Synthesis and Application of Albumin Nanoparticles Loaded with Prussian Blue Nanozymes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Instrumentation
2.2. Synthesis of Prussian Blue Nanozymes
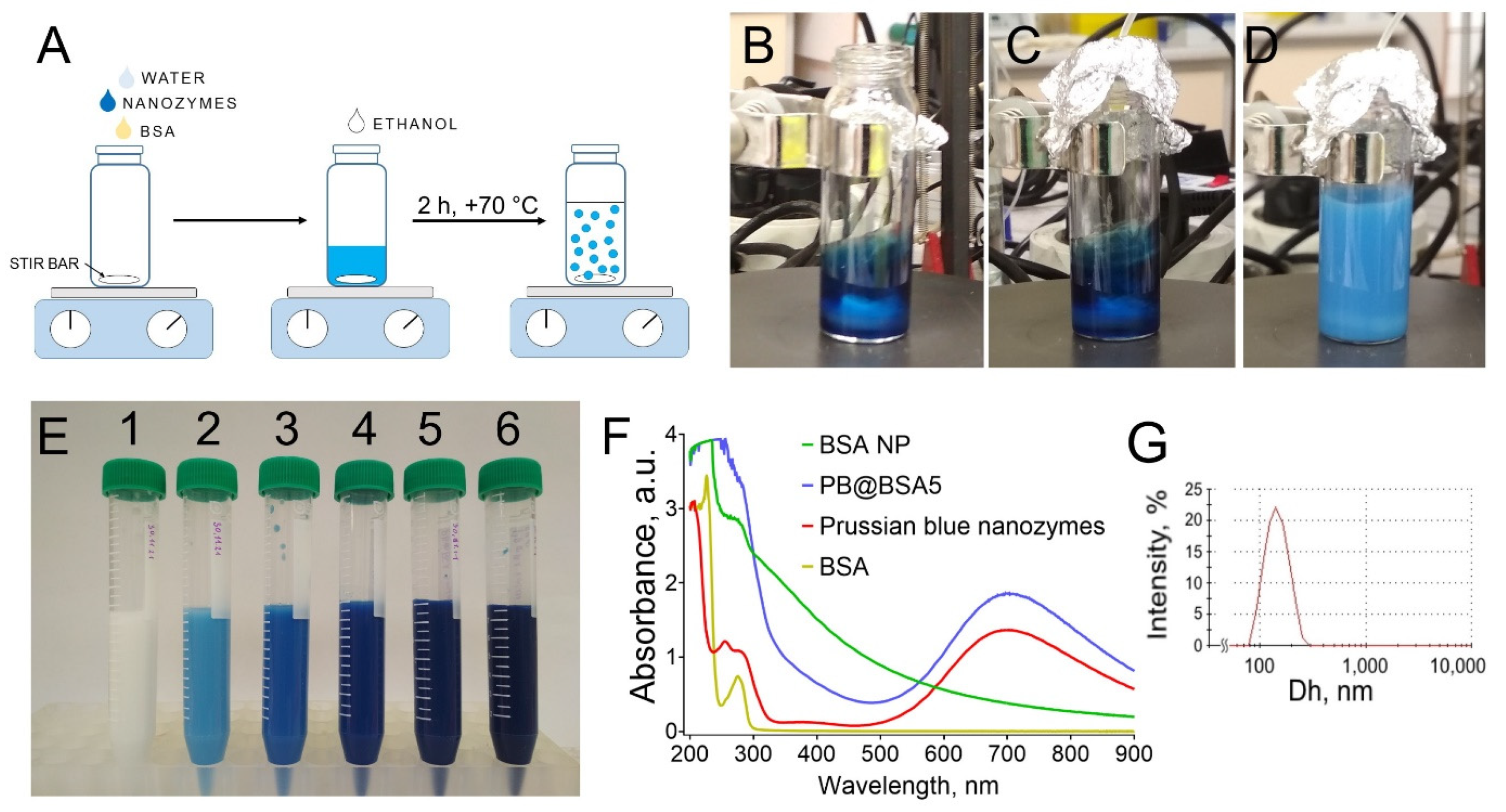
2.3. Preparation of Prussian Blue Nanozyme-Loaded BSA Nanoparticles (PB@BSA)
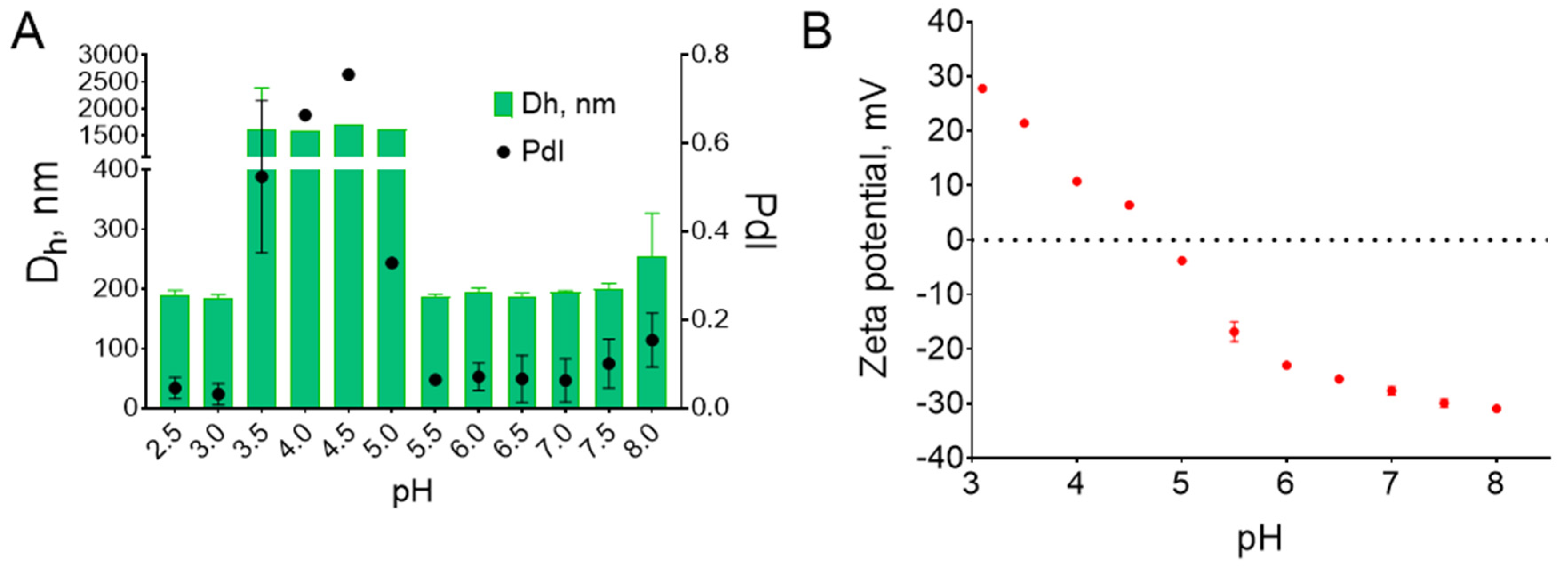
2.4. Characterization of PB@BSA Nanoparticles
2.5. Loading Efficiency of Prussian Blue Nanozymes
2.6. Functionalization of PB@BSA Nanoparticles with Antibodies, BSA, and Streptavidin
2.7. PSA Immunoassay
3. Results and Discussion
3.1. Synthesis of Nanozyme-Loaded Albumin Nanoparticles: Properties and Reproducibility
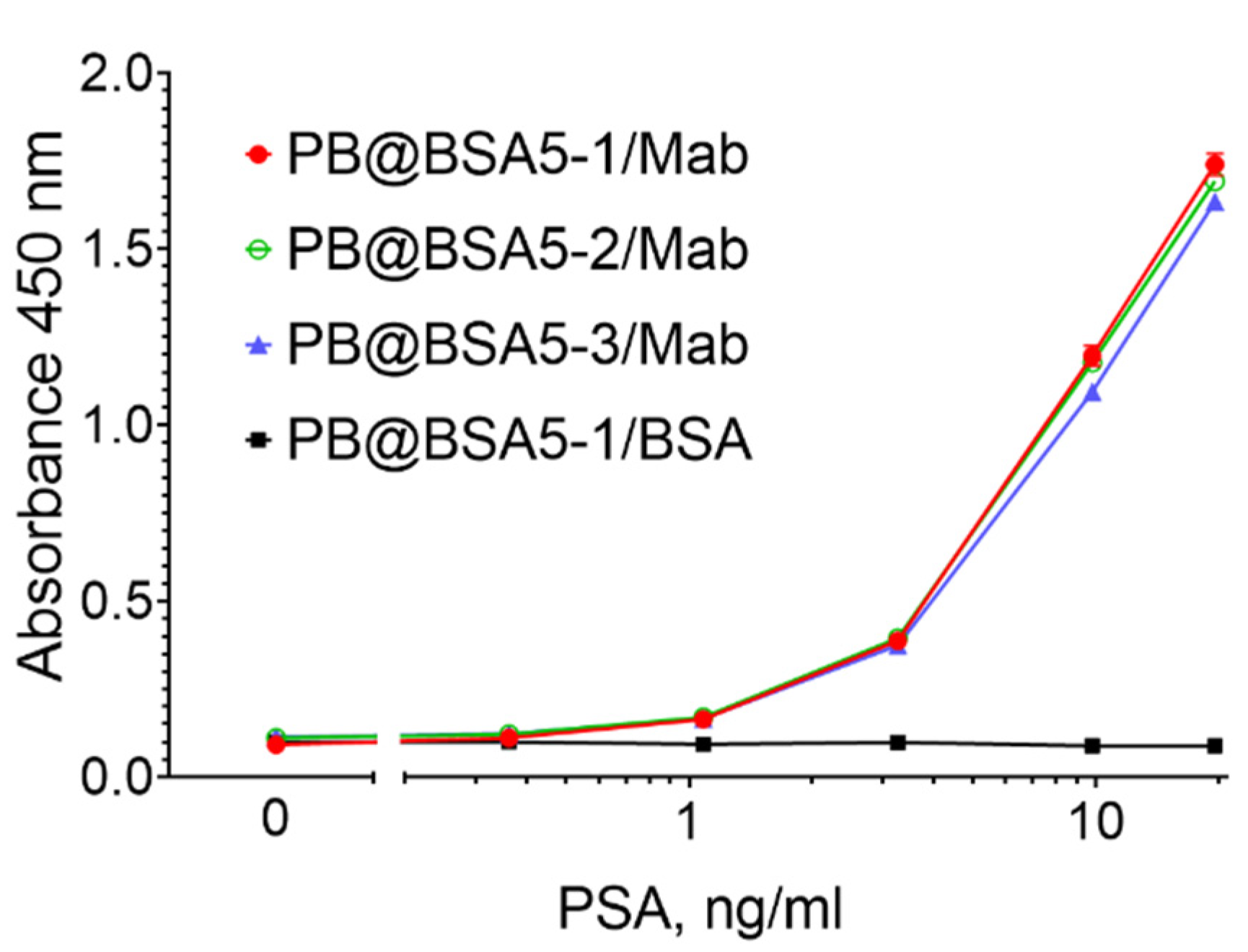
3.2. Functionalization of PB@BSA Nanoparticles
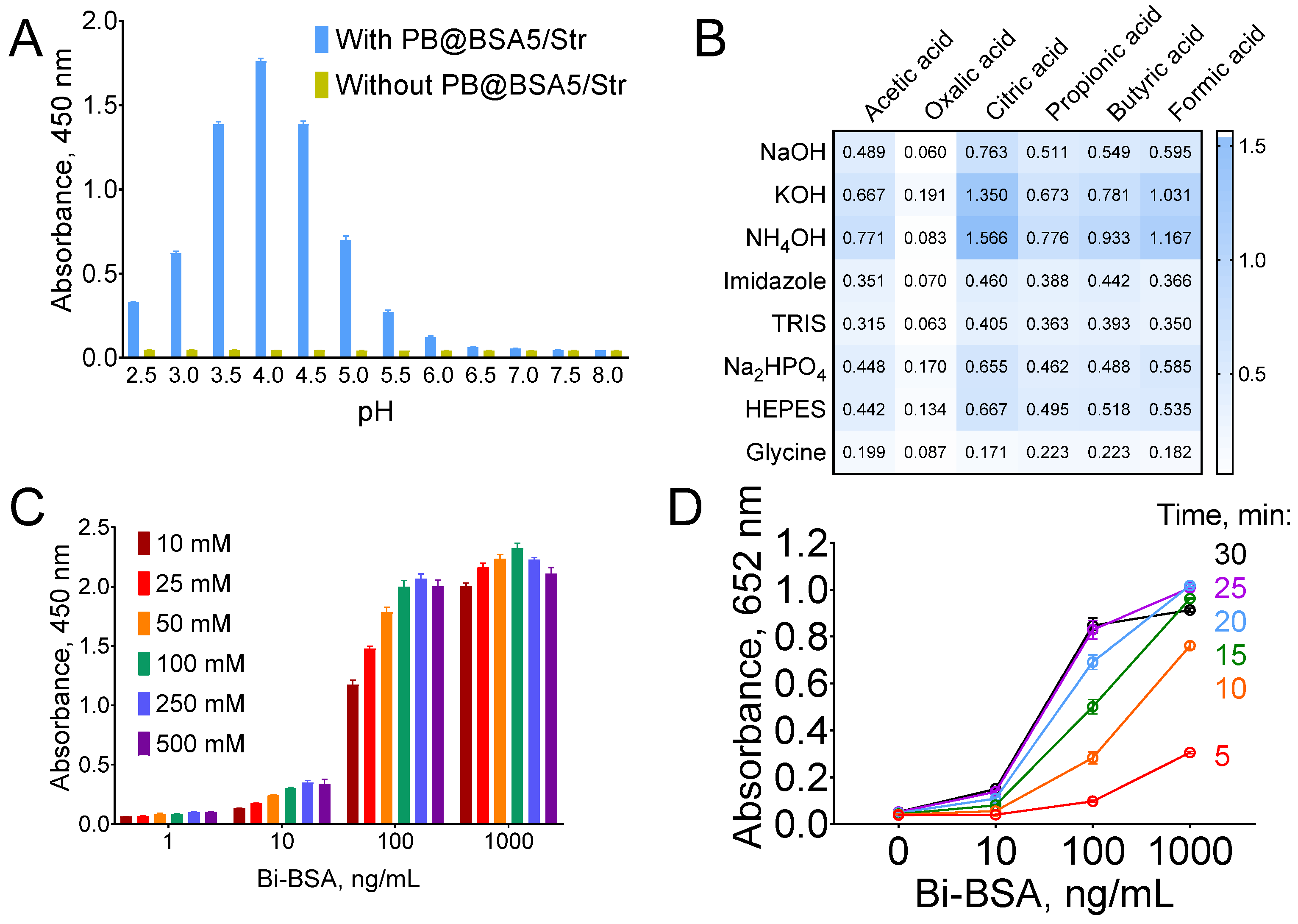
3.3. Optimization of Substrate Buffer
3.4. PSA Assay: Optimization and Reproducibility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Das, B.; Franco, J.L.; Logan, N.; Balasubramanian, P.; Kim, M.I.; Cao, C. Nanozymes in Point-of-Care Diagnosis: An Emerging Futuristic Approach for Biosensing. Nano-Micro Lett. 2021, 13, 193. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, D.; Wang, Y.; Li, H.; Zhang, Y.; Chen, H.; Li, X.; Huo, M. Nanozymes-recent development and biomedical applications. J. Nanobiotechnol. 2022, 20, 92. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Wang, Q.; Lin, A.; Wei, H. Nanozyme-Enabled Analytical Chemistry. Anal. Chem. 2022, 94, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhou, Y.; Chandrawati, R. Metal and Metal Oxide Nanoparticles to Enhance the Performance of Enzyme-Linked Immunosorbent Assay (ELISA). ACS Appl. Nano Mater. 2020, 3, 1–21. [Google Scholar] [CrossRef]
- Kraft, A. Some considerations on the structure, composition, and properties of Prussian blue: A contribution to the current discussion. Ionics 2021, 27, 2289–2305. [Google Scholar] [CrossRef]
- Farka, Z.; Čunderlová, V.; Horáčková, V.; Pastucha, M.; Mikušová, Z.; Hlaváček, A.; Skládal, P. Prussian Blue Nanoparticles as a Catalytic Label in a Sandwich Nanozyme-Linked Immunosorbent Assay. Anal. Chem. 2018, 90, 2348–2354. [Google Scholar] [CrossRef]
- He, Q.; Yang, H.; Chen, Y.; Shen, D.; Cui, X.; Zhang, C.; Xiao, H.; Eremin, S.A.; Fang, Y.; Zhao, S. Prussian blue nanoparticles with peroxidase-mimicking properties in a dual immunoassays for glycocholic acid. J. Pharm. Biomed. 2020, 187, 113317. [Google Scholar] [CrossRef]
- Tian, M.; Xie, W.; Zhang, T.; Liu, Y.; Lu, Z.; Li, C.M.; Liu, Y. A sensitive lateral flow immunochromatographic strip with prussian blue nanoparticles mediated signal generation and cascade amplification. Sens. Actuators B Chem. 2020, 309, 127728. [Google Scholar] [CrossRef]
- Shokouhimehr, M.; Soehnlen, E.S.; Hao, J.; Griswold, M.; Flask, C.; Fan, X.; Basilion, J.P.; Basu, S.; Huang, S.D. Dual purpose Prussian blue nanoparticles for cellular imaging and drug delivery: A new generation of T1-weighted MRI contrast and small molecule delivery agents. J. Mater. Chem. 2010, 20, 5251–5259. [Google Scholar] [CrossRef]
- Qin, Z.; Li, Y.; Gu, N. Progress in Applications of Prussian Blue Nanoparticles in Biomedicine. Adv. Healthc. Mater. 2018, 7, 1800347. [Google Scholar] [CrossRef]
- Weber, C.; Coester, C.; Kreuter, J.; Langer, K. Desolvation process and surface characterisation of protein nanoparticles. Int. J. Pharm. 2000, 194, 91–102. [Google Scholar] [CrossRef]
- Peralta, D.V.; He, J.; Wheeler, D.A.; Zhang, J.Z.; Tarr, M.A. Encapsulating gold nanomaterials into size-controlled human serum albumin nanoparticles for cancer therapy platforms. J. Microencapsul. 2014, 31, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Tang, Q.; Xue, W.; Xiang, J.; Zhang, L.; Wang, X. The preparation and characterization of folate-conjugated human serum albumin magnetic cisplatin nanoparticles. J. Biomed. Res. 2010, 24, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Angelova, N.; Yordanov, G. Entrapment of β-FeO(OH) nanoparticles in human serum albumin: Preparation, characterization and hemocompatibility. Colloids Surf. A Physicochem. Eng. Asp. 2017, 516, 317–324. [Google Scholar] [CrossRef]
- Bhushan, B.; Gopinath, P. Antioxidant nanozyme: A facile synthesis and evaluation of the reactive oxygen species scavenging potential of nanoceria encapsulated albumin nanoparticles. J. Mater. Chem. B 2015, 3, 4843–4852. [Google Scholar] [CrossRef]
- Karyakin, A.A. Advances of Prussian blue and its analogues in (bio)sensors. Curr. Opin. Electrochem. 2017, 5, 92–98. [Google Scholar] [CrossRef]
- Galisteo-González, F.; Molina-Bolívar, J.A. Systematic study on the preparation of BSA nanoparticles. Colloids Surf. B 2014, 123, 286–292. [Google Scholar] [CrossRef]
- Liu, X.; Meng, H. Consideration for the scale-up manufacture of nanotherapeutics—A critical step for technology transfer. VIEW 2021, 2, 20200190. [Google Scholar] [CrossRef]
- Spada, A.; Emami, J.; Tuszynski, J.A.; Lavasanifar, A. The Uniqueness of Albumin as a Carrier in Nanodrug Delivery. Mol. Pharm. 2021, 18, 1862–1894. [Google Scholar] [CrossRef]
- Wacker, M.; Zensi, A.; Kufleitner, J.; Ruff, A.; Schütz, J.; Stockburger, T.; Marstaller, T.; Vogel, V. A toolbox for the upscaling of ethanolic human serum albumin (HSA) desolvation. Int. J. Pharm. 2011, 414, 225–232. [Google Scholar] [CrossRef]
- Yedomon, B.; Fessi, H.; Charcosset, C. Preparation of Bovine Serum Albumin (BSA) nanoparticles by desolvation using a membrane contactor: A new tool for large scale production. Eur. J. Pharm. Biopharm. 2013, 85, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Jahanban-Esfahlan, A.; Dastmalchi, S.; Davaran, S. A simple improved desolvation method for the rapid preparation of albumin nanoparticles. Int. J. Biol. Macromol. 2016, 91, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, Y.; Zhao, S.; Shao, T.; Cheng, Y. Human serum albumin (HSA) nanoparticles stabilized with intermolecular disulfide bonds. Chem. Commun. 2013, 49, 2234. [Google Scholar] [CrossRef]
- Drozd, M.; Pietrzak, M.; Parzuchowski, P.G.; Malinowska, E. Pitfalls and capabilities of various hydrogen donors in evaluation of peroxidase-like activity of gold nanoparticles. Anal. Bioanal. Chem. 2016, 408, 8505–8513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.-J.J.; Yang, J.; Chong, K.; Ma, Q.; Li, M.; Zhang, F.; Moon, W.J.; Zhang, G.; Liu, J. Good’s buffers have various affinities to gold nanoparticles regulating fluorescent and colorimetric DNA sensing. Chem. Sci. 2020, 11, 6795–6804. [Google Scholar] [CrossRef]
- Shan, Z.; Lu, M.; Wang, L.; MacDonald, B.; MacInnis, J.; Mkandawire, M.; Zhang, X.; Oakes, K.D. Chloride accelerated Fenton chemistry for the ultrasensitive and selective colorimetric detection of copper. Chem. Commun. 2016, 52, 2087–2090. [Google Scholar] [CrossRef]
- Pham, X.-H.; Hahm, E.; Huynh, K.-H.; Son, B.S.; Kim, H.-M.; Jun, B.-H. Sensitive Colorimetric Detection of Prostate Specific Antigen Using a Peroxidase-Mimicking Anti-PSA Antibody Coated Au Nanoparticle. Biochip J. 2020, 14, 158–168. [Google Scholar] [CrossRef]
- Komkova, M.A.; Karyakina, E.E.; Karyakin, A.A. Catalytically synthesized Prussian Blue nanoparticles defeating natural enzyme peroxidase. J. Am. Chem. Soc. 2018, 140, 11302–11307. [Google Scholar] [CrossRef]
- Wang, S.; Yan, H.; Wang, Y.; Wang, N.; Lin, Y.; Li, M. Hollow Prussian Blue nanocubes as peroxidase mimetic and enzyme carriers for colorimetric determination of ethanol. Microchim. Acta 2019, 186, 738. [Google Scholar] [CrossRef]
- Stable Indicator Solutions for Detection of Peroxidatic Activity. Available online: https://patents.google.com/patent/WO1990002339A1/en (accessed on 7 April 2022).
- Duffy, M.J. Biomarkers for prostate cancer: Prostate-specific antigen and beyond. Clin. Chem. Lab. Med. 2020, 58, 326–339. [Google Scholar] [CrossRef] [Green Version]


| Nanoparticle Type | Dh, nm | PdI | ZP, mV | Loading Capacity, μg/mg | Encapsulation Efficiency, % | Yield, % |
|---|---|---|---|---|---|---|
| BSA NP | 234 ± 3 | 0.078 ± 0.006 | −19.1 ± 0.9 | 0.0 | 0.0 | 79.8 |
| PB@BSA1 | 237 ± 53 | 0.166 ± 0.073 | −17.8 ± 1.4 | 7.6 | 40.6 | 66.4 |
| PB@BSA2 | 169 ± 5 | 0.175 ± 0.007 | −16.8 ± 0.8 | 24.7 | 74.5 | 73.6 |
| PB@BSA3 | 123 ± 1 | 0.176 ± 0.008 | −19.2 ± 1.7 | 51.1 | 84.3 | 78.6 |
| PB@BSA4 | 131 ± 18 | 0.131 ± 0.090 | −17.9 ± 0.8 | 101.7 | 91.4 | 81.8 |
| PB@BSA5 | 140 ± 1 | 0.040 ± 0.026 | −17.8 ± 0.4 | 186.5 | 96.3 | 86.1 |
| Batch | Dh, nm | PdI | ZP, mV | Loading Capacity, μg/mg | Encapsulation Efficiency, % | Yield, % |
|---|---|---|---|---|---|---|
| PB@BSA5-1 | 168 ± 1 | 0.104 ± 0.007 | −18.2 ± 1.6 | 192 | 99.7 | 86.7 |
| PB@BSA5-2 | 167 ± 1 | 0.106 ± 0.011 | −21.0 ± 1.0 | 191 | 99.8 | 90.6 |
| PB@BSA5-3 | 161 ± 1 | 0.178 ± 0.017 | −17.5 ± 0.9 | 187 | 99.6 | 91.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khramtsov, P.; Kropaneva, M.; Bochkova, M.; Timganova, V.; Kiselkov, D.; Zamorina, S.; Rayev, M. Synthesis and Application of Albumin Nanoparticles Loaded with Prussian Blue Nanozymes. Colloids Interfaces 2022, 6, 29. https://doi.org/10.3390/colloids6020029
Khramtsov P, Kropaneva M, Bochkova M, Timganova V, Kiselkov D, Zamorina S, Rayev M. Synthesis and Application of Albumin Nanoparticles Loaded with Prussian Blue Nanozymes. Colloids and Interfaces. 2022; 6(2):29. https://doi.org/10.3390/colloids6020029
Chicago/Turabian StyleKhramtsov, Pavel, Maria Kropaneva, Maria Bochkova, Valeria Timganova, Dmitriy Kiselkov, Svetlana Zamorina, and Mikhail Rayev. 2022. "Synthesis and Application of Albumin Nanoparticles Loaded with Prussian Blue Nanozymes" Colloids and Interfaces 6, no. 2: 29. https://doi.org/10.3390/colloids6020029
APA StyleKhramtsov, P., Kropaneva, M., Bochkova, M., Timganova, V., Kiselkov, D., Zamorina, S., & Rayev, M. (2022). Synthesis and Application of Albumin Nanoparticles Loaded with Prussian Blue Nanozymes. Colloids and Interfaces, 6(2), 29. https://doi.org/10.3390/colloids6020029






