%) | |
| Patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Months after last positive HCV RT-PCR | 2.3 | 1.3 | 3.3 | 2.8 | 5.3 | 3.5 | 3.5 | 3.0 | 2.0 | 3.5 | 2.3 |
| Decline of antibody titer | yes | no | no | yes | yes | no | yes | yes | yes | yes | yes |
| core g4a | 32 | 290 | 135 | 13 | 62 | n/r | 79 | 74 | 60 | 73 | 53 |
| c22 g1a | 41 | 309 | 108 | 35 | 46 | 117 | 85 | 92 | 93 | 74 | n/r |
| core g1b | 35 | 238 | 127 | 26 | 53 | 91 | 82 | 87 | 78 | 58 | 94 |
| NS3 g4a | 56 | 115 | 118 | 36 | 83 | 101 | 82 | 100 | 91 | 88 | 101 |
| NS3 g1a | 39 | 290 | 123 | 8 | 62 | 92 | 52 | 85 | 62 | 60 | 100 |
| NS3 g1b | 33 | 431 | 85 | 6 | 71 | 89 | 50 | 94 | 60 | 73 | 100 |
| 5-1-1 g4a | n/r | 806 | 270 | 14 | 91 | 154 | 80 | 85 | 99 | 98 | 82 |
| c100 g4a | 140 | 123 | 309 | 14 | n/r | 111 | 394 | 100 | 102 | 77 | 70 |
| NS4 n/a | 115 | 196 | 266 | 26 | 119 | 109 | 209 | 98 | 77 | 75 | 95 |
| NS5A g4a | n/r | n/r | n/r | 15 | n/r | n/r | 47 | 67 | 209 | 70 | n/r |
| NS5B g4a | n/r | n/r | n/r | 0 | 102 | 92 | n/r | 70 | 101 | 79 | 61 |
| NS5 g1 | 63 | 144 | 176 | 9 | n/r | 111 | 71 | 43 | 269 | 46 | 122 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filomena, A.; Göpfert, J.C.; Duffy, D.; Pol, S.; Abdel-Hamid, M.; Esmat, G.; Fontanet, A.; Albert, M.L.; Joos, T.O.; Schneiderhan-Marra, N. Study of the Humoral Immune Response towards HCV Genotype 4 Using a Bead-Based Multiplex Serological Assay. High-Throughput 2017, 6, 15. https://doi.org/10.3390/ht6040015
Filomena A, Göpfert JC, Duffy D, Pol S, Abdel-Hamid M, Esmat G, Fontanet A, Albert ML, Joos TO, Schneiderhan-Marra N. Study of the Humoral Immune Response towards HCV Genotype 4 Using a Bead-Based Multiplex Serological Assay. High-Throughput. 2017; 6(4):15. https://doi.org/10.3390/ht6040015
Chicago/Turabian StyleFilomena, Angela, Jens C. Göpfert, Darragh Duffy, Stanislas Pol, Mohamed Abdel-Hamid, Gamal Esmat, Arnaud Fontanet, Matthew L. Albert, Thomas O. Joos, and Nicole Schneiderhan-Marra. 2017. "Study of the Humoral Immune Response towards HCV Genotype 4 Using a Bead-Based Multiplex Serological Assay" High-Throughput 6, no. 4: 15. https://doi.org/10.3390/ht6040015
APA StyleFilomena, A., Göpfert, J. C., Duffy, D., Pol, S., Abdel-Hamid, M., Esmat, G., Fontanet, A., Albert, M. L., Joos, T. O., & Schneiderhan-Marra, N. (2017). Study of the Humoral Immune Response towards HCV Genotype 4 Using a Bead-Based Multiplex Serological Assay. High-Throughput, 6(4), 15. https://doi.org/10.3390/ht6040015






